Comprehensive Analysis of DNA Methyltransferases Expression in Primary and Relapsed Ovarian Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. TNM Plot
2.2. TCGA
2.3. Proteomic Data Commons
2.4. GEPIA 2
2.5. Survival Analysis
2.6. Tissue Cohort
Patients
2.7. Imunohistochemistry
2.8. Evaluation of Immunohistochemical Stains
2.9. Statistical Analysis
3. Results
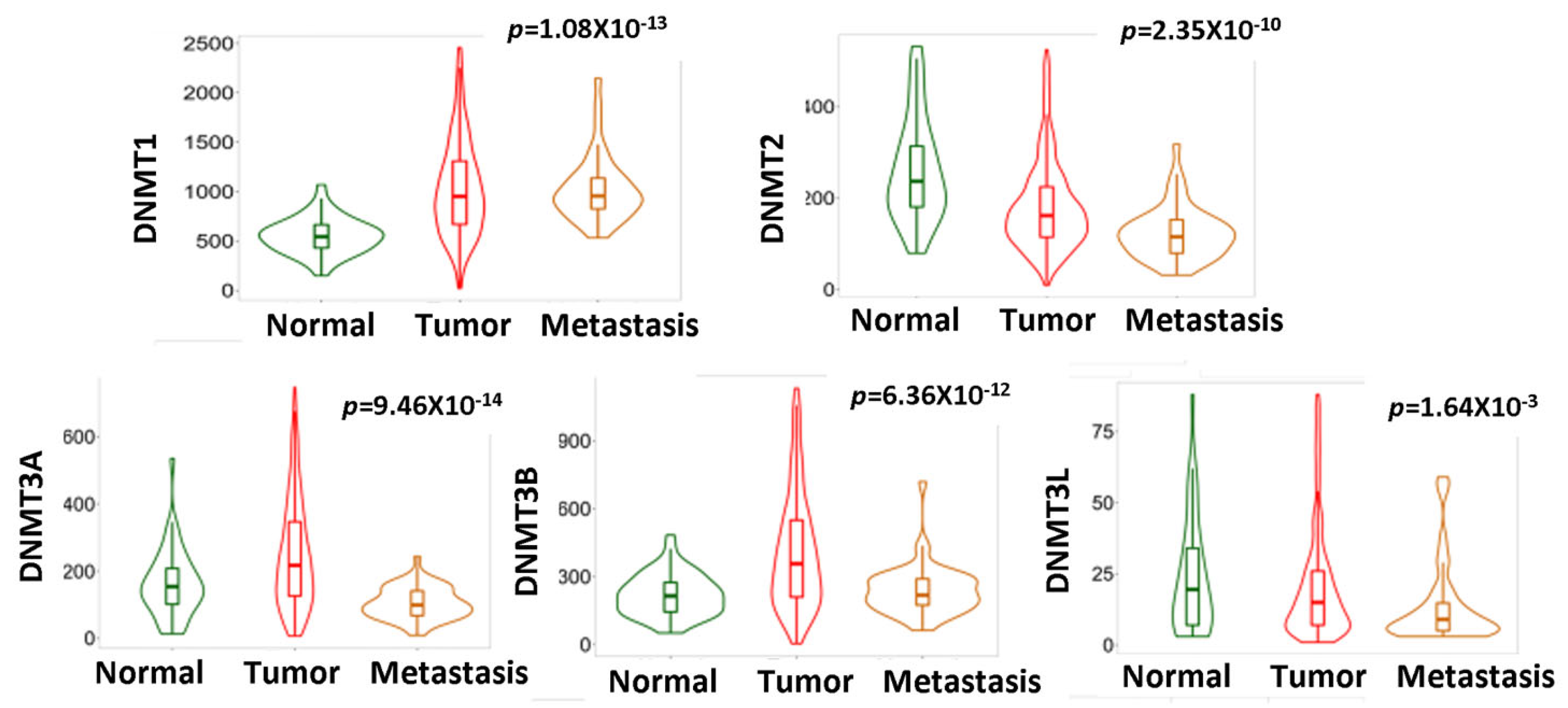
3.1. Aberrant DNMTs Expression Correlates with OΕC Development
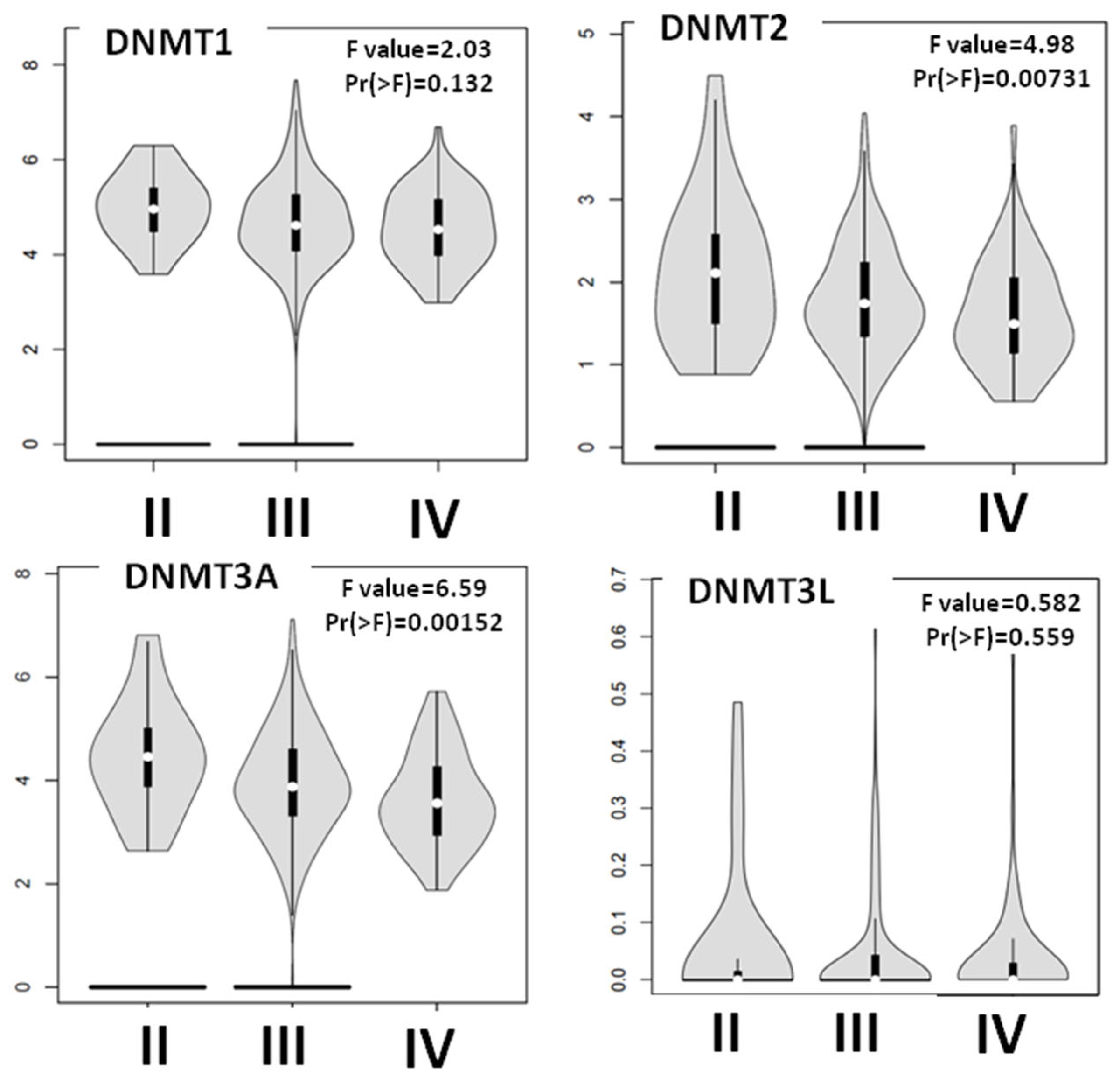
3.2. DNMTs Expression Is Altered with the Progression of EOC
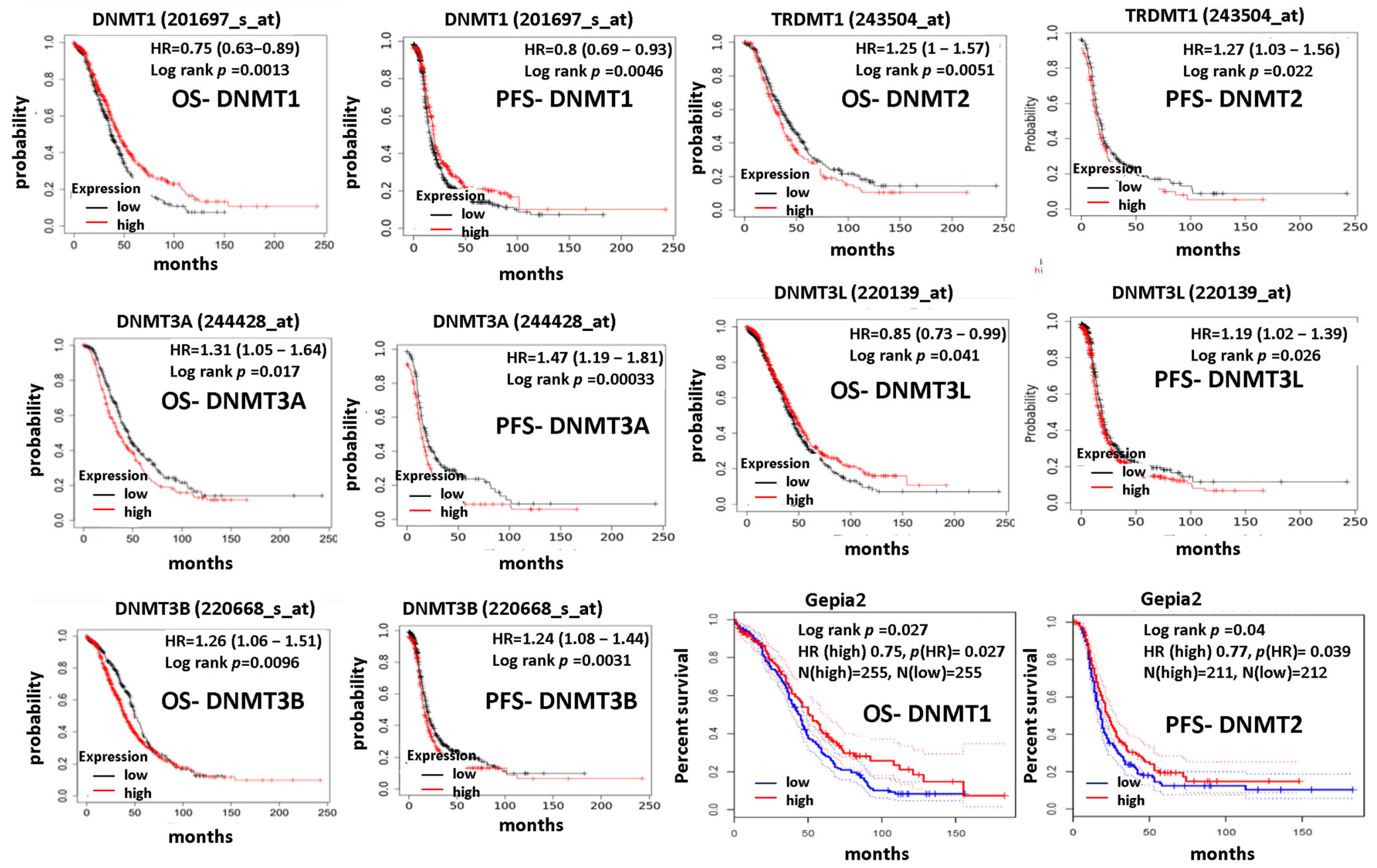
3.3. DNMTs Expression Correlates with Patients’ Prognoses
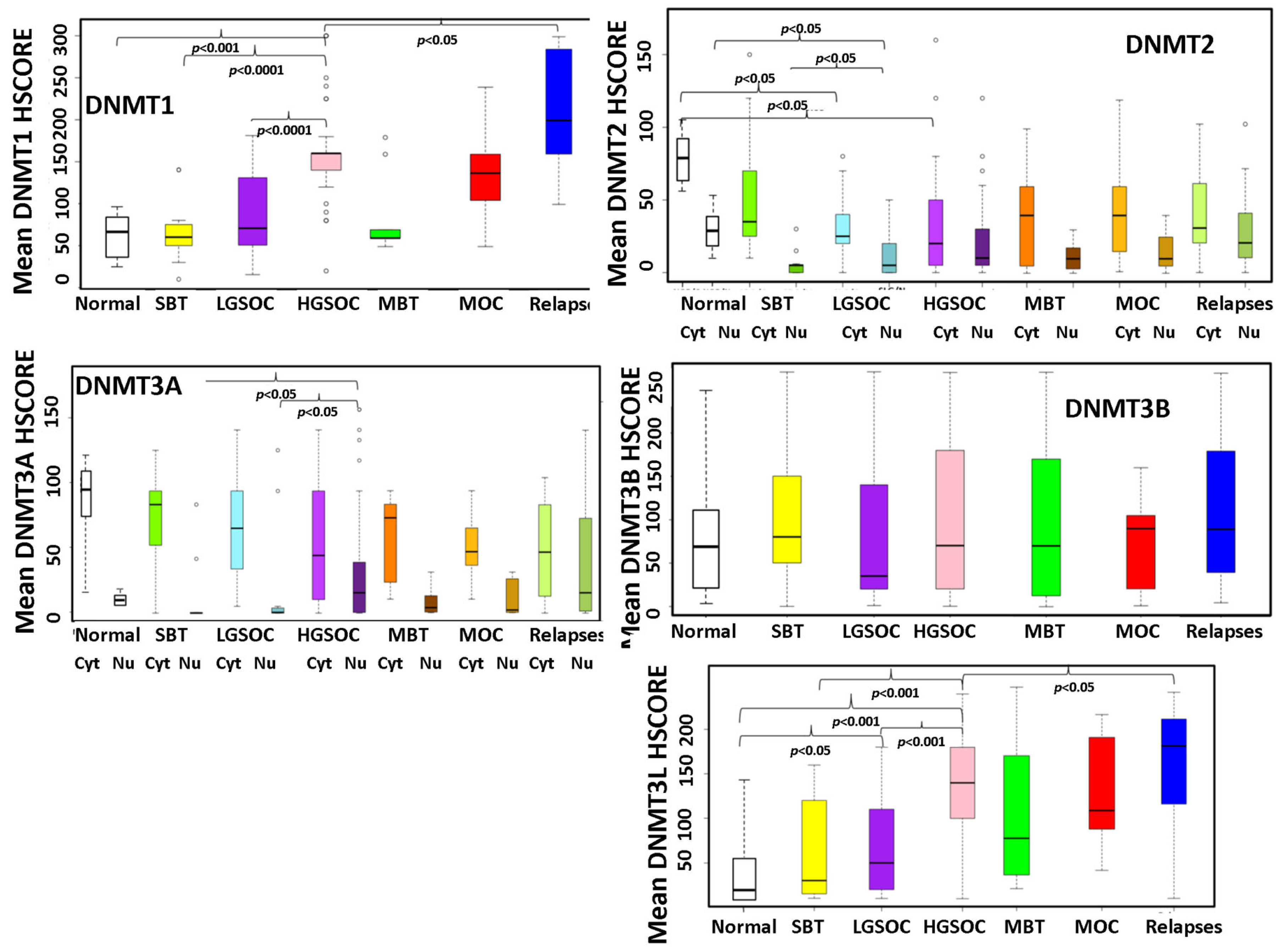
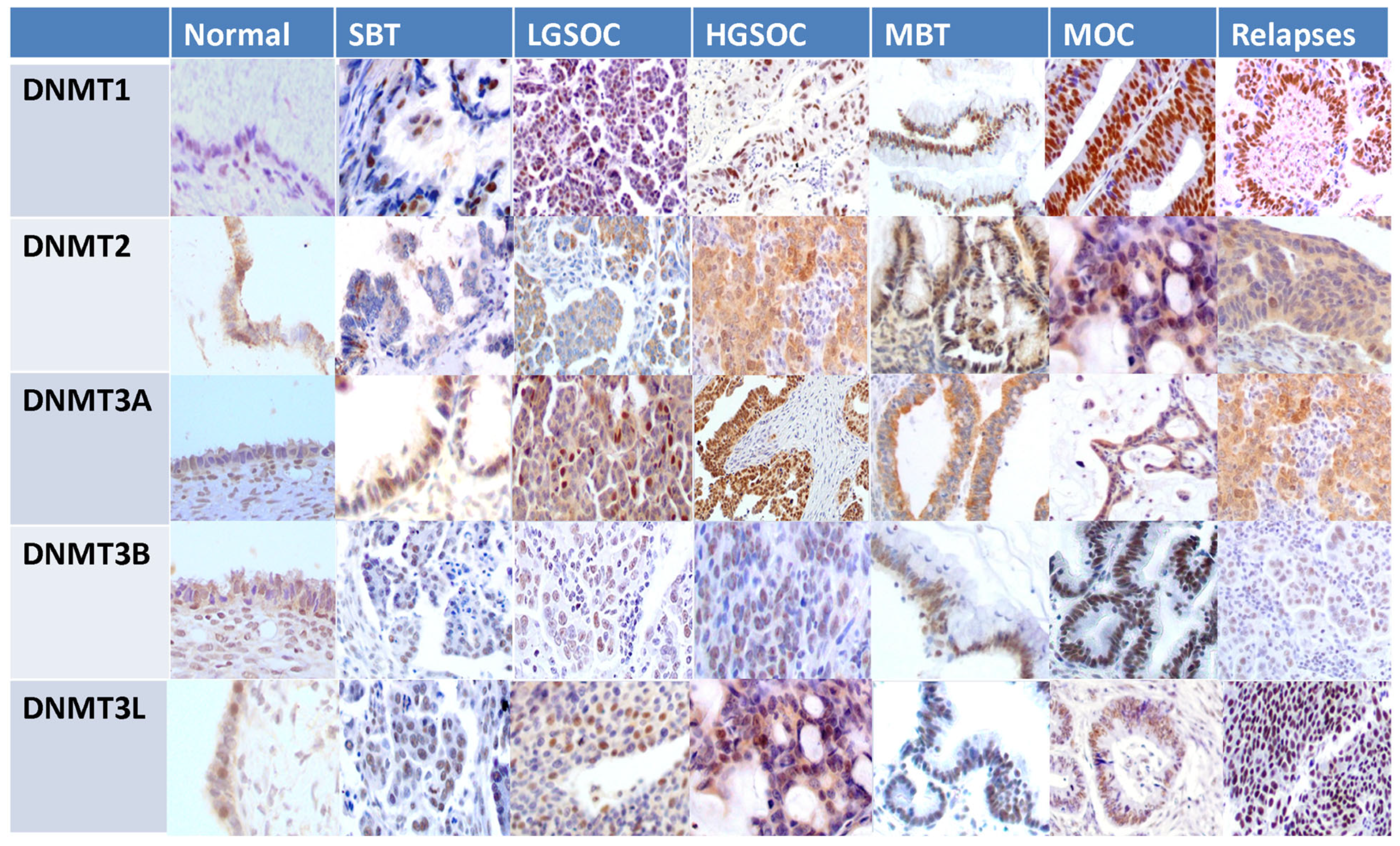
3.4. Expression of DNMTs in a Cohort of EOC Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cabasag, C.J.; Fagan, P.J.; Ferlay, J.; Vignat, J.; Laversanne, M.; Liu, L.; van der Aa, M.A.; Bray, F.; Soerjomataram, I. Ovarian cancer today and tomorrow: A global assessment by world region and Human Development Index using GLOBOCAN 2020. Int. J. Cancer 2022, 151, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Lheureux, S.; Gourley, C.; Vergote, I.; Oza, A.M. Epithelial ovarian cancer. Lancet 2019, 393, 1240–1253. [Google Scholar] [CrossRef] [PubMed]
- La Vecchia, C. Ovarian cancer: Epidemiology and risk factors. Eur. J. Cancer Prev. 2017, 26, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Romero, I.; Leskelä, S.; Mies, B.P.; Velasco, A.P.; Palacios, J. Morphological and molecular heterogeneity of epithelial ovarian cancer: Therapeutic implications. Eur. J. Cancer Suppl. 2020, 15, 1–15. [Google Scholar] [CrossRef]
- Jasen, P. From the “Silent Killer” to the “Whispering Disease”: Ovarian Cancer and the Uses of Metaphor. Med. Hist. 2009, 53, 489–512. [Google Scholar] [CrossRef]
- Hatano, Y.; Hatano, K.; Tamada, M.; Morishige, K.; Tomita, H.; Yanai, H.; Hara, A. A Comprehensive Review of Ovarian Serous Carcinoma. Adv. Anat. Pathol. 2019, 26, 329–339. [Google Scholar] [CrossRef]
- Ramus, S.J.; Gayther, S.A. The Contribution of BRCA1 and BRCA2 to Ovarian Cancer. Mol. Oncol. 2009, 3, 138–150. [Google Scholar] [CrossRef]
- Quesada, S.; Fabbro, M.; Solassol, J. Toward More Comprehensive Homologous Recombination Deficiency Assays in Ovarian Cancer, Part 1: Technical Considerations. Cancers 2022, 14, 1132. [Google Scholar] [CrossRef]
- Cole, A.J.; Dwight, T.; Gill, A.J.; Dickson, K.-A.; Zhu, Y.; Clarkson, A.; Gard, G.B.; Maidens, J.; Valmadre, S.; Clifton-Bligh, R.; et al. Assessing mutant p53 in primary high-grade serous ovarian cancer using immunohistochemistry and massively parallel sequencing. Sci. Rep. 2016, 6, 26191. [Google Scholar] [CrossRef]
- Ricciardi, E.; Baert, T.; Ataseven, B.; Heitz, F.; Prader, S.; Bommert, M.; Schneider, S.; du Bois, A.; Harter, P. Low-grade Serous Ovarian Carcinoma. Geburtshilfe Frauenheilkd. 2018, 78, 972–976. [Google Scholar] [CrossRef]
- Lapke, N.; Chen, C.-H.; Chang, T.-C.; Chao, A.; Lu, Y.-J.; Lai, C.-H.; Tan, K.T.; Chen, H.-C.; Lu, H.-Y.; Chen, S.-J. Genetic alterations and their therapeutic implications in epithelial ovarian cancer. BMC Cancer 2021, 21, 499. [Google Scholar] [CrossRef] [PubMed]
- Nonomura, Y.; Nakayama, K.; Nakamura, K.; Razia, S.; Yamashita, H.; Ishibashi, T.; Ishikawa, M.; Sato, S.; Nakayama, S.; Otsuki, Y.; et al. Ovarian Endometrioid and Clear Cell Carcinomas with Low Prevalence of Microsatellite Instability: A Unique Subset of Ovarian Carcinomas Could Benefit from Combination Therapy with Immune Checkpoint Inhibitors and Other Anticancer Agents. Healthcare 2022, 10, 694. [Google Scholar] [CrossRef] [PubMed]
- Lisio, M.-A.; Fu, L.; Goyeneche, A.; Gao, Z.; Telleria, C. High-Grade Serous Ovarian Cancer: Basic Sciences, Clinical and Therapeutic Standpoints. Int. J. Mol. Sci. 2019, 20, 952. [Google Scholar] [CrossRef] [PubMed]
- Matulonis, U.A.; Sood, A.K.; Fallowfield, L.; Howitt, B.E.; Sehouli, J.; Karlan, B.Y. Ovarian cancer. Nat. Rev. Dis. Prim. 2016, 2, 16061. [Google Scholar] [CrossRef]
- Yang, Q.; Yang, Y.; Zhou, N.; Tang, K.; Lau, W.B.; Lau, B.; Wang, W.; Xu, L.; Yang, Z.; Huang, S.; et al. Epigenetics in ovarian cancer: Premise, properties, and perspectives. Mol. Cancer 2018, 17, 109. [Google Scholar] [CrossRef]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef]
- Jin, B.; Li, Y.; Robertson, K.D. DNA Methylation: Superior or Subordinate in the Epigenetic Hierarchy? Genes Cancer 2011, 2, 607–617. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef]
- Jones, P.A.; Taylor, S.M.; Wilson, V.L. Inhibition of DNA Methylation by 5-Azacytidine. In Modified Nucleosides and Cancer; Springer: Berlin/Heidelberg, Germany, 1983; pp. 202–211. [Google Scholar]
- Feinberg, A.P.; Vogelstein, B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983, 301, 89–92. [Google Scholar] [CrossRef]
- Ehrlich, M. DNA hypomethylation in cancer cells. Epigenomics 2009, 1, 239–259. [Google Scholar] [CrossRef]
- Rodriguez, J.; Frigola, J.; Vendrell, E.; Risques, R.-A.; Fraga, M.F.; Morales, C.; Moreno, V.; Esteller, M.; Capellà, G.; Ribas, M.; et al. Chromosomal Instability Correlates with Genome-wide DNA Demethylation in Human Primary Colorectal Cancers. Cancer Res. 2006, 66, 8462–9468. [Google Scholar] [CrossRef] [PubMed]
- Costello, J.F.; Frühwald, M.C.; Smiraglia, D.J.; Rush, L.J.; Robertson, G.P.; Gao, X.; Wright, F.A.; Feramisco, J.D.; Peltomäki, P.; Lang, J.C.; et al. Aberrant CpG-island methylation has non-random and tumour-type–specific patterns. Nat. Genet. 2000, 24, 132–138. [Google Scholar] [CrossRef] [PubMed]
- Melki, J.R.; Vincent, P.C.; Clark, S.J. Concurrent DNA hypermethylation of multiple genes in acute myeloid leukemia. Cancer Res. 1999, 59, 3730–3740. [Google Scholar] [PubMed]
- Kanwal, R.; Gupta, K.; Gupta, S. Cancer Epigenetics: An Introduction. In Methods in Molecular Biology; Verma, M., Ed.; Humana Press: New York, NY, USA, 2015; pp. 3–25. [Google Scholar]
- Jeltsch, A.; Ehrenhofer-Murray, A.; Jurkowski, T.P.; Lyko, F.; Reuter, G.; Ankri, S.; Nellen, W.; Schaefer, M.; Helm, M. Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation. RNA Biol. 2017, 14, 1108–1123. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, C.; Wu, C.; Cui, W.; Wang, L. DNA methyltransferases in cancer: Biology, paradox, aberrations, and targeted therapy. Cancers 2020, 12, 2123. [Google Scholar] [CrossRef]
- Cheray, M.; Pacaud, R.; Hervouet, E.; Vallette, F.; Cartron, P.-F. DNMT Inhibitors in Cancer, Current Treatments and Future Promising Approach: Inhibition of Specific DNMT-Including Complexes. Epigenet. Diagn. Ther. 2015, 1, 37–48. [Google Scholar] [CrossRef]
- Blecua, P.; Martinez-Verbo, L.; Esteller, M. The DNA methylation landscape of hematological malignancies: An update. Mol. Oncol. 2020, 14, 1616–1639. [Google Scholar] [CrossRef]
- Hu, C.; Liu, X.; Zeng, Y.; Liu, J.; Wu, F. DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: Mechanism and clinical application. Clin. Epigenet. 2021, 13, 166. [Google Scholar] [CrossRef]
- Giri, A.K.; Aittokallio, T. DNMT Inhibitors Increase Methylation in the Cancer Genome. Front. Pharmacol. 2019, 10, 385. [Google Scholar] [CrossRef]
- Hentze, J.L.; Høgdall, C.K.; Høgdall, E.V. Methylation and ovarian cancer: Can DNA methylation be of diagnostic use? (Review). Mol. Clin. Oncol. 2019, 10, 323–330. [Google Scholar] [CrossRef]
- Reid, B.M.; Fridley, B.L. DNA Methylation in Ovarian Cancer Susceptibility. Cancers 2020, 13, 108. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, D.; Thombre, R.; Dhar, A.; Anant, S. DNA Methyltransferases: A Novel Target for Prevention and Therapy. Front. Oncol. 2014, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Song, Z.; Fu, Y.; Yu, Z.; Zhao, L.; Zhao, H.; Yao, W.; Huang, D.; Mi, X.; Wang, E.; et al. Clinicopathological Significance and Prognostic Value of DNA Methyltransferase 1, 3a, and 3b Expressions in Sporadic Epithelial Ovarian Cancer. PLoS ONE 2012, 7, e40024. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Yan, X.; Gao, Y.; Liao, Q. Expression of DNA methyltransferase 1, 3A and 3B mRNA in the epithelial ovarian carcinoma. Zhonghua Fu Chan Ke Za Zhi 2005, 40, 770–774. [Google Scholar]
- Ehrlich, M.; Woods, C.B.; Yu, M.C.; Dubeau, L.; Yang, F.; Campan, M.; Weisenberger, D.J.; Long, T.; Youn, B.; Fiala, E.S.; et al. Quantitative analysis of associations between DNA hypermethylation, hypomethylation, and DNMT RNA levels in ovarian tumors. Oncogene 2006, 25, 2636–2645. [Google Scholar] [CrossRef]
- Dedeurwaerdere, F.; Claes, K.B.; Van Dorpe, J.; Rottiers, I.; Van der Meulen, J.; Breyne, J.; Swaerts, K.; Martens, G. Comparison of microsatellite instability detection by immunohistochemistry and molecular techniques in colorectal and endometrial cancer. Sci. Rep. 2021, 11, 12880. [Google Scholar] [CrossRef]
- Ness, N.; Andersen, S.; Khanehkenari, M.R.; Nordbakken, C.V.; Valkov, A.; Paulsen, E.-E.; Nordby, Y.; Bremnes, R.M.; Donnem, T.; Busund, L.-T.; et al. The prognostic role of immune checkpoint markers programmed cell death protein 1 (PD-1) and programmed death ligand 1 (PD-L1) in a large, multicenter prostate cancer cohort. Oncotarget 2017, 8, 26789–26801. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Berek, J.S.; Renz, M.; Kehoe, S.; Kumar, L.; Friedlander, M. Cancer of the ovary, fallopian tube, and peritoneum: 2021 update. Int. J. Gynecol. Obstet. 2021, 155, 61–85. [Google Scholar] [CrossRef]
- Schacht, V.; Kern, J.S. Basics of immunohistochemistry. J. Investig. Dermatol. 2015, 135, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Roh, J.; Park, C.S. Immunohistochemistry for Pathologists: Protocols, Pitfalls, and Tips. J. Pathol. Transl. Med. 2016, 50, 411–418. [Google Scholar] [CrossRef] [PubMed]
- O’Hurley, G.; Sjöstedt, E.; Rahman, A.; Li, B.; Kampf, C.; Pontén, F.; Gallagher, W.M.; Lindskog, C. Garbage in, garbage out: A critical evaluation of strategies used for validation of immunohistochemical biomarkers. Mol. Oncol. 2014, 8, 783–798. [Google Scholar] [CrossRef] [PubMed]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between differentially expressed mRNA and mRNA-protein correlations in a xenograft model system. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef] [PubMed]
- Maier, T.; Güell, M.; Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009, 583, 3966–3973. [Google Scholar] [CrossRef]
- Greenbaum, D.; Colangelo, C.; Williams, K.; Gerstein, M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003, 4, 117. [Google Scholar] [CrossRef]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- Fagerberg, L.; Hallström, B.M.; Oksvold, P.; Kampf, C.; Djureinovic, D.; Odeberg, J.; Habuka, M.; Tahmasebpoor, S.; Danielsson, A.; Edlund, K.; et al. Analysis of the Human Tissue-specific Expression by Genome-wide Integration of Transcriptomics and Antibody-based Proteomics. Mol. Cell. Proteom. 2014, 13, 397–406. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, J. DNA methyltransferases and their roles in tumorigenesis. Biomark. Res. 2017, 5, 1. [Google Scholar] [CrossRef]
- Tzelepi, V.; Logotheti, S.; Efstathiou, E.; Troncoso, P.; Aparicio, A.; Sakellakis, M.; Hoang, A.; Perimenis, P.; Melachrinou, M.; Logothetis, C.; et al. Epigenetics and prostate cancer: Defining the timing of DNA methyltransferase deregulation during prostate cancer progression. Pathology 2020, 52, 218–227. [Google Scholar] [CrossRef]
- Liu, P.; Yang, F.; Zhang, L.; Hu, Y.; Chen, B.; Wang, J.; Su, L.; Wu, M.; Chen, W. Emerging role of different DNA methyltransferases in the pathogenesis of cancer. Front. Pharmacol. 2022, 13, 958146. [Google Scholar] [CrossRef]
- Papakonstantinou, E.; Androutsopoulos, G.; Logotheti, S.; Adonakis, G.; Maroulis, I.; Tzelepi, V. DNA Methylation in Epithelial Ovarian Cancer: Current Data and Future Perspectives. Curr. Mol. Pharmacol. 2021, 14, 1013–1027. [Google Scholar] [CrossRef] [PubMed]
- Ahluwalia, A.; Hurteau, J.A.; Bigsby, R.M.; Nephew, K.P. DNA Methylation in Ovarian Cancer. Gynecol. Oncol. 2001, 82, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Zhen, S.; Tuo, X.; Chang, S.; Yang, X.; Zhou, Y.; Chen, W.; Zhao, L.; Li, X. Downregulation of DNMT3A Attenuates the Warburg Effect, Proliferation, and Invasion via Promoting the Inhibition of miR-603 on HK2 in Ovarian Cancer. Technol. Cancer Res. Treat. 2022, 21, 153303382211106. [Google Scholar] [CrossRef] [PubMed]
- Del Castillo Falconi, V.M.; Díaz-Chávez, J.; Torres-Arciga, K.; Luna-Maldonado, F.; Gudiño-Gomez, A.A.; Pedroza-Torres, A.; Castro-Hernández, C.; Cantú de León, D.; Herrera, L.A. Expression of DNA Methyltransferase 3B Isoforms Is Associated with DNA Satellite 2 Hypomethylation and Clinical Prognosis in Advanced High-Grade Serous Ovarian Carcinoma. Int. J. Mol. Sci. 2022, 23, 12759. [Google Scholar] [CrossRef]
- Gu, Y.; Yang, P.; Shao, Q.; Liu, X.; Xia, S.; Zhang, M.; Xu, H.; Shao, Q. Investigation of the expression patterns and correlation of DNA methyltransferases and class I histone deacetylases in ovarian cancer tissues. Oncol. Lett. 2013, 5, 452–458. [Google Scholar] [CrossRef] [PubMed]
- Kinney, S.R.M.; Moser, M.T.; Pascual, M.; Greally, J.M.; Foster, B.A.; Karpf, A.R. Opposing roles of Dnmt1 in early- and late-stage murine prostate cancer. Mol. Cell. Biol. 2010, 30, 4159–4174. [Google Scholar] [CrossRef]
- Antunez, C.A.V.; Chayeb, L.T.; Rodriguez-Secura, M.Á.; Álvarez, G.S.L.; Garcia-Cuellar, C.M.; Trevino, S.V. DNA methyltransferases 3a and 3b are differentially expressed in the early stages of a rat liver carcinogenesis model. Oncol. Rep. 2014, 32, 2093–2103. [Google Scholar] [CrossRef][Green Version]
- Agoston, A.T.; Argani, P.; Yegnasubramanian, S.; De Marzo, A.M.; Ansari-Lari, M.A.; Hicks, J.L.; Davidson, N.E.; Nelson, W.G. Increased Protein Stability Causes DNA Methyltransferase 1 Dysregulation in Breast Cancer. J. Biol. Chem. 2005, 280, 18302–18310. [Google Scholar] [CrossRef]
- Lavoie, G.; St-Pierre, Y. Phosphorylation of human DNMT1: Implication of cyclin-dependent kinases. Biochem. Biophys. Res. Commun. 2011, 409, 187–192. [Google Scholar] [CrossRef]
- Veland, N.; Lu, Y.; Hardikar, S.; Gaddis, S.; Zeng, Y.; Liu, B.; Estecio, M.R.; Takata, Y.; Lin, K.; Tomida, M.W.; et al. DNMT3L facilitates DNA methylation partly by maintaining DNMT3A stability in mouse embryonic stem cells. Nucleic Acids Res. 2019, 47, 152–167. [Google Scholar] [CrossRef]
- Takashima, S.; Takehashi, M.; Lee, J.; Chuma, S.; Okano, M.; Hata, K.; Suetake, I.; Nakatsuji, N.; Miyoshi, H.; Tajima, S.; et al. Abnormal DNA Methyltransferase Expression in Mouse Germline Stem Cells Results in Spermatogenic Defects1. Biol. Reprod. 2009, 81, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Minami, K.; Chano, T.; Kawakami, T.; Ushida, H.; Kushima, R.; Okabe, H.; Okada, Y.; Okamoto, K. DNMT3L Is a Novel Marker and Is Essential for the Growth of Human Embryonal Carcinoma. Clin. Cancer Res. 2010, 16, 2751–2759. [Google Scholar] [CrossRef] [PubMed]
- Elhardt, W.; Shanmugam, R.; Jurkowski, T.P.; Jeltsch, A. Somatic cancer mutations in the DNMT2 tRNA methyltransferase alter its catalytic properties. Biochimie 2015, 112, 66–72. [Google Scholar] [CrossRef]
- He, M.; Fan, J.; Jiang, R.; Tang, W.-X.; Wang, Z.-W. Expression of DNMTs and MBD2 in GIST. Biomed. Rep. 2013, 1, 223–227. [Google Scholar] [CrossRef]
- Li, X.; Meng, Y. Expression and prognostic characteristics of m 5 C regulators in low-grade glioma. J. Cell. Mol. Med. 2021, 25, 1383–1393. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Tao, Z.; Zhao, Y.; Li, L.; Zheng, J.; Li, Z.; Chen, X. 5-methylcytosine RNA methyltransferases and their potential roles in cancer. J. Transl. Med. 2022, 20, 214. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Yang, H.; Zhu, X.; Yadav, T.; Ouyang, J.; Truesdell, S.S.; Tan, J.; Wang, Y.; Duan, M.; Wei, L.; et al. m5C modification of mRNA serves a DNA damage code to promote homologous recombination. Nat. Commun. 2020, 11, 2834. [Google Scholar] [CrossRef]
- Cao, Y.-L.; Zhuang, T.; Xing, B.-H.; Li, N.; Li, Q. Exosomal DNMT1 mediates cisplatin resistance in ovarian cancer. Cell Biochem. Funct. 2017, 35, 296–303. [Google Scholar] [CrossRef]
- Kobayashi, Y.; Absher, D.M.; Gulzar, Z.G.; Young, S.R.; McKenney, J.K.; Peehl, D.M.; Brooks, J.D.; Myers, R.M.; Sherlock, G. DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer. Genome Res. 2011, 21, 1017–1027. [Google Scholar] [CrossRef]
- Gomez, S.; Cox, O.L.; Walker, R.R.; Rentia, U.; Hadley, M.; Arthofer, E.; Diab, N.; Grundy, E.E.; Kanholm, T.; McDonald, J.I.; et al. Inhibiting DNA methylation and RNA editing upregulates immunogenic RNA to transform the tumor microenvironment and prolong survival in ovarian cancer. J. Immunother. Cancer 2022, 10, e004974. [Google Scholar] [CrossRef]
- Moufarrij, S.; Srivastava, A.; Gomez, S.; Hadley, M.; Palmer, E.; Austin, P.T.; Chisholm, S.; Diab, N.; Roche, K.; Yu, A.; et al. Combining DNMT and HDAC6 inhibitors increases anti-tumor immune signaling and decreases tumor burden in ovarian cancer. Sci. Rep. 2020, 10, 3470. [Google Scholar] [CrossRef] [PubMed]
- Matei, D.; Fang, F.; Shen, C.; Schilder, J.; Arnold, A.; Zeng, Y.; Berry, W.A.; Huang, T.; Nephew, K.P. Epigenetic resensitization to platinum in ovarian cancer. Cancer Res. 2012, 72, 2197–2205. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Sun, H.; Li, X.; Lin, F.; Wang, Z.; Wang, X. Ovarian cancer: Epigenetics, drug resistance, and progression. Cancer Cell Int. 2021, 21, 434. [Google Scholar] [CrossRef] [PubMed]
- Abbotts, R.; Topper, M.J.; Biondi, C.; Fontaine, D.; Goswami, R.; Stojanovic, L.; Choi, E.Y.; McLaughlin, L.; Kogan, A.A.; Xia, L.; et al. DNA methyltransferase inhibitors induce a BRCAness phenotype that sensitizes NSCLC to PARP inhibitor and ionizing radiation. Proc. Natl. Acad. Sci. USA 2019, 116, 22609–22618. [Google Scholar] [CrossRef] [PubMed]


| SBT Ν = 12 | LGSOC Ν = 17 | HGSOC Ν = 60 | MBT Ν = 12 | MOC Ν = 7 | Total N = 108 | |
|---|---|---|---|---|---|---|
| Mean Age | 38 | 63 | 57 | 41 | 51 | 55 |
| Figo Stage | ||||||
| 1 | 12 | 7 | 13 | 12 | 4 | 48 |
| 2 | 2 | 4 | 1 | 7 | ||
| 3 | 8 | 41 | 2 | 51 | ||
| 4 | 2 | 2 | ||||
| T stage | ||||||
| T1 | 12 | 7 | 13 | 12 | 4 | 48 |
| T2 | 0 | 2 | 7 | 0 | 1 | 10 |
| T3 | 0 | 8 | 40 | 0 | 2 | 50 |
| N Stage | ||||||
| N0 | 12 | 15 | 40 | 12 | 7 | 86 |
| N1 | 0 | 2 | 20 | 0 | 0 | 22 |
| Antibody | Clone | Source | Antigen Retrieval | Dilution- Incubation | Positive Control |
|---|---|---|---|---|---|
| DNMT1 | Mouse monoclonal H-12 | Santa Cruz, CA, USA | 1mM EDTA-NaOH pH8 | 1:50/2 h/room temperature | Lymph node |
| DNMT2 | Rabbit polyclonal | Santa Cruz, CA, USA | 10mM citrate buffer pH6 | 1:150/2 h/room temperature | Placenta |
| DNMT3a | Mouse monoclonal 64B1446 | Abcam, UK | 1mM EDTA-NaOH pH8 | 1:250/1 h/room temperature | Testicle |
| DNMT3B | Mouse monoclonal 52A1018 | Abcam, UK | 1mM EDTA-NaOH pH8 | 1:150/1 h/room temperature | Lymph node |
| DNMT3L | Rabbit polyclonal | Novus Biologicals, Littleton, CO, USA | 1mM EDTA-NaOH pH8 | 1:150/2 h/room temperature | Testicle |
| N | DNMT1 | p Value | DNMT2 Nucleus | p Value | DNMT2 Cytoplasm | p Value | ||
|---|---|---|---|---|---|---|---|---|
| Non-neoplastic (non) | 20 | 70 | vs. SHGOC p = 0.04 vs. MOC p = 0.04 | 30 | 85 | |||
| Histologic type | SBT | 12 | 68.3 | vs. HGSOC < 0.0001 | 6.3 | vs. non p = 0.03 | 52.9 | |
| LGSOC | 17 | 89.6 | vs. HGSOC < 0.0001 | 11.9 | vs. non p = 0.03 | 32.6 | vs. non p = 0.045 | |
| HGSOC | 60 | 159.6 | 21.2 | 31.3 | vs. non p = 0.045 | |||
| MBT | 12 | 82 | 10.9 | 41.1 | ||||
| MOC | 7 | 136.8 | vs. non p = 0.042 | 15.7 | 44.4 | |||
| FIGO Stage | 1 | 48 | 100.2 | 15.2 | 47.8 | |||
| 2 | 7 | 132.5 | 7.5 | 70 | ||||
| 3 | 51 | 147.5 | 17.7 | 28.9 | ||||
| 4 | 2 | 85 | 5 | 60 | ||||
| Relapses | 26 | 210.4 | vs. HGSOC p = 0.02 | 28.2 | 36.2 |
| N | DNMT3a Nucleus | p Value | DNMT3a Cytoplasm | p Value | DNMT3b | p Value | DNMT3L | p Value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Non- neoplastic (non) | 20 | 10 | vs. HGSOC p = 0.02 | 95 | 70 | 20 | vs. LGSOC < 0.005 vs. HGSOC < 0.001 | |||
| Histologic type | SBT | 12 | 20.2 | vs. HGSOC p = 0.02 | 138.3 | 103.3 | 62.5 | vs. HGSOC < 0.001 | ||
| LGSOC | 17 | 28.06 | vs. HGSOC < p = 0.02 | 123.75 | 71.1 | 68.1 | vs. HGSOC < 0.001 | |||
| HGSOC | 60 | 58.3 | 102.6 | 95.1 | 133.1 | |||||
| MBT | 12 | 14 | 109.1 | 104.1 | 100.8 | |||||
| MOC | 7 | 21.1 | vs. non p = 0.04 | 96.2 | 71.7 | 127.8 | vs. non p = 0.045 | |||
| FIGO Stage | 1 | 48 | 25.4 | 113.3 | 94.9 | 94.4 | ||||
| 2 | 7 | 37.8 | 153.3 | 82.5 | 55.8 | |||||
| 3 | 51 | 56.6 | 102.5 | 90.6 | 126.5 | |||||
| 4 | 2 | 7.5 | 70 | 60 | 60 | |||||
| Relapses | 26 | 67 | 87.8 | 108.7 | 157.3 | vs. SHGOC p = 0.032 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papakonstantinou, E.; Pappa, I.; Androutsopoulos, G.; Adonakis, G.; Maroulis, I.; Tzelepi, V. Comprehensive Analysis of DNA Methyltransferases Expression in Primary and Relapsed Ovarian Carcinoma. Cancers 2023, 15, 4950. https://doi.org/10.3390/cancers15204950
Papakonstantinou E, Pappa I, Androutsopoulos G, Adonakis G, Maroulis I, Tzelepi V. Comprehensive Analysis of DNA Methyltransferases Expression in Primary and Relapsed Ovarian Carcinoma. Cancers. 2023; 15(20):4950. https://doi.org/10.3390/cancers15204950
Chicago/Turabian StylePapakonstantinou, Efthymia, Ioanna Pappa, Georgios Androutsopoulos, Georgios Adonakis, Ioannis Maroulis, and Vasiliki Tzelepi. 2023. "Comprehensive Analysis of DNA Methyltransferases Expression in Primary and Relapsed Ovarian Carcinoma" Cancers 15, no. 20: 4950. https://doi.org/10.3390/cancers15204950
APA StylePapakonstantinou, E., Pappa, I., Androutsopoulos, G., Adonakis, G., Maroulis, I., & Tzelepi, V. (2023). Comprehensive Analysis of DNA Methyltransferases Expression in Primary and Relapsed Ovarian Carcinoma. Cancers, 15(20), 4950. https://doi.org/10.3390/cancers15204950






