Histone and DNA Methylation as Epigenetic Regulators of DNA Damage Repair in Gastric Cancer and Emerging Therapeutic Opportunities
Abstract
Simple Summary
Abstract
1. Introduction
2. Methylation: A Key Epigenetic Alteration in GC
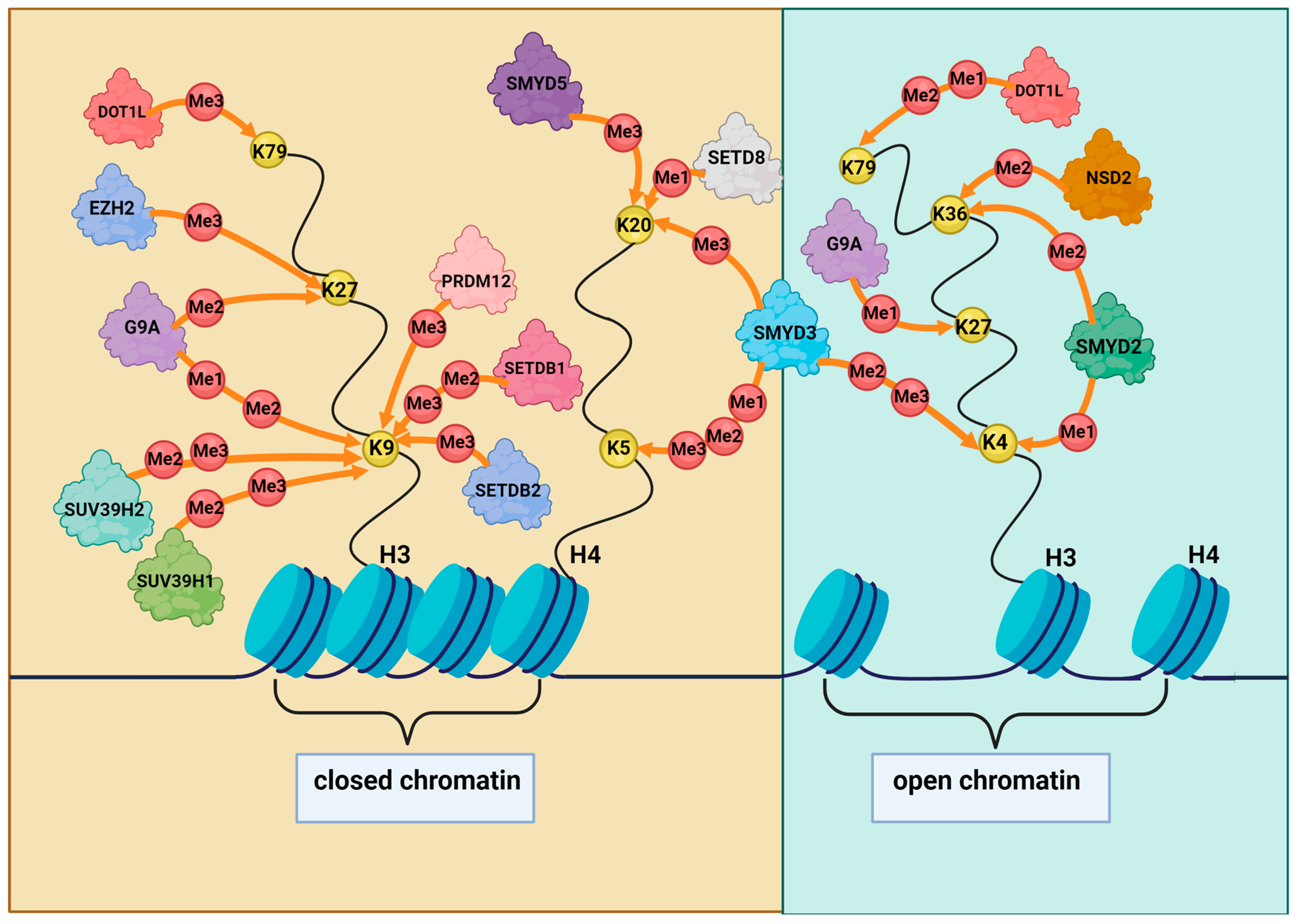
2.1. Histone Methylation and Its Role in DDR in GC
2.2. Methylation of Non-Histone DDR Proteins by HMTs in GC
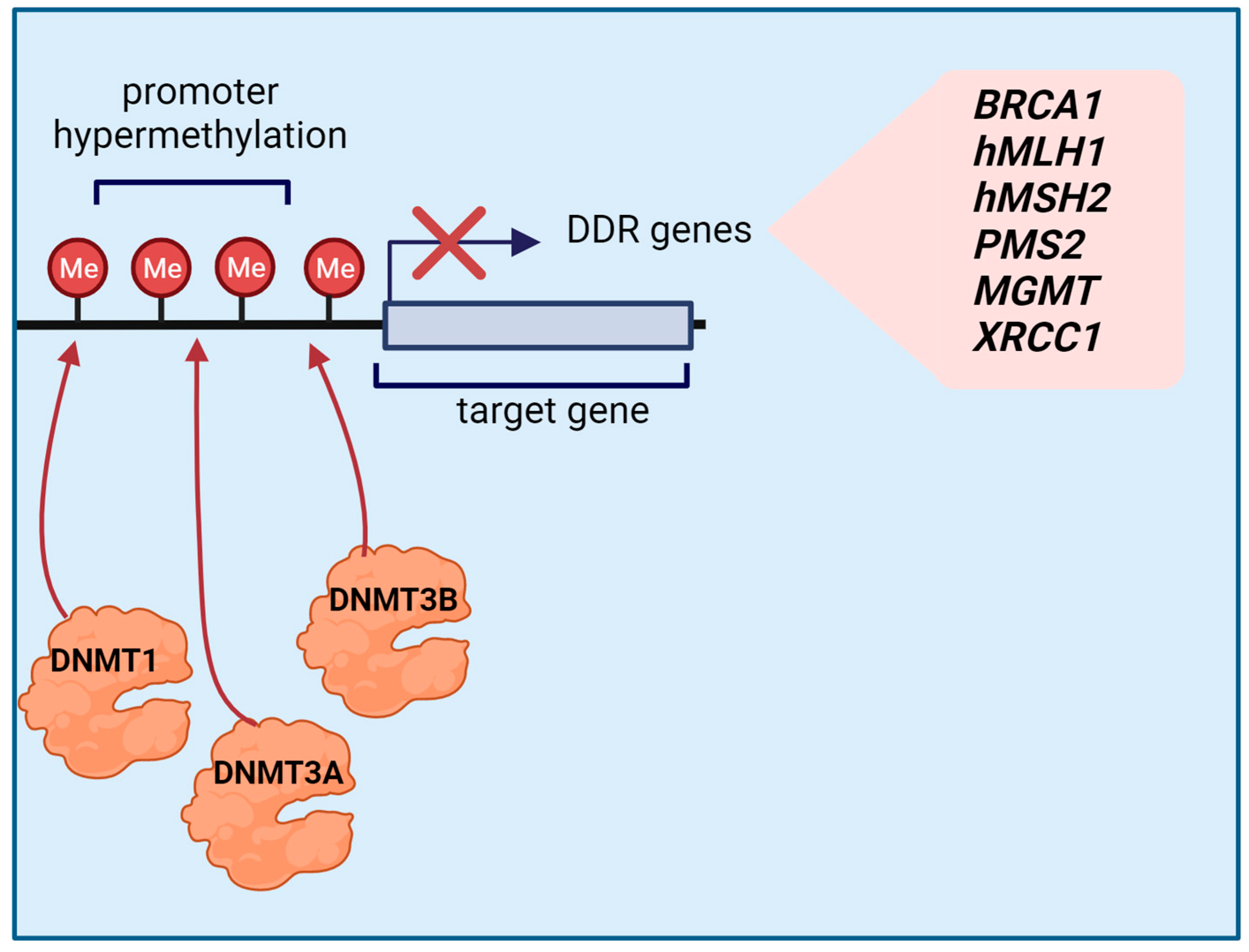
2.3. Effects of Promoter Methylation on the Expression of Major DNA Repair Genes in GC
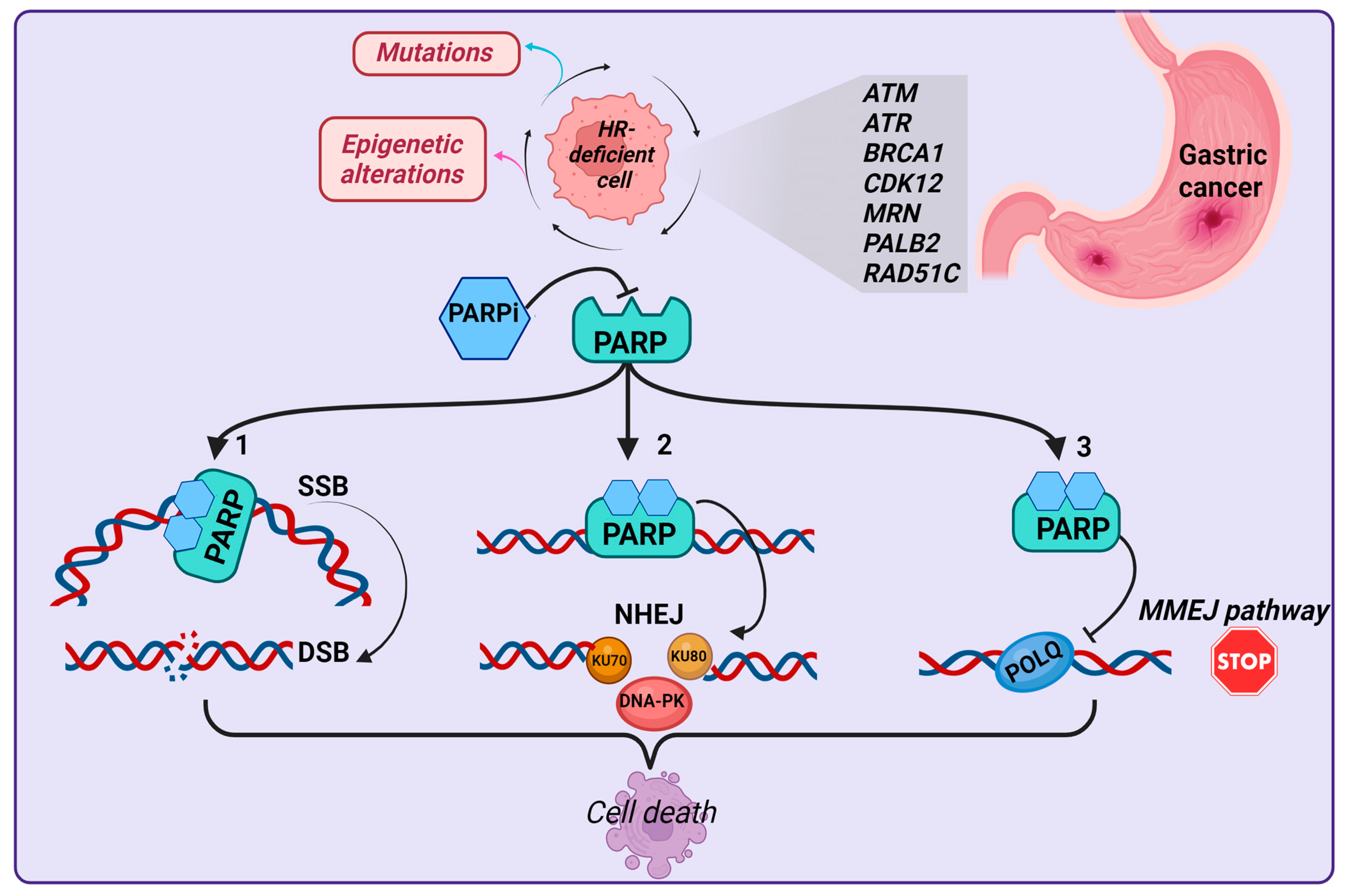
3. HR: The Predominant DSB Repair Pathway
HRD as a Common Feature in GC
4. Current Therapeutic Strategies Targeting DDR Proteins in GC
5. Conclusions and Future Directions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Santos, J.C.; Ribeiro, M.L. Epigenetic Regulation of DNA Repair Machinery in Helicobacter pylori-Induced Gastric Carcinogenesis. World J. Gastroenterol. 2015, 21, 9021–9037. [Google Scholar] [CrossRef]
- Lorant, K.; Roland, K.; Bianca, O.; Sorin, Z. Histopathological Lauren Classification of Gastric Carcinoma with Biopsy Specimen and a Histological Difference with Dysplasia. Clin. Med. Investig. 2019, 4. [Google Scholar] [CrossRef]
- Berlth, F.; Bollschweiler, E.; Drebber, U.; Hoelscher, A.H.; Moenig, S. Pathohistological Classification Systems in Gastric Cancer: Diagnostic Relevance and Prognostic Value. World J. Gastroenterol. 2014, 20, 5679–5684. [Google Scholar] [CrossRef]
- Hamilton, J.P.; Meltzer, S.J. A Review of the Genomics of Gastric Cancer. Clin. Gastroenterol. Hepatol. 2006, 4, 416–425. [Google Scholar] [CrossRef]
- Canale, M.; Casadei-Gardini, A.; Ulivi, P.; Arechederra, M.; Berasain, C.; Lollini, P.-L.; Fernández-Barrena, M.G.; Avila, M.A. Epigenetic Mechanisms in Gastric Cancer: Potential New Therapeutic Opportunities. Int. J. Mol. Sci. 2020, 21, 5500. [Google Scholar] [CrossRef]
- Puneet; Kazmi, H.R.; Kumari, S.; Tiwari, S.; Khanna, A.; Narayan, G. Epigenetic Mechanisms and Events in Gastric Cancer-Emerging Novel Biomarkers. Pathol. Oncol. Res. 2018, 24, 757–770. [Google Scholar] [CrossRef]
- Oliveira, C.; Pinheiro, H.; Figueiredo, J.; Seruca, R.; Carneiro, F. Familial Gastric Cancer: Genetic Susceptibility, Pathology, and Implications for Management. Lancet Oncol. 2015, 16, e60–e70. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, R.C.; Hardwick, R.; Huntsman, D.; Carneiro, F.; Guilford, P.; Blair, V.; Chung, D.C.; Norton, J.; Ragunath, K.; Van Krieken, J.H.; et al. Hereditary Diffuse Gastric Cancer: Updated Consensus Guidelines for Clinical Management and Directions for Future Research. J. Med. Genet. 2010, 47, 436–444. [Google Scholar] [CrossRef]
- Singh, A.D.; Gupta, A.; Mehta, N.; Heald, B.; Macaron, C.; Liska, D.; Bhatt, A.; Burke, C.A. Occurrence of Gastric Cancer in Patients with Juvenile Polyposis Syndrome: A Systematic Review and Meta-Analysis. Gastrointest. Endosc. 2023, 97, 407–414.e1. [Google Scholar] [CrossRef] [PubMed]
- Forte, G.; Cariola, F.; De Marco, K.; Manghisi, A.; Guglielmi, F.A.; Armentano, R.; Lippolis, G.; Giorgio, P.; Simone, C.; Disciglio, V. A Novel STK11 Gene Mutation (c.388dupG, p.Glu130Glyfs∗33) in a Peutz-Jeghers Family and Evidence of Higher Gastric Cancer Susceptibility Associated with Alterations in STK11 Region Aa 107-170. Genes Dis. 2022, 9, 288–291. [Google Scholar] [CrossRef] [PubMed]
- Disciglio, V.; Fasano, C.; Cariola, F.; Forte, G.; Grossi, V.; Sanese, P.; Signorile, M.L.; Resta, N.; Lotesoriere, C.; Stella, A.; et al. Gastric Polyposis and Desmoid Tumours as a New Familial Adenomatous Polyposis Clinical Variant Associated with APC Mutation at the Extreme 3′-End. J. Med. Genet. 2020, 57, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Bentrem, D. Environmental and Genetic Risk Factors for Gastric Cancer. J. Surg. Oncol. 2022, 125, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Choi, M.; Kipps, T.; Kurzrock, R. ATM Mutations in Cancer: Therapeutic Implications. Mol. Cancer Ther. 2016, 15, 1781–1791. [Google Scholar] [CrossRef]
- Calin, G.; Ranzani, G.N.; Amadori, D.; Herlea, V.; Matei, I.; Barbanti-Brodano, G.; Negrini, M. Somatic Frameshift Mutations in the Bloom Syndrome BLM Gene Are Frequent in Sporadic Gastric Carcinomas with Microsatellite Mutator Phenotype. BMC Genet. 2001, 2, 14. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, H.; Wakai, T.; Nagahashi, M.; Shimada, Y.; Hanyu, T.; Kano, Y.; Muneoka, Y.; Ishikawa, T.; Takizawa, K.; Tajima, Y.; et al. Pathogenic Germline BRCA1/2 Mutations and Familial Predisposition to Gastric Cancer. JCO Precis. Oncol. 2018, 2, PO.18.00097. [Google Scholar] [CrossRef]
- Ghojazadeh, M.; Somi, M.H.; Naseri, A.; Salehi-Pourmehr, H.; Hassannezhad, S.; Hajikamanaj Olia, A.; Kafshdouz, L.; Nikniaz, Z. Systematic Review and Meta-Analysis of TP53, HER2/ERBB2, KRAS, APC, and PIK3CA Genes Expression Pattern in Gastric Cancer. Middle East J. Dig. Dis. 2022, 14, 335–345. [Google Scholar] [CrossRef]
- Ebrahimi, V.; Soleimanian, A.; Ebrahimi, T.; Azargun, R.; Yazdani, P.; Eyvazi, S.; Tarhriz, V. Epigenetic Modifications in Gastric Cancer: Focus on DNA Methylation. Gene 2020, 742, 144577. [Google Scholar] [CrossRef]
- Gong, F.; Miller, K.M. Histone Methylation and the DNA Damage Response. Mutat. Res. Rev. Mutat. Res. 2019, 780, 37–47. [Google Scholar] [CrossRef]
- Wang, M.; Xie, C. DNA Damage Repair and Current Therapeutic Approaches in Gastric Cancer: A Comprehensive Review. Front. Genet 2022, 13, 931866. [Google Scholar] [CrossRef]
- Helena, J.M.; Joubert, A.M.; Grobbelaar, S.; Nolte, E.M.; Nel, M.; Pepper, M.S.; Coetzee, M.; Mercier, A.E. Deoxyribonucleic Acid Damage and Repair: Capitalizing on Our Understanding of the Mechanisms of Maintaining Genomic Integrity for Therapeutic Purposes. Int. J. Mol. Sci. 2018, 19, 1148. [Google Scholar] [CrossRef]
- Shibata, A.; Jeggo, P.A. DNA Double-Strand Break Repair in a Cellular Context. Clin. Oncol. 2014, 26, 243–249. [Google Scholar] [CrossRef]
- Wu, X.; Wang, Q.; Liu, P.; Sun, L.; Wang, Y. Potential Value of the Homologous Recombination Deficiency Signature We Developed in the Prognosis and Drug Sensitivity of Gastric Cancer. Front. Genet. 2022, 13, 1026871. [Google Scholar] [CrossRef]
- Young, K.; Starling, N.; Cunningham, D. Targeting Deficient DNA Damage Repair in Gastric Cancer. Expert Opin. Pharmacother. 2016, 17, 1757–1766. [Google Scholar] [CrossRef]
- Necula, L.; Matei, L.; Dragu, D.; Neagu, A.I.; Mambet, C.; Nedeianu, S.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Recent Advances in Gastric Cancer Early Diagnosis. World J. Gastroenterol. 2019, 25, 2029–2044. [Google Scholar] [CrossRef]
- Karakaidos, P.; Karagiannis, D.; Rampias, T. Resolving DNA Damage: Epigenetic Regulation of DNA Repair. Molecules 2020, 25, 2496. [Google Scholar] [CrossRef] [PubMed]
- Abdelfatah, E.; Kerner, Z.; Nanda, N.; Ahuja, N. Epigenetic Therapy in Gastrointestinal Cancer: The Right Combination. Ther. Adv. Gastroenterol. 2016, 9, 560–579. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, C.; Garcia-Gonzalez, M.A.; Machado, J.C. Molecular Pathogenesis of Gastric Cancer. Helicobacter 2013, 18 (Suppl. 1), 28–33. [Google Scholar] [CrossRef]
- Padmanabhan, N.; Ushijima, T.; Tan, P. How to Stomach an Epigenetic Insult: The Gastric Cancer Epigenome. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Nobili, S.; Bruno, L.; Landini, I.; Napoli, C.; Bechi, P.; Tonelli, F.; Rubio, C.A.; Mini, E.; Nesi, G. Genomic and Genetic Alterations Influence the Progression of Gastric Cancer. World J. Gastroenterol. 2011, 17, 290–299. [Google Scholar] [CrossRef]
- Tang, S.-Y.; Zhou, P.-J.; Meng, Y.; Zeng, F.-R.; Deng, G.-T. Gastric Cancer: An Epigenetic View. World J. Gastrointest. Oncol. 2022, 14, 90–109. [Google Scholar] [CrossRef]
- Michalak, E.M.; Burr, M.L.; Bannister, A.J.; Dawson, M.A. The Roles of DNA, RNA and Histone Methylation in Ageing and Cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 573–589. [Google Scholar] [CrossRef] [PubMed]
- Qu, Y.; Dang, S.; Hou, P. Gene Methylation in Gastric Cancer. Clin. Chim. Acta 2013, 424, 53–65. [Google Scholar] [CrossRef]
- Calcagno, D.Q.; Gigek, C.O.; Chen, E.S.; Burbano, R.R.; Smith, M.d.A.C. DNA and Histone Methylation in Gastric Carcinogenesis. World J. Gastroenterol. 2013, 19, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.-Y.; Gu, J.-L.; Zhen, T.-M. Recent Advances of Histone Modification in Gastric Cancer. J. Cancer Res. Ther. 2014, 10 (Suppl. 4), 240–245. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.; Song, J.-J.; Lee, J.; Kim, M.Y. Epigenetics: An Emerging Player in Gastric Cancer. World J. Gastroenterol. 2014, 20, 6433–6447. [Google Scholar] [CrossRef]
- Petermann, E.; Lan, L.; Zou, L. Sources, Resolution and Physiological Relevance of R-Loops and RNA-DNA Hybrids. Nat. Rev. Mol. Cell Biol. 2022, 23, 521–540. [Google Scholar] [CrossRef] [PubMed]
- García-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604–618. [Google Scholar] [CrossRef]
- Mackay, R.P.; Xu, Q.; Weinberger, P.M. R-Loop Physiology and Pathology: A Brief Review. DNA Cell Biol. 2020, 39, 1914–1925. [Google Scholar] [CrossRef]
- Shih, H.-T.; Chen, W.-Y.; Wang, H.-Y.; Chao, T.; Huang, H.-D.; Chou, C.-H.; Chang, Z.-F. DNMT3b Protects Centromere Integrity by Restricting R-Loop-Mediated DNA Damage. Cell Death Dis. 2022, 13, 546. [Google Scholar] [CrossRef] [PubMed]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell 2019, 73, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.-T.; Hawley, B.R.; Skalka, G.L.; Baldock, R.A.; Smith, E.M.; Bader, A.S.; Malewicz, M.; Watts, F.Z.; Wilczynska, A.; Bushell, M. Drosha Drives the Formation of DNA:RNA Hybrids around DNA Break Sites to Facilitate DNA Repair. Nat. Commun. 2018, 9, 532. [Google Scholar] [CrossRef]
- Sanz, L.A.; Hartono, S.R.; Lim, Y.W.; Steyaert, S.; Rajpurkar, A.; Ginno, P.A.; Xu, X.; Chédin, F. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol. Cell 2016, 63, 167–178. [Google Scholar] [CrossRef]
- Al-Hadid, Q.; Yang, Y. R-Loop: An Emerging Regulator of Chromatin Dynamics. Acta Biochim. Biophys. Sin. 2016, 48, 623–631. [Google Scholar] [CrossRef]
- Sharma, A.K.; Hendzel, M.J. The Relationship between Histone Posttranslational Modification and DNA Damage Signaling and Repair. Int. J. Radiat. Biol. 2019, 95, 382–393. [Google Scholar] [CrossRef]
- Mulero-Navarro, S.; Esteller, M. Epigenetic Biomarkers for Human Cancer: The Time Is Now. Crit. Rev. Oncol. Hematol. 2008, 68, 1–11. [Google Scholar] [CrossRef]
- Wong, M.; Polly, P.; Liu, T. The Histone Methyltransferase DOT1L: Regulatory Functions and a Cancer Therapy Target. Am. J. Cancer Res. 2015, 5, 2823–2837. [Google Scholar]
- Albert, M.; Helin, K. Histone Methyltransferases in Cancer. Semin. Cell Dev. Biol. 2010, 21, 209–220. [Google Scholar] [CrossRef]
- He, L.-J.; Cai, M.-Y.; Xu, G.-L.; Li, J.-J.; Weng, Z.-J.; Xu, D.-Z.; Luo, G.-Y.; Zhu, S.-L.; Xie, D. Prognostic Significance of Overexpression of EZH2 and H3k27me3 Proteins in Gastric Cancer. Asian Pac. J. Cancer Prev. 2012, 13, 3173–3178. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Liu, N.; Song, X.; Chen, L.; Wang, M.; Xiao, G.; Li, T.; Wang, Z.; Zhang, Y. EZH2: An Accomplice of Gastric Cancer. Cancers 2023, 15, 425. [Google Scholar] [CrossRef]
- Matsukawa, Y.; Semba, S.; Kato, H.; Ito, A.; Yanagihara, K.; Yokozaki, H. Expression of the Enhancer of Zeste Homolog 2 Is Correlated with Poor Prognosis in Human Gastric Cancer. Cancer Sci. 2006, 97, 484–491. [Google Scholar] [CrossRef]
- Chen, Z.; Yuan, L.; Li, X.; Yu, J.; Xu, Z. BMP2 Inhibits Cell Proliferation by Downregulating EZH2 in Gastric Cancer. Cell Cycle 2022, 21, 2298–2308. [Google Scholar] [CrossRef]
- Zhao, K.; He, J.; Wang, Y.-F.; Jin, S.-D.; Fan, Y.; Fang, N.; Qian, J.; Xu, T.-P.; Guo, R.-H. EZH2-Mediated Epigenetic Suppression of EphB3 Inhibits Gastric Cancer Proliferation and Metastasis by Affecting E-Cadherin and Vimentin Expression. Gene 2019, 686, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Wang, Z.; Lu, W.; Jiang, H.; Lu, J.; Qiu, J.; Ye, G. EZH2 Promotes Gastric Cancer Cells Proliferation by Repressing P21 Expression. Pathol. Res. Pract. 2019, 215, 152374. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zheng, Y.; Yang, L. The Oncogenic Potential of SUV39H2: A Comprehensive and Perspective View. J. Cancer 2019, 10, 721–729. [Google Scholar] [CrossRef]
- Reyes, D.A.; Sarría, V.M.S.; Salazar-Viedma, M.; D’Afonseca, V. Histone Methyltransferases Useful in Gastric Cancer Research. Cancer Inform. 2021, 20, 11769351211039862. [Google Scholar] [CrossRef]
- Yang, J.; Xu, P.; Chen, Z.; Zhang, X.; Xia, Y.; Fang, L.; Xie, L.; Li, B.; Xu, Z. N6-Methyadenosine Modified SUV39H2 Regulates Homologous Recombination through Epigenetic Repression of DUSP6 in Gastric Cancer. Cancer Lett. 2023, 558, 216092. [Google Scholar] [CrossRef]
- Cai, L.; Ma, X.; Huang, Y.; Zou, Y.; Chen, X. Aberrant Histone Methylation and the Effect of Suv39H1 SiRNA on Gastric Carcinoma. Oncol. Rep. 2014, 31, 2593–2600. [Google Scholar] [CrossRef]
- Song, Z.; Wei, Z.; Wang, Q.; Zhang, X.; Tao, X.; Wu, N.; Liu, X.; Qian, J. The Role of DOT1L in the Proliferation and Prognosis of Gastric Cancer. Biosci. Rep. 2020, 40, BSR20193515. [Google Scholar] [CrossRef]
- Kari, V.; Raul, S.K.; Henck, J.M.; Kitz, J.; Kramer, F.; Kosinsky, R.L.; Übelmesser, N.; Mansour, W.Y.; Eggert, J.; Spitzner, M.; et al. The Histone Methyltransferase DOT1L Is Required for Proper DNA Damage Response, DNA Repair, and Modulates Chemotherapy Responsiveness. Clin. Epigenet. 2019, 11, 4. [Google Scholar] [CrossRef]
- Komatsu, S.; Ichikawa, D.; Hirajima, S.; Nagata, H.; Nishimura, Y.; Kawaguchi, T.; Miyamae, M.; Okajima, W.; Ohashi, T.; Konishi, H.; et al. Overexpression of SMYD2 Contributes to Malignant Outcome in Gastric Cancer. Br. J. Cancer 2015, 112, 357–364. [Google Scholar] [CrossRef]
- Liu, D.; Liu, M.; Wang, W.; Li, X.; Shi, E.; Zhang, C.; Wang, Y.; Zhang, Y.; Wang, L.; Wang, X. SMYD Family Members Serve as Potential Prognostic Markers and Correlate with Immune Infiltrates in Gastric Cancer. J. Oncol. 2023, 2023, 6032864. [Google Scholar] [CrossRef]
- Bernard, B.J.; Nigam, N.; Burkitt, K.; Saloura, V. SMYD3: A Regulator of Epigenetic and Signaling Pathways in Cancer. Clin. Epigenet. 2021, 13, 45. [Google Scholar] [CrossRef]
- Dong, Q.-Q.; Wang, Q.-T.; Wang, L.; Jiang, Y.-X.; Liu, M.-L.; Hu, H.-J.; Liu, Y.; Zhou, H.; He, H.-P.; Zhang, T.-C.; et al. SMYD3-Associated Pathway Is Involved in the Anti-Tumor Effects of Sulforaphane on Gastric Carcinoma Cells. Food Sci. Biotechnol. 2018, 27, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Deng, J.; Luo, X.; Pan, Y.; Zhang, L.; Zhang, R.; Liang, H. Overexpression of SMYD3 Was Associated with Increased STAT3 Activation in Gastric Cancer. Med. Oncol. 2015, 32, 404. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, Y.; Kong, F.; Xin, W.; Li, X.; Liang, H.; Jia, Y. Elevated Levels of SET and MYND Domain-Containing Protein 3 Are Correlated with Overexpression of Transforming Growth Factor-Β1 in Gastric Cancer. J. Am. Coll. Surg. 2015, 221, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liu, H.; Luo, X.; Deng, J.; Pan, Y.; Liang, H. Overexpression of SMYD3 and Matrix Metalloproteinase-9 Are Associated with Poor Prognosis of Patients with Gastric Cancer. Tumor Biol. 2015, 36, 4377–4386. [Google Scholar] [CrossRef]
- Fasano, C.; Lepore Signorile, M.; De Marco, K.; Forte, G.; Sanese, P.; Grossi, V.; Simone, C. Identifying Novel SMYD3 Interactors on the Trail of Cancer Hallmarks. Comput. Struct. Biotechnol. J. 2022, 20, 1860–1875. [Google Scholar] [CrossRef]
- Hu, L.; Zang, M.; Wang, H.-X.; Zhang, B.-G.; Wang, Z.-Q.; Fan, Z.-Y.; Wu, H.; Li, J.-F.; Su, L.-P.; Yan, M.; et al. G9A Promotes Gastric Cancer Metastasis by Upregulating ITGB3 in a SET Domain-Independent Manner. Cell Death Dis. 2018, 9, 278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Wei, S.; Hu, J.; Xiong, Z. Upregulated Expression of G9a Is Correlated with Poor Prognosis of Gastric Cancer Patients. Medicine 2019, 98, e18212. [Google Scholar] [CrossRef]
- Yin, C.; Ke, X.; Zhang, R.; Hou, J.; Dong, Z.; Wang, F.; Zhang, K.; Zhong, X.; Yang, L.; Cui, H. G9a Promotes Cell Proliferation and Suppresses Autophagy in Gastric Cancer by Directly Activating MTOR. FASEB J. 2019, 33, 14036–14050. [Google Scholar] [CrossRef]
- Lin, X.; Huang, Y.; Zou, Y.; Chen, X.; Ma, X. Depletion of G9a Gene Induces Cell Apoptosis in Human Gastric Carcinoma. Oncol. Rep. 2016, 35, 3041–3049. [Google Scholar] [CrossRef]
- Wang, L.; Chen, J.; Zuo, Q.; Wu, C.; Yu, T.; Zheng, P.; Huang, H.; Deng, J.; Fang, L.; Liu, H.; et al. Calreticulin Enhances Gastric Cancer Metastasis by Dimethylating H3K9 in the E-Cadherin Promoter Region Mediating by G9a. Oncogenesis 2022, 11, 29. [Google Scholar] [CrossRef]
- Yang, Q.; Zhu, Q.; Lu, X.; Du, Y.; Cao, L.; Shen, C.; Hou, T.; Li, M.; Li, Z.; Liu, C.; et al. G9a Coordinates with the RPA Complex to Promote DNA Damage Repair and Cell Survival. Proc. Natl. Acad. Sci. USA 2017, 114, E6054–E6063. [Google Scholar] [CrossRef] [PubMed]
- Lazaro-Camp, V.J.; Salari, K.; Meng, X.; Yang, S. SETDB1 in Cancer: Overexpression and Its Therapeutic Implications. Am. J. Cancer Res. 2021, 11, 1803–1827. [Google Scholar]
- Sp1-Induced SETDB1 Overexpression Transcriptionally Inhibits HPGD in a β-Catenin-Dependent Manner and Promotes the Proliferation and Metastasis of Gastric Cancer-PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/36316108/ (accessed on 13 June 2023).
- Shang, W.; Wang, Y.; Liang, X.; Li, T.; Shao, W.; Liu, F.; Cui, X.; Wang, Y.; Lv, L.; Chai, L.; et al. SETDB1 Promotes Gastric Carcinogenesis and Metastasis via Upregulation of CCND1 and MMP9 Expression. J. Pathol. 2021, 253, 148–159. [Google Scholar] [CrossRef]
- Nishikawaji, T.; Akiyama, Y.; Shimada, S.; Kojima, K.; Kawano, T.; Eishi, Y.; Yuasa, Y.; Tanaka, S. Oncogenic Roles of the SETDB2 Histone Methyltransferase in Gastric Cancer. Oncotarget 2016, 7, 67251–67265. [Google Scholar] [CrossRef]
- Choi, Y.J.; Oh, H.R.; Choi, M.R.; Gwak, M.; An, C.H.; Chung, Y.J.; Yoo, N.J.; Lee, S.H. Frameshift Mutation of a Histone Methylation-Related Gene SETD1B and Its Regional Heterogeneity in Gastric and Colorectal Cancers with High Microsatellite Instability. Hum. Pathol. 2014, 45, 1674–1681. [Google Scholar] [CrossRef] [PubMed]
- Piao, L.; Che, N.; Li, H.; Li, M.; Feng, Y.; Liu, X.; Kim, S.; Jin, Y.; Xuan, Y. SETD8 Promotes Stemness Characteristics and Is a Potential Prognostic Biomarker of Gastric Adenocarcinoma. Exp. Mol. Pathol. 2020, 117, 104560. [Google Scholar] [CrossRef]
- Nourbakhsh, N.; Emadi-Baygi, M.; Salehi, R.; Nikpour, P. Gene Expression Analysis of Two Epithelial-Mesenchymal Transition-Related Genes: Long Noncoding RNA-ATB and SETD8 in Gastric Cancer Tissues. Adv. Biomed. Res. 2018, 7, 42. [Google Scholar] [CrossRef]
- Alagoz, M.; Katsuki, Y.; Ogiwara, H.; Ogi, T.; Shibata, A.; Kakarougkas, A.; Jeggo, P. SETDB1, HP1 and SUV39 Promote Repositioning of 53BP1 to Extend Resection during Homologous Recombination in G2 Cells. Nucleic Acids Res. 2015, 43, 7931–7944. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, L.; Sun, J.; Hu, X.; Kalvakolanu, D.V.; Ren, H.; Guo, B. Roles for the Methyltransferase SETD8 in DNA Damage Repair. Clin. Epigenet. 2022, 14, 34. [Google Scholar] [CrossRef]
- Meng, X.; Zhao, Y.; Liu, J.; Wang, L.; Dong, Z.; Zhang, T.; Gu, X.; Zheng, Z. Comprehensive Analysis of Histone Modification-Associated Genes on Differential Gene Expression and Prognosis in Gastric Cancer. Exp. Ther. Med. 2019, 18, 2219–2230. [Google Scholar] [CrossRef]
- Shen, D.-D.; Pang, J.-R.; Bi, Y.-P.; Zhao, L.-F.; Li, Y.-R.; Zhao, L.-J.; Gao, Y.; Wang, B.; Wang, N.; Wei, L.; et al. LSD1 Deletion Decreases Exosomal PD-L1 and Restores T-Cell Response in Gastric Cancer. Mol. Cancer 2022, 21, 75. [Google Scholar] [CrossRef]
- Fang, R.; Xu, J.; Lin, H.; Xu, X.; Tian, F. The Histone Demethylase Lysine-Specific Demethylase-1-Mediated Epigenetic Silence of KLF2 Contributes to Gastric Cancer Cell Proliferation, Migration, and Invasion. Tumor Biol. 2017, 39, 1010428317698356. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Tian, X.; Sui, C.-G.; Jiang, Y.-H.; Liu, Y.-P.; Meng, F.-D. Interference of Lysine-Specific Demethylase 1 Inhibits Cellular Invasion and Proliferation in Vivo in Gastric Cancer MKN-28 Cells. Biomed. Pharmacother. 2016, 82, 498–508. [Google Scholar] [CrossRef]
- Gale, M.; Sayegh, J.; Cao, J.; Norcia, M.; Gareiss, P.; Hoyer, D.; Merkel, J.S.; Yan, Q. Screen-Identified Selective Inhibitor of Lysine Demethylase 5A Blocks Cancer Cell Growth and Drug Resistance. Oncotarget 2016, 7, 39931–39944. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, L.; Song, P.; Geng, X.; Liang, X.; Zhou, M.; Wang, Y.; Chen, C.; Jia, J.; Zeng, J. Critical Role of Histone Demethylase RBP2 in Human Gastric Cancer Angiogenesis. Mol. Cancer 2014, 13, 81. [Google Scholar] [CrossRef]
- Zeng, J.; Ge, Z.; Wang, L.; Li, Q.; Wang, N.; Björkholm, M.; Jia, J.; Xu, D. The Histone Demethylase RBP2 Is Overexpressed in Gastric Cancer and Its Inhibition Triggers Senescence of Cancer Cells. Gastroenterology 2010, 138, 981–992. [Google Scholar] [CrossRef]
- Liang, X.; Zeng, J.; Wang, L.; Shen, L.; Ma, X.; Li, S.; Wu, Y.; Ma, L.; Ci, X.; Guo, Q.; et al. Histone Demethylase RBP2 Promotes Malignant Progression of Gastric Cancer through TGF-Β1-(p-Smad3)-RBP2-E-Cadherin-Smad3 Feedback Circuit. Oncotarget 2015, 6, 17661–17674. [Google Scholar] [CrossRef][Green Version]
- Wang, Z.; Tang, F.; Qi, G.; Yuan, S.; Zhang, G.; Tang, B.; He, S. KDM5B Is Overexpressed in Gastric Cancer and Is Required for Gastric Cancer Cell Proliferation and Metastasis. Am. J. Cancer Res. 2015, 5, 87–100. [Google Scholar]
- Zhao, L.-F.; Qi, F.-Y.; Zhang, J.-G.; Pang, J.-R.; Ren, H.-M.; Shen, D.-D.; Zhao, L.-J.; Qi, L.; Liu, H.-M.; Zheng, Y.-C. Identification of the Upstream Regulators of KDM5B in Gastric Cancer. Life Sci. 2022, 298, 120458. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Wu, W.; Cheng, G.; Qian, M.; Hu, K.; Yin, G.; Wang, S. Enhancement of Proliferation and Invasion of Gastric Cancer Cell by KDM5C Via Decrease in P53 Expression. Technol. Cancer Res. Treat. 2017, 16, 141–149. [Google Scholar] [CrossRef]
- Huang, Y.; Liu, Y.; Yu, L.; Chen, J.; Hou, J.; Cui, L.; Ma, D.; Lu, W. Histone Demethylase KDM2A Promotes Tumor Cell Growth and Migration in Gastric Cancer. Tumor Biol. 2015, 36, 271–278. [Google Scholar] [CrossRef]
- Li, S.; Wu, L.; Wang, Q.; Li, Y.; Wang, X. KDM4B Promotes Epithelial-Mesenchymal Transition through up-Regulation of ZEB1 in Pancreatic Cancer. Acta Biochim. Biophys. Sin. 2015, 47, 997–1004. [Google Scholar] [CrossRef]
- Jing, J.-C.; Feng, Z.; Chen, Z.-H.; Ji, B.-N.; Hong, J.; Tang, N.; Yu, J.L.; Wang, S.-Y. KDM4B Promotes Gastric Cancer Metastasis by Regulating MiR-125b-Mediated Activation of Wnt Signaling. J. Cell. Biochem. 2019, 120, 7897–7906. [Google Scholar] [CrossRef]
- Wang, S.; Wang, Y.; Zhu, H.; Chen, M.; Zhang, L. Expression Pattern of Histone Lysine-Specific Demethylase 6B in Gastric Cancer. Oncol. Lett. 2021, 21, 491. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, Y.; Yang, Z.; Cui, X.; Zheng, L.; Fu, Y.; Shao, W.; Zhang, L.; Yang, Q.; Jia, J. KDM6B Promotes Gastric Carcinogenesis and Metastasis via Upregulation of CXCR4 Expression. Cell Death Dis. 2022, 13, 1068. [Google Scholar] [CrossRef] [PubMed]
- Biggar, K.K.; Li, S.S.-C. Non-Histone Protein Methylation as a Regulator of Cellular Signalling and Function. Nat. Rev. Mol. Cell Biol. 2015, 16, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Berger, S.L. The Emerging Field of Dynamic Lysine Methylation of Non-Histone Proteins. Curr. Opin. Genet. Dev. 2008, 18, 152–158. [Google Scholar] [CrossRef]
- Lakin, N.D.; Jackson, S.P. Regulation of P53 in Response to DNA Damage. Oncogene 1999, 18, 7644–7655. [Google Scholar] [CrossRef]
- Huang, J.; Perez-Burgos, L.; Placek, B.J.; Sengupta, R.; Richter, M.; Dorsey, J.A.; Kubicek, S.; Opravil, S.; Jenuwein, T.; Berger, S.L. Repression of P53 Activity by Smyd2-Mediated Methylation. Nature 2006, 444, 629–632. [Google Scholar] [CrossRef]
- Shi, X.; Kachirskaia, I.; Yamaguchi, H.; West, L.E.; Wen, H.; Wang, E.W.; Dutta, S.; Appella, E.; Gozani, O. Modulation of P53 Function by SET8-Mediated Methylation at Lysine 382. Mol. Cell 2007, 27, 636–646. [Google Scholar] [CrossRef]
- Huang, J.; Dorsey, J.; Chuikov, S.; Zhang, X.; Jenuwein, T.; Reinberg, D.; Berger, S.L. G9a and Glp Methylate Lysine 373 in the Tumor Suppressor P53. J. Biol. Chem. 2010, 285, 9636–9641. [Google Scholar] [CrossRef]
- Cho, H.-S.; Hayami, S.; Toyokawa, G.; Maejima, K.; Yamane, Y.; Suzuki, T.; Dohmae, N.; Kogure, M.; Kang, D.; Neal, D.E.; et al. RB1 Methylation by SMYD2 Enhances Cell Cycle Progression through an Increase of RB1 Phosphorylation. Neoplasia 2012, 14, 476–486. [Google Scholar] [CrossRef]
- Piao, L.; Kang, D.; Suzuki, T.; Masuda, A.; Dohmae, N.; Nakamura, Y.; Hamamoto, R. The Histone Methyltransferase SMYD2 Methylates PARP1 and Promotes Poly(ADP-Ribosyl)Ation Activity in Cancer Cells. Neoplasia 2014, 16, 257–264.e2. [Google Scholar] [CrossRef]
- Huang, P.H.; Cook, R.; Mittnacht, S. RB in DNA Repair. Oncotarget 2015, 6, 20746–20747. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhou, T.; Wang, Z.; Yi, F.; Li, C.; Guo, W.; Xu, H.; Cui, H.; Dong, X.; Liu, J.; et al. Post-Translational Modifications of PCNA in Control of DNA Synthesis and DNA Damage Tolerance-the Implications in Carcinogenesis. Int. J. Biol. Sci. 2021, 17, 4047–4059. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Chang, Y.; Yang, J.; Wei, Q. Post-Translational Modifications of Proliferating Cell Nuclear Antigen: A Key Signal Integrator for DNA Damage Response (Review). Oncol. Lett. 2014, 7, 1363–1369. [Google Scholar] [CrossRef]
- Wang, P.; Zhao, L.; Rui, Y.; Ding, Y. SMYD3 Regulates Gastric Cancer Progression and Macrophage Polarization through EZH2 Methylation. Cancer Gene Ther. 2023, 30, 575–581. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, Q. The Roles of EZH2 in Cancer and Its Inhibitors. Med. Oncol. 2023, 40, 167. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Q.-T.; Liu, Y.-P.; Dong, Q.-Q.; Hu, H.-J.; Miao, Z.; Li, S.; Liu, Y.; Zhou, H.; Zhang, T.-C.; et al. ATM Signaling Pathway Is Implicated in the SMYD3-Mediated Proliferation and Migration of Gastric Cancer Cells. J. Gastric Cancer 2017, 17, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Sanese, P.; Fasano, C.; Buscemi, G.; Bottino, C.; Corbetta, S.; Fabini, E.; Silvestri, V.; Valentini, V.; Disciglio, V.; Forte, G.; et al. Targeting SMYD3 to Sensitize Homologous Recombination-Proficient Tumors to PARP-Mediated Synthetic Lethality. iScience 2020, 23, 101604. [Google Scholar] [CrossRef]
- Sanese, P.; Fasano, C.; Simone, C. Playing on the Dark Side: SMYD3 Acts as a Cancer Genome Keeper in Gastrointestinal Malignancies. Cancers 2021, 13, 4427. [Google Scholar] [CrossRef] [PubMed]
- Usui, G.; Matsusaka, K.; Mano, Y.; Urabe, M.; Funata, S.; Fukayama, M.; Ushiku, T.; Kaneda, A. DNA Methylation and Genetic Aberrations in Gastric Cancer. Digestion 2021, 102, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Rong, H.; Xu, J.; Cao, R.; Li, S.; Gao, Y.; Cheng, B.; Zhou, T. DNA Methylation: An Important Biomarker and Therapeutic Target for Gastric Cancer. Front. Genet. 2022, 13, 823905. [Google Scholar] [CrossRef]
- Sogutlu, F.; Pekerbas, M.; Biray Avci, C. Epigenetic Signatures in Gastric Cancer: Current Knowledge and Future Perspectives. Expert Rev. Mol. Diagn. 2022, 22, 1063–1075. [Google Scholar] [CrossRef]
- Ksiâa, F.; Ziadi, S.; Dhiab, M.B.; Gacem, R.B.; Trimeche, M. Increased DNA Methyltransferase 1 Protein Expression Correlates Significantly with Intestinal Histological Type and Gender in Gastric Carcinomas. Adv. Med. Sci. 2015, 60, 50–57. [Google Scholar] [CrossRef]
- Yang, J.; Wei, X.; Wu, Q.; Xu, Z.; Gu, D.; Jin, Y.; Shen, Y.; Huang, H.; Fan, H.; Chen, J. Clinical Significance of the Expression of DNA Methyltransferase Proteins in Gastric Cancer. Mol. Med. Rep. 2011, 4, 1139–1143. [Google Scholar] [CrossRef]
- Ding, W.-J.; Fang, J.-Y.; Chen, X.-Y.; Peng, Y.-S. The Expression and Clinical Significance of DNA Methyltransferase Proteins in Human Gastric Cancer. Dig. Dis. Sci. 2008, 53, 2083–2089. [Google Scholar] [CrossRef] [PubMed]
- Etoh, T.; Kanai, Y.; Ushijima, S.; Nakagawa, T.; Nakanishi, Y.; Sasako, M.; Kitano, S.; Hirohashi, S. Increased DNA Methyltransferase 1 (DNMT1) Protein Expression Correlates Significantly with Poorer Tumor Differentiation and Frequent DNA Hypermethylation of Multiple CpG Islands in Gastric Cancers. Am. J. Pathol. 2004, 164, 689–699. [Google Scholar] [CrossRef]
- Fang, J.-Y.; Cheng, Z.-H.; Chen, Y.-X.; Lu, R.; Yang, L.; Zhu, H.-Y.; Lu, L.-G. Expression of Dnmt1, Demethylase, MeCP2 and Methylation of Tumor-Related Genes in Human Gastric Cancer. World J. Gastroenterol. 2004, 10, 3394–3398. [Google Scholar] [CrossRef]
- Kataoka, I.; Funata, S.; Nagahama, K.; Isogaya, K.; Takeuchi, H.; Abe, N.; Shibahara, J. DNMT3A Overexpression Is Associated with Aggressive Behavior and Enteroblastic Differentiation of Gastric Adenocarcinoma. Ann. Diagn. Pathol. 2020, 44, 151456. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Meissner, A. DNA Methylation: Roles in Mammalian Development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Bernal, C.; Vargas, M.; Ossandón, F.; Santibáñez, E.; Urrutia, J.; Luengo, V.; Zavala, L.F.; Backhouse, C.; Palma, M.; Argandoña, J.; et al. DNA Methylation Profile in Diffuse Type Gastric Cancer: Evidence for Hypermethylation of the BRCA1 Promoter Region in Early-Onset Gastric Carcinogenesis. Biol. Res. 2008, 41, 303–315. [Google Scholar] [CrossRef]
- Yoshida, K.; Miki, Y. Role of BRCA1 and BRCA2 as Regulators of DNA Repair, Transcription, and Cell Cycle in Response to DNA Damage. Cancer Sci. 2004, 95, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Keijzers, G.; Rasmussen, L.J. DNA Mismatch Repair and Its Many Roles in Eukaryotic Cells. Mutat. Res. Rev. Mutat. Res. 2017, 773, 174–187. [Google Scholar] [CrossRef]
- Yanagisawa, Y.; Akiyama, Y.; Iida, S.; Ito, E.; Nomizu, T.; Sugihara, K.; Yuasa, Y.; Maruyama, K. Methylation of the HMLH1 Promoter in Familial Gastric Cancer with Microsatellite Instability. Int. J. Cancer 2000, 85, 50–53. [Google Scholar] [CrossRef]
- Moura Lima, E.; Ferreira Leal, M.; Cardoso Smith, M.d.A.; Rodríguez Burbano, R.; Pimentel de Assumpção, P.; Bello, M.J.; Rey, J.A.; Ferreira de Lima, F.; Casartelli, C. DNA Mismatch Repair Gene Methylation in Gastric Cancer in Individuals from Northern Brazil. Biocell 2008, 32, 237–243. [Google Scholar] [CrossRef]
- Guan, J.; Lu, C.; Jin, Q.; Lu, H.; Chen, X.; Tian, L.; Zhang, Y.; Ortega, J.; Zhang, J.; Siteni, S.; et al. MLH1 Deficiency-Triggered DNA Hyperexcision by Exonuclease 1 Activates the CGAS-STING Pathway. Cancer Cell 2021, 39, 109–121.e5. [Google Scholar] [CrossRef]
- Jiang, M.; Jia, K.; Wang, L.; Li, W.; Chen, B.; Liu, Y.; Wang, H.; Zhao, S.; He, Y.; Zhou, C. Alterations of DNA Damage Repair in Cancer: From Mechanisms to Applications. Ann. Transl. Med. 2020, 8, 1685. [Google Scholar] [CrossRef]
- Conde-Pérezprina, J.C.; León-Galván, M.Á.; Konigsberg, M. DNA Mismatch Repair System: Repercussions in Cellular Homeostasis and Relationship with Aging. Oxid. Med. Cell. Longev. 2012, 2012, 728430. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Cao, T.; Han, Y.-D.; Huang, F.-S. Relationships between MGMT Promoter Methylation and Gastric Cancer: A Meta-Analysis. OncoTargets Ther. 2016, 9, 6049–6057. [Google Scholar] [CrossRef]
- Nissar, B.; Kadla, S.A.; Wani, K.A.; Shah, I.A.; Ganai, B.A. Promoter CpG Island Hypermethylation and down Regulation of XRCC1 Gene Can Augment in the Gastric Carcinogenesis Events. Mol. Biol. Rep. 2021, 48, 405–412. [Google Scholar] [CrossRef]
- Jin, B.; Robertson, K.D. DNA Methyltransferases, DNA Damage Repair, and Cancer. Adv. Exp. Med. Biol. 2013, 754, 3–29. [Google Scholar] [CrossRef]
- Mortusewicz, O.; Schermelleh, L.; Walter, J.; Cardoso, M.C.; Leonhardt, H. Recruitment of DNA Methyltransferase I to DNA Repair Sites. Proc. Natl. Acad. Sci. USA 2005, 102, 8905–8909. [Google Scholar] [CrossRef]
- Palii, S.S.; Van Emburgh, B.O.; Sankpal, U.T.; Brown, K.D.; Robertson, K.D. DNA Methylation Inhibitor 5-Aza-2′-Deoxycytidine Induces Reversible Genome-Wide DNA Damage That Is Distinctly Influenced by DNA Methyltransferases 1 and 3B. Mol. Cell. Biol. 2008, 28, 752–771. [Google Scholar] [CrossRef] [PubMed]
- Ha, K.; Lee, G.E.; Palii, S.S.; Brown, K.D.; Takeda, Y.; Liu, K.; Bhalla, K.N.; Robertson, K.D. Rapid and Transient Recruitment of DNMT1 to DNA Double-Strand Breaks Is Mediated by Its Interaction with Multiple Components of the DNA Damage Response Machinery. Hum. Mol. Genet. 2011, 20, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Rothenberg, E.; Ramsden, D.A.; Lieber, M.R. The Molecular Basis and Disease Relevance of Non-Homologous DNA End Joining. Nat. Rev. Mol. Cell Biol. 2020, 21, 765–781. [Google Scholar] [CrossRef]
- Wang, Y.; Zheng, K.; Huang, Y.; Xiong, H.; Su, J.; Chen, R.; Zou, Y. PARP Inhibitors in Gastric Cancer: Beacon of Hope. J. Exp. Clin. Cancer Res. 2021, 40, 211. [Google Scholar] [CrossRef]
- Sun, Y.; McCorvie, T.J.; Yates, L.A.; Zhang, X. Structural Basis of Homologous Recombination. Cell. Mol. Life Sci. 2020, 77, 3–18. [Google Scholar] [CrossRef]
- Yamane, A.; Robbiani, D.F.; Resch, W.; Bothmer, A.; Nakahashi, H.; Oliveira, T.; Rommel, P.C.; Brown, E.J.; Nussenzweig, A.; Nussenzweig, M.C.; et al. RPA Accumulation during Class Switch Recombination Represents 5′-3′ DNA-End Resection during the S-G2/M Phase of the Cell Cycle. Cell Rep. 2013, 3, 138–147. [Google Scholar] [CrossRef]
- Toh, M.; Ngeow, J. Homologous Recombination Deficiency: Cancer Predispositions and Treatment Implications. Oncologist 2021, 26, e1526–e1537. [Google Scholar] [CrossRef] [PubMed]
- Lordick, F.; Shitara, K.; Janjigian, Y.Y. New Agents on the Horizon in Gastric Cancer. Ann. Oncol. 2017, 28, 1767–1775. [Google Scholar] [CrossRef]
- Zhang, Z.-Z.; Liu, Y.J.C.; Yin, X.-L.; Zhan, P.; Gu, Y.; Ni, X.-Z. Loss of BRCA1 Expression Leads to Worse Survival in Patients with Gastric Carcinoma. World J. Gastroenterol. 2013, 19, 1968–1974. [Google Scholar] [CrossRef]
- Molinaro, E.; Andrikou, K.; Casadei-Gardini, A.; Rovesti, G. BRCA in Gastrointestinal Cancers: Current Treatments and Future Perspectives. Cancers 2020, 12, 3346. [Google Scholar] [CrossRef]
- Kim, H.S.; Kim, M.A.; Hodgson, D.; Harbron, C.; Wellings, R.; O’Connor, M.J.; Womack, C.; Yin, X.; Bang, Y.-J.; Im, S.-A.; et al. Concordance of ATM (Ataxia Telangiectasia Mutated) Immunohistochemistry between Biopsy or Metastatic Tumor Samples and Primary Tumors in Gastric Cancer Patients. Pathobiology 2013, 80, 127–137. [Google Scholar] [CrossRef]
- Fan, Y.; Ying, H.; Wu, X.; Chen, H.; Hu, Y.; Zhang, H.; Wu, L.; Yang, Y.; Mao, B.; Zheng, L. The Mutational Pattern of Homologous Recombination (HR)-Associated Genes and Its Relevance to the Immunotherapeutic Response in Gastric Cancer. Cancer Biol. Med. 2020, 17, 1002–1013. [Google Scholar] [CrossRef]
- Liu, M.; Fan, H.; Li, T.; Sihong, L.; Qiao, S.; Bi, J. Low Expression of CDK12 in Gastric Cancer Is Correlated with Advanced Stage and Poor Outcome. Pathol. Res. Pract. 2020, 216, 152962. [Google Scholar] [CrossRef]
- Bian, L.; Meng, Y.; Zhang, M.; Li, D. MRE11-RAD50-NBS1 Complex Alterations and DNA Damage Response: Implications for Cancer Treatment. Mol. Cancer 2019, 18, 169. [Google Scholar] [CrossRef] [PubMed]
- Koustas, E.; Karamouzis, M.V.; Sarantis, P.; Schizas, D.; Papavassiliou, A.G. Inhibition of C-MET Increases the Antitumour Activity of PARP Inhibitors in Gastric Cancer Models. J. Cell Mol. Med. 2020, 24, 10420–10431. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wei, M.; Xu, J.; Hua, J.; Liang, C.; Meng, Q.; Zhang, Y.; Liu, J.; Zhang, B.; Yu, X.; et al. PARP Inhibitors in Pancreatic Cancer: Molecular Mechanisms and Clinical Applications. Mol. Cancer 2020, 19, 49. [Google Scholar] [CrossRef] [PubMed]
- Alhusaini, A.; Cannon, A.; Maher, S.G.; Reynolds, J.V.; Lynam-Lennon, N. Therapeutic Potential of PARP Inhibitors in the Treatment of Gastrointestinal Cancers. Biomedicines 2021, 9, 1024. [Google Scholar] [CrossRef]
- Jackson, S.E.; Chester, J.D. Personalised Cancer Medicine. Int. J. Cancer 2015, 137, 262–266. [Google Scholar] [CrossRef] [PubMed]
- D’Andrea, A.D. Mechanisms of PARP Inhibitor Sensitivity and Resistance. DNA Repair 2018, 71, 172–176. [Google Scholar] [CrossRef]
- Rose, M.; Burgess, J.T.; O’Byrne, K.; Richard, D.J.; Bolderson, E. PARP Inhibitors: Clinical Relevance, Mechanisms of Action and Tumor Resistance. Front. Cell Dev. Biol. 2020, 8, 564601. [Google Scholar] [CrossRef] [PubMed]
- Deeks, E.D. Olaparib: First Global Approval. Drugs 2015, 75, 231–240. [Google Scholar] [CrossRef] [PubMed]
- Bang, Y.-J.; Xu, R.-H.; Chin, K.; Lee, K.-W.; Park, S.H.; Rha, S.Y.; Shen, L.; Qin, S.; Xu, N.; Im, S.-A.; et al. Olaparib in Combination with Paclitaxel in Patients with Advanced Gastric Cancer Who Have Progressed Following First-Line Therapy (GOLD): A Double-Blind, Randomised, Placebo-Controlled, Phase 3 Trial. Lancet Oncol. 2017, 18, 1637–1651. [Google Scholar] [CrossRef]
- AbbVie (Prior Sponsor, Abbott) A Phase 1 Open-Label, Dose-Escalation Study of Veliparib in Combination with Bimonthly FOLFIRI in Subjects with Advanced Solid Tumors. 2021. Available online: https://clinicaltrials.gov/study/NCT01123876 (accessed on 21 April 2023).
- Pfizer. A Phase 1, First In Human, Single-Arm, Open-Label Study of Once a Day, Orally Administered Talazoparib (Bmn 673) in Patients with Advanced or Recurrent Solid Tumors. 2018. Available online: https://clinicaltrials.gov/study/NCT01286987 (accessed on 21 April 2023).
- Zai Lab (Shanghai) Co., Ltd. A Multicenter, Open-Label, Single-Arm, Phase Ib Dose Escalation and Multi-Cohort Expansion Clinical Study to Assess the Safety and Antitumor Activity of Niraparib in Combination with MGD013 in Patients with Advanced or Metastatic Solid Tumor Who Failed Prior Treatment. 2022. Available online: https://clinicaltrials.gov/study/NCT04178460 (accessed on 21 April 2023).
- BeiGene. PARALLEL 303: A Phase 2, Double-Blind, Randomized Study of BGB-290 Versus Placebo as Maintenance Therapy in Patients with Inoperable Locally Advanced or Metastatic Gastric Cancer That Responded to Platinum-Based First-Line Chemotherapy. 2023. Available online: https://clinicaltrials.gov/study/NCT03427814 (accessed on 21 April 2023).
- Clovis Oncology, Inc. A Phase 2 Multicenter, Open-Label Study of Rucaparib as Treatment for Solid Tumors Associated with Deleterious Mutations in Homologous Recombination Repair Genes. 2022. Available online: https://clinicaltrials.gov/study/NCT04171700 (accessed on 21 April 2023).
- Jin, N.; Xia, Y.; Gao, Q. Combined PARP Inhibitors and Small Molecular Inhibitors in Solid Tumor Treatment (Review). Int. J. Oncol. 2023, 62, 28. [Google Scholar] [CrossRef]
- Jones, G.N.; Rooney, C.; Griffin, N.; Roudier, M.; Young, L.A.; Garcia-Trinidad, A.; Hughes, G.D.; Whiteaker, J.R.; Wilson, Z.; Odedra, R.; et al. PRAD50: A Novel and Clinically Applicable Pharmacodynamic Biomarker of Both ATM and ATR Inhibition Identified Using Mass Spectrometry and Immunohistochemistry. Br. J. Cancer 2018, 119, 1233–1243. [Google Scholar] [CrossRef]
- AstraZeneca. A Phase I, Open-Label Study to Assess the Safety, Tolerability, Pharmacokinetics and Preliminary Efficacy of Ascending Doses of AZD0156 Monotherapy or in Combination with Either Cytotoxic Chemotherapies or Novel Anti-Cancer Agents in Patients with Advanced Malignancies. 2022. Available online: https://clinicaltrials.gov/study/NCT02588105 (accessed on 4 September 2023).
- AstraZeneca. A Modular Phase I, Open-Label, Multicentre Study to Assess the Safety, Tolerability, Pharmacokinetics and Preliminary Anti-Tumour Activity of Ceralasertib in Combination with Cytotoxic Chemotherapy and/or DNA Damage Repair/Novel Anti-Cancer Agents in Patients with Advanced Solid Malignancies. 2023. Available online: https://clinicaltrials.gov/study/NCT02264678 (accessed on 4 September 2023).
- Kim, H.-Y.; Cho, Y.; Kang, H.; Yim, Y.-S.; Kim, S.-J.; Song, J.; Chun, K.-H. Targeting the WEE1 Kinase as a Molecular Targeted Therapy for Gastric Cancer. Oncotarget 2016, 7, 49902–49916. [Google Scholar] [CrossRef]
- Do, K.; Wilsker, D.; Ji, J.; Zlott, J.; Freshwater, T.; Kinders, R.J.; Collins, J.; Chen, A.P.; Doroshow, J.H.; Kummar, S. Phase I Study of Single-Agent AZD1775 (MK-1775), a Wee1 Kinase Inhibitor, in Patients with Refractory Solid Tumors. J. Clin. Oncol. 2015, 33, 3409–3415. [Google Scholar] [CrossRef]
| Altered DDR Gene or Protein | Function in HR | Study Design | Correlation | References |
|---|---|---|---|---|
| BRCA1 | Crucial component of the HR pathway, it recruits BRCA2 complexes to DSB sites | In vivo | Has been associated with reduced survival time and patients with diffused Lauren type, higher tumor grades, and advanced clinical stage | [146,148] |
| ATM | Key player that starts the HR pathway by phosphorylating various DDR proteins | In vitro and in vivo | Has been linked to poor differentiation, lymph node metastasis, and decreased survival | [149,150] |
| ATR | Important kinase that orchestrates the HR repair pathway | In vivo | May promote cancer development | [146] |
| PALB2 | BRCA2 complex interactor | In vivo | Potential valuable prognostic marker | [19,151] |
| RAD51C | BRCA2 interactor that plays a pivotal role in presynaptic filament assembly | In vivo | Potential valuable prognostic marker | [19,151] |
| CDK12 | Affects DNA repair and contributes to the maintenance of genomic integrity | In vitro and in vivo | Has been associated with worse prognosis | [152] |
| MRN | One of the first sensors and responders to DSBs | In vivo | Has been associated with cancer development and worse overall patient survival | [146,153] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Marco, K.; Sanese, P.; Simone, C.; Grossi, V. Histone and DNA Methylation as Epigenetic Regulators of DNA Damage Repair in Gastric Cancer and Emerging Therapeutic Opportunities. Cancers 2023, 15, 4976. https://doi.org/10.3390/cancers15204976
De Marco K, Sanese P, Simone C, Grossi V. Histone and DNA Methylation as Epigenetic Regulators of DNA Damage Repair in Gastric Cancer and Emerging Therapeutic Opportunities. Cancers. 2023; 15(20):4976. https://doi.org/10.3390/cancers15204976
Chicago/Turabian StyleDe Marco, Katia, Paola Sanese, Cristiano Simone, and Valentina Grossi. 2023. "Histone and DNA Methylation as Epigenetic Regulators of DNA Damage Repair in Gastric Cancer and Emerging Therapeutic Opportunities" Cancers 15, no. 20: 4976. https://doi.org/10.3390/cancers15204976
APA StyleDe Marco, K., Sanese, P., Simone, C., & Grossi, V. (2023). Histone and DNA Methylation as Epigenetic Regulators of DNA Damage Repair in Gastric Cancer and Emerging Therapeutic Opportunities. Cancers, 15(20), 4976. https://doi.org/10.3390/cancers15204976








