Post-Translational Modifications of Histone Variants in the Absence and Presence of a Methionine-Depleting Enzyme in Normal and Cancer Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. MGL Expression and Purification
2.2. HT-29 and Hs27 Culture Conditions
2.3. Histone PTM Preparation and Analysis by Mass Spectrometry
2.4. Statistical Analysis and Data Availability
3. Results
3.1. Differences on PTMs of Histone Variants between Cancer and Normal Cell Lines
3.2. Effect of MGL-Mediated Methionine Depletion on PTMs of Histone Variants in HT-29 and Hs27 Cell Lines
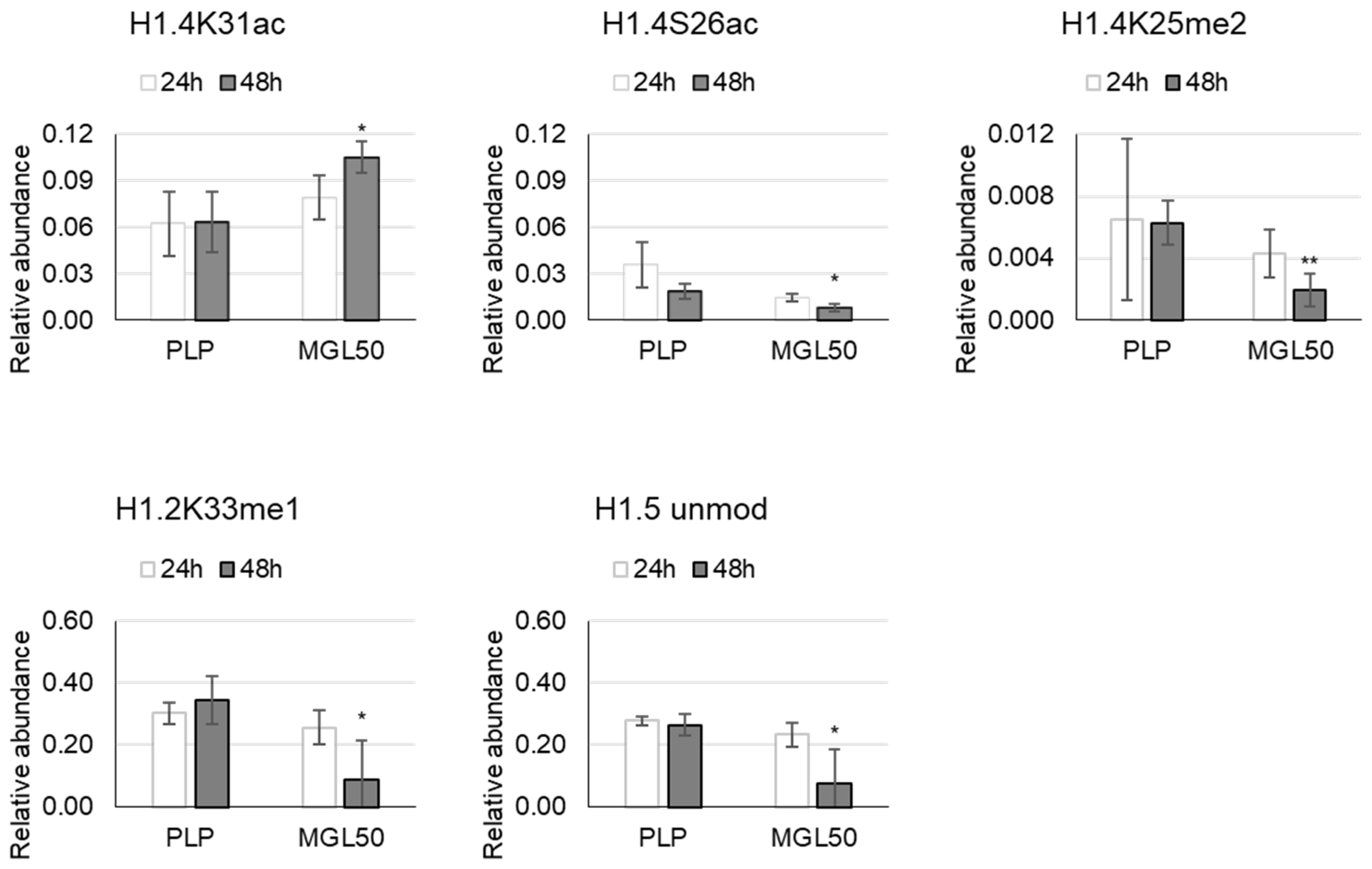
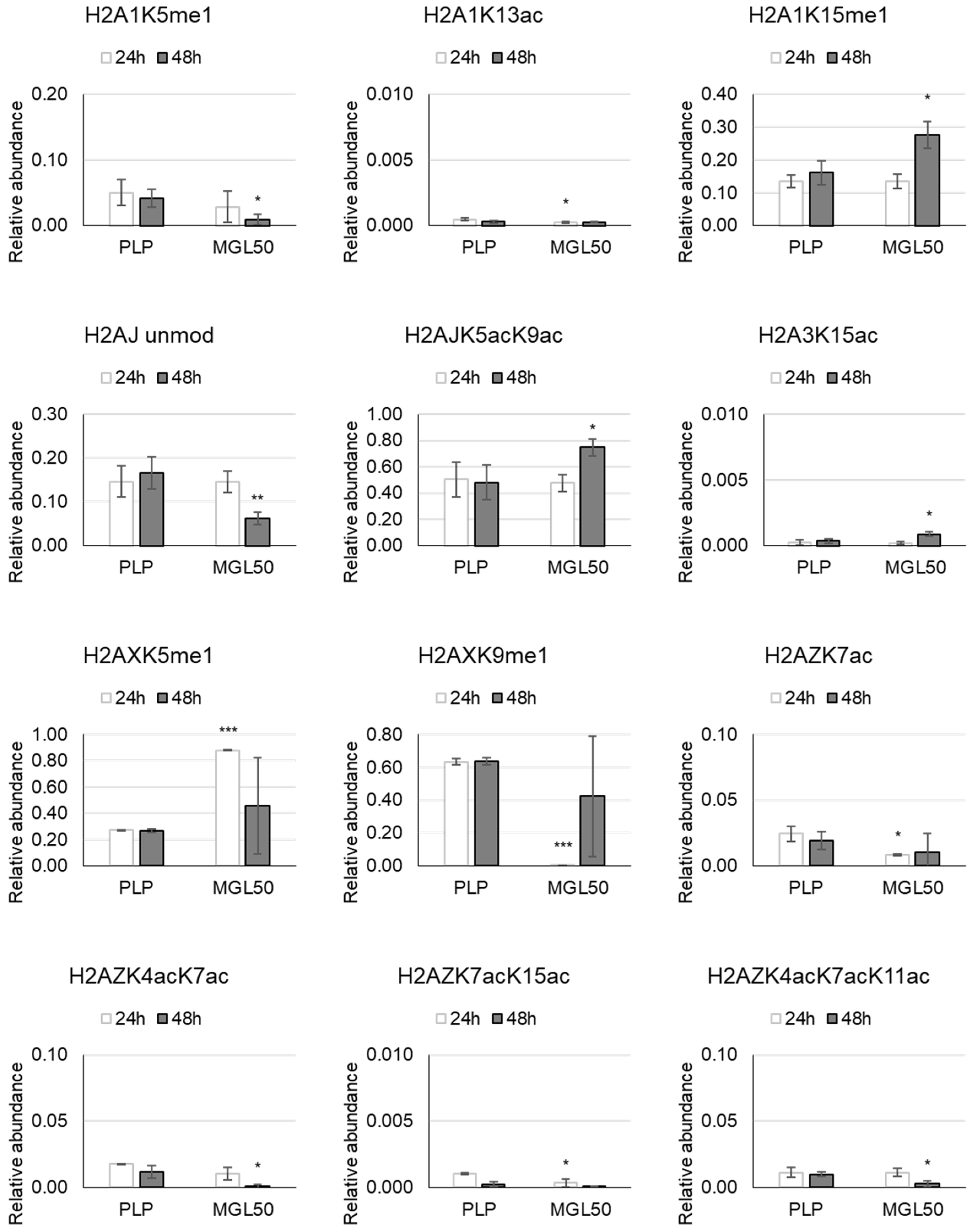
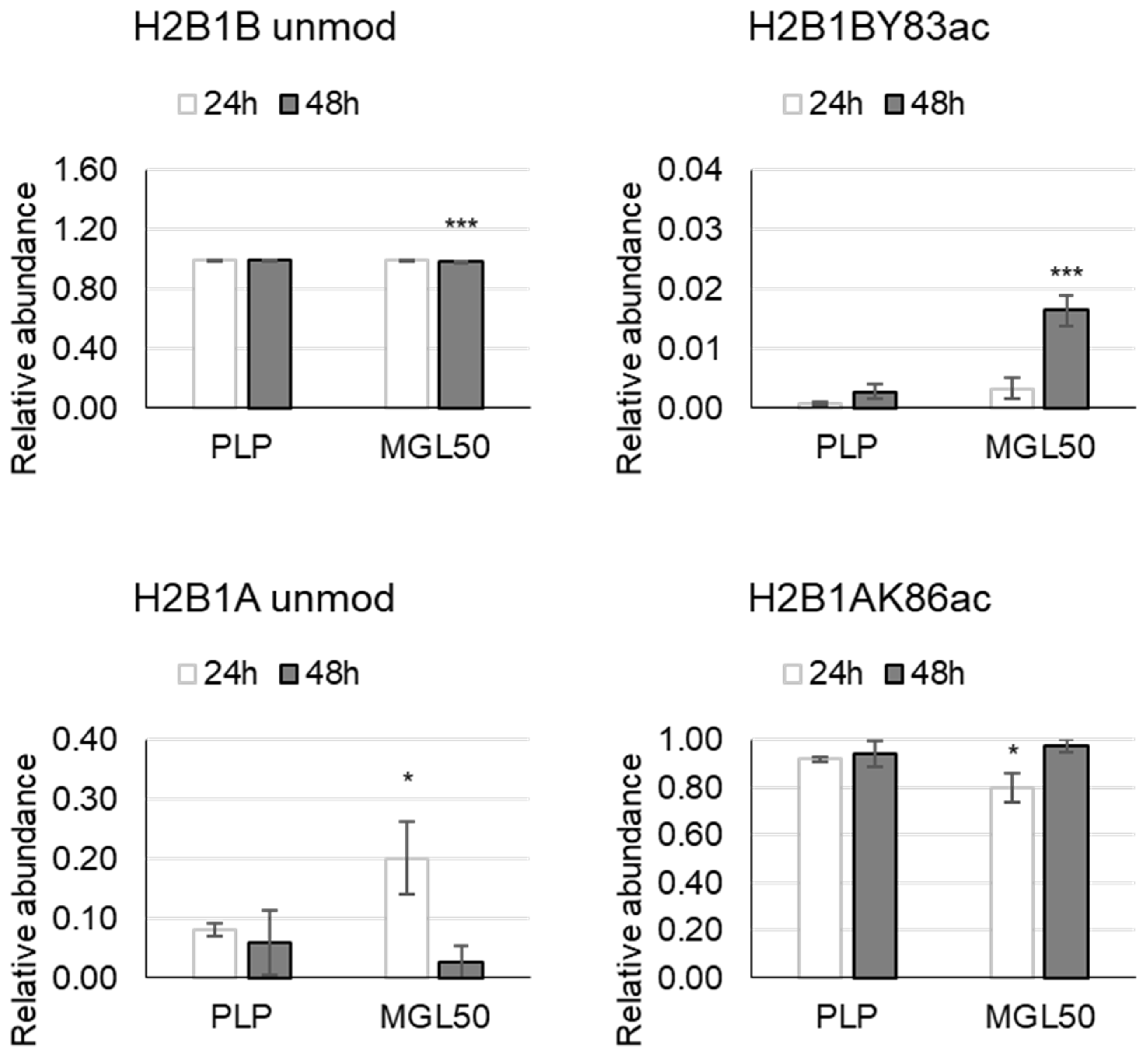
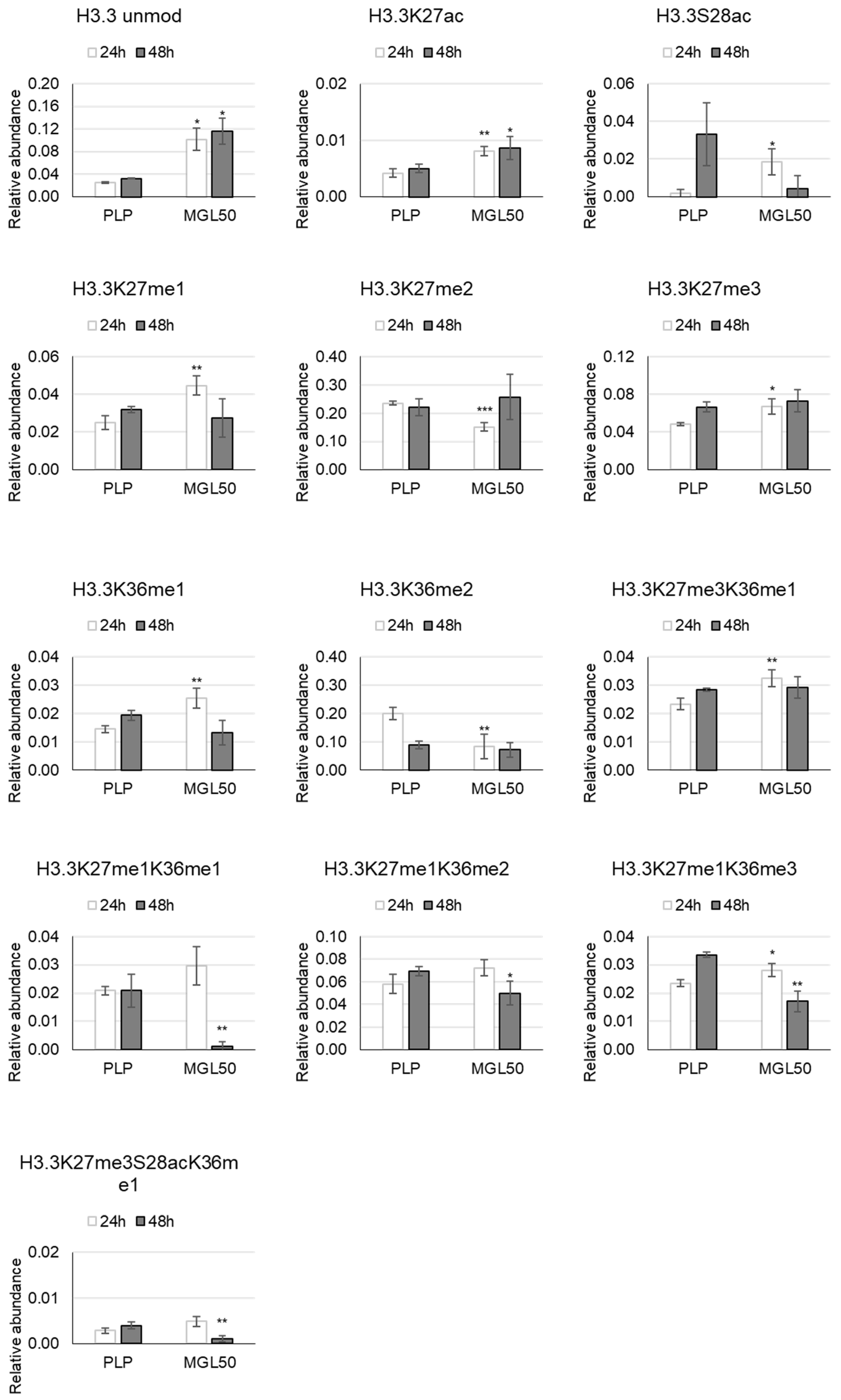
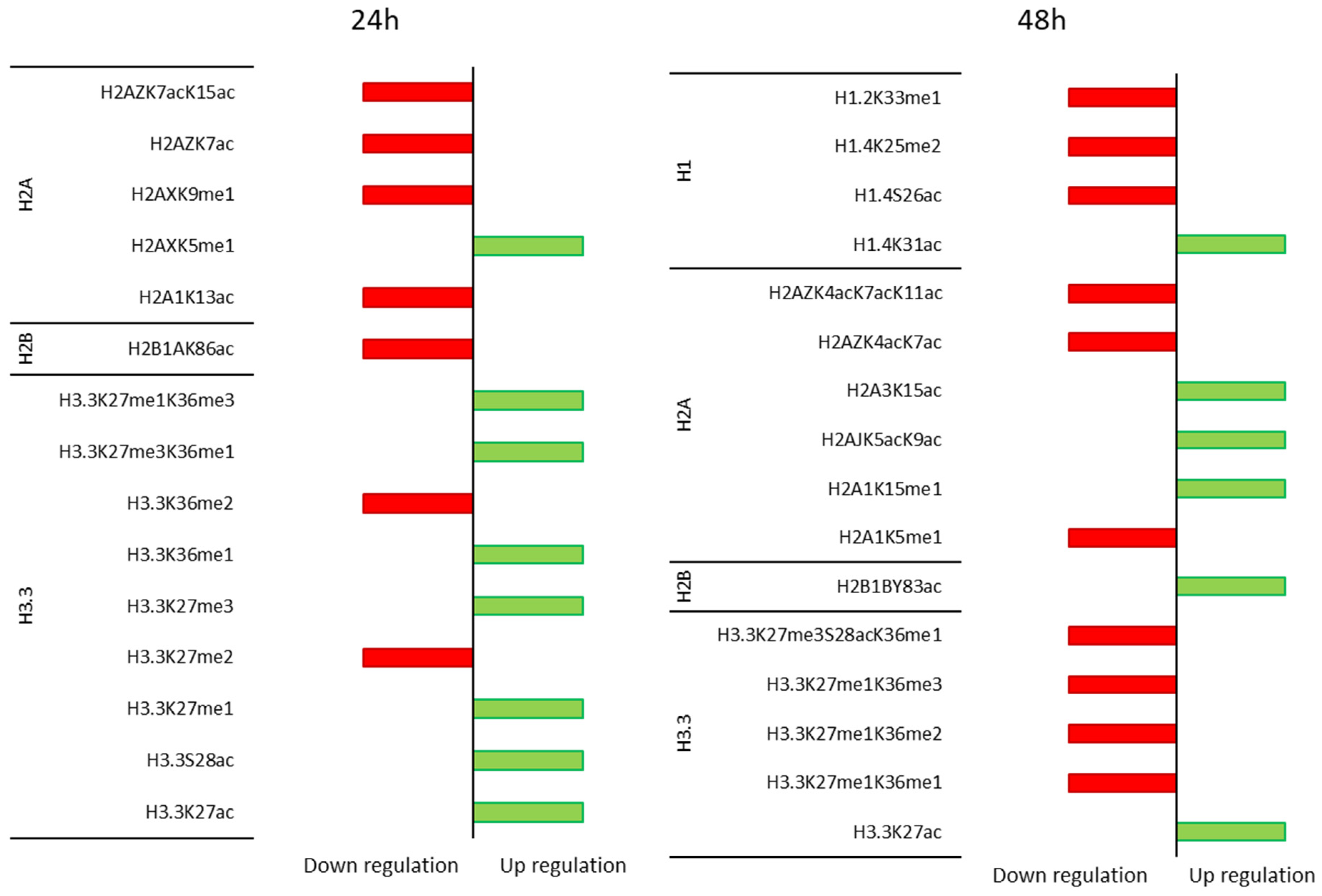
3.3. Time Course of the Effect of MGL-Mediated Methionine Depletion on PTMs of Histone Variants in HT-29 Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Endicott, M.; Jones, M.; Hull, J. Amino acid metabolism as a therapeutic target in cancer: A review. Amino Acids 2021, 53, 1169–1179. [Google Scholar] [CrossRef]
- Kuo, M.T.; Chen, H.H.W.; Feun, L.G.; Savaraj, N. Targeting the Proline-Glutamine-Asparagine-Arginine Metabolic Axis in Amino Acid Starvation Cancer Therapy. Pharmaceuticals 2021, 14, 72. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, P. Methionine Dependence of Cancer. Biomolecules 2020, 10, 568. [Google Scholar] [CrossRef] [PubMed]
- Cavuoto, P.; Fenech, M.F. A review of methionine dependency and the role of methionine restriction in cancer growth control and life-span extension. Cancer Treat. Rev. 2012, 38, 726–736. [Google Scholar] [CrossRef]
- Hoffman, R.M. Development of recombinant methioninase to target the general cancer-specific metabolic defect of methionine dependence: A 40-year odyssey. Expert Opin. Biol. Ther. 2015, 15, 21. [Google Scholar] [CrossRef]
- Raboni, S.; Montalbano, S.; Stransky, S.; Garcia, B.A.; Buschini, A.; Bettati, S.; Sidoli, S.; Mozzarelli, A. A Key Silencing Histone Mark on Chromatin Is Lost When Colorectal Adenocarcinoma Cells Are Depleted of Methionine by Methionine γ-Lyase. Front. Mol. Biosci. 2021, 8, 735303. [Google Scholar] [CrossRef]
- Aminabad, N.S.; Farshbaf, M.; Akbarzadeh, A. Recent Advances of Gold Nanoparticles in Biomedical Applications: State of the Art. Cell Biochem. Biophys. 2019, 77, 123–137. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, K.; Han, Q.; Li, S.; Tan, Y.; Igarashi, K.; Murakami, T.; Unno, M.; Hoffman, R.M. Efficacy of Recombinant Methioninase (rMETase) on Recalcitrant Cancer Patient-Derived Orthotopic Xenograft (PDOX) Mouse Models: A Review. Cells 2019, 8, 410. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Xu, M.; Tan, X.; Wang, X.; Saikawa, Y.; Nagahama, T.; Sun, X.; Lenz, M.; Hoffman, R.M. Overexpression and large-scale production of recombinant L-methionine-alpha-deamino-gammamercaptomethane- lyase for novel anticancer therapy. Protein Expr. Purif. 1997, 9, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Raboni, S.; Revtovich, S.; Demitri, N.; Giabbai, B.; Storici, P.; Cocconcelli, C.; Faggiano, S.; Rosini, E.; Pollegioni, L.; Galati, S.; et al. Engineering methionine γ-lyase from Citrobacter freundii for anticancer activity. Biochim. Biophys. Acta Proteins Proteom. 2018, 1866, 1260–1270. [Google Scholar] [CrossRef]
- Kulikova, V.V.; Morozova, E.A.; Revtovich, S.V.; Kotlov, M.I.; Anufrieva, N.V.; Bazhulina, N.P.; Raboni, S.; Faggiano, S.; Gabellieri, E.; Cioni, P.; et al. Gene cloning, characterization, and cytotoxic activity of methionine γ-lyase from Clostridium novyi. IUBMB Life 2017, 69, 668–676. [Google Scholar] [CrossRef]
- Morozova, E.A.; Kulikova, V.V.; Faggiano, S.; Raboni, S.; Gabellieri, E.; Cioni, P.; Anufrieva, N.V.; Revtovich, S.V.; Demidkina, T.; Mozzarelli, A. Soluble and Nanoporous Silica Gel-Entrapped C. freundii Methionine γ-Lyase. J. Nanosci. Nanotechnol. 2018, 18, 2210–2219. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Xu, M.; Hoffman, R.M. Broad selective efficacy of recombinant methioninase and polyethylene glycol-modified recombinant methioninase on cancer cells In Vitro. Anticancer Res. 2010, 30, 1041–1046. [Google Scholar] [PubMed]
- Xin, L.; Caot, J.Q.; Liu, C.; Zeng, F.; Cheng, H.; Hu, X.Y.; Shao, J.H. Evaluation of rMETase-Loaded Stealth PLGA/Liposomes Modified with Anti-CAGE scFV for Treatment of Gastric Carcinoma. J. Biomed. Nanotechnol. 2015, 11, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Gay, F.; Aguera, K.; Sénéchal, K.; Tainturier, A.; Berlier, W.; Maucort-Boulch, D.; Honnorat, J.; Horand, F.; Godfrin, Y.; Bourgeaux, V. Methionine tumor starvation by erythrocyte-encapsulated methionine gamma-lyase activity controlled with per os vitamin B6. Cancer Med. 2017, 6, 1437–1452. [Google Scholar] [CrossRef] [PubMed]
- Miki, K.; Al-Refaie, W.; Xu, M.; Jiang, P.; Tan, Y.; Bouvet, M.; Zhao, M.; Gupta, A.; Chishima, T.; Shimada, H.; et al. Methioninase gene therapy of human cancer cells is synergistic with recombinant methioninase treatment. Cancer Res. 2000, 60, 2696–2702. [Google Scholar]
- Wanders, D.; Hobson, K.; Ji, X. Methionine Restriction and Cancer Biology. Nutrients 2020, 12, 684. [Google Scholar] [CrossRef]
- Kulis, M.; Esteller, M. DNA methylation and cancer. Adv. Genet. 2010, 70, 27–56. [Google Scholar] [CrossRef] [PubMed]
- Parkhitko, A.A.; Jouandin, P.; Mohr, S.E.; Perrimon, N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell 2019, 18, e13034. [Google Scholar] [CrossRef]
- Ye, C.; Sutter, B.M.; Wang, Y.; Kuang, Z.; Tu, B.P. A Metabolic Function for Phospholipid and Histone Methylation. Mol. Cell 2017, 66, 180–193.e8. [Google Scholar] [CrossRef]
- Lu, S.; Epner, D.E. Molecular mechanisms of cell cycle block by methionine restriction in human prostate cancer cells. Nutr. Cancer 2000, 38, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017, 358, 813–818. [Google Scholar] [CrossRef]
- Hoffman, R.M.; Jacobsen, S.J. Reversible growth arrest in simian virus 40-transformed human fibroblasts. Proc. Natl. Acad. Sci. USA 1980, 77, 7306–7310. [Google Scholar] [CrossRef]
- Mentch, S.J.; Mehrmohamadi, M.; Huang, L.; Liu, X.; Gupta, D.; Mattocks, D.; Gómez Padilla, P.; Ables, G.; Bamman, M.M.; Thalacker-Mercer, A.E.; et al. Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab. 2015, 22, 861–873. [Google Scholar] [CrossRef]
- Tang, X.; Keenan, M.M.; Wu, J.; Lin, C.A.; Dubois, L.; Thompson, J.W.; Freedland, S.J.; Murphy, S.K.; Chi, J.T. Comprehensive profiling of amino acid response uncovers unique methionine-deprived response dependent on intact creatine biosynthesis. PLoS Genet. 2015, 11, e1005158. [Google Scholar] [CrossRef]
- Dai, Z.; Mentch, S.J.; Gao, X.; Nichenametla, S.N.; Locasale, J.W. Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nat. Commun. 2018, 9, 1955. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Sanderson, S.M.; Dai, Z.; Reid, M.A.; Cooper, D.E.; Lu, M.; Richie, J.P.; Ciccarella, A.; Calcagnotto, A.; Mikhael, P.G.; et al. Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 2019, 572, 397–401. [Google Scholar] [CrossRef]
- Haws, S.A.; Yu, D.; Ye, C.; Wille, C.K.; Nguyen, L.C.; Krautkramer, K.A.; Tomasiewicz, J.L.; Yang, S.E.; Miller, B.R.; Liu, W.H.; et al. Methyl-Metabolite Depletion Elicits Adaptive Responses to Support Heterochromatin Stability and Epigenetic Persistence. Mol. Cell 2020, 78, 210–223.e8. [Google Scholar] [CrossRef] [PubMed]
- Machover, D.; Rossi, L.; Hamelin, J.; Desterke, C.; Goldschmidt, E.; Chadefaux-Vekemans, B.; Bonnarme, P.; Briozzo, P.; Kopečný, D.; Pierigè, F.; et al. Effects in Cancer Cells of the Recombinant l-Methionine Gamma-Lyase from Brevibacterium aurantiacum. Encapsulation in Human Erythrocytes for Sustained l-Methionine Elimination. J. Pharmacol. Exp. Ther. 2019, 369, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, J.; Han, Q.; Inubushi, S.; Sugisawa, N.; Hamada, K.; Nishino, H.; Miyake, K.; Kumamoto, T.; Matsuyama, R.; Bouvet, M.; et al. Histone methylation status of H3K4me3 and H3K9me3 under methionine restriction is unstable in methionine-addicted cancer cells, but stable in normal cells. Biochem. Biophys. Res. Commun. 2020, 533, 1034–1038. [Google Scholar] [CrossRef]
- Xiao, Z.; Locasale, J.W. Epigenomic links from metabolism-methionine and chromatin architecture. Curr. Opin. Chem. Biol. 2021, 63, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521. [Google Scholar] [CrossRef] [PubMed]
- Di Cerbo, V.; Schneider, R. Cancers with wrong HATs: The impact of acetylation. Brief Funct. Genom. 2013, 12, 231–243. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, Z.; Jia, J.; Du, T.; Zhang, N.; Tang, Y.; Fang, Y.; Fang, D. Overview of Histone Modification. Adv. Exp. Med. Biol. 2021, 1283, 1–16. [Google Scholar] [CrossRef]
- Black, J.C.; Van Rechem, C.; Whetstine, J.R. Histone lysine methylation dynamics: Establishment, regulation, and biological impact. Mol. Cell. 2012, 48, 491–507. [Google Scholar] [CrossRef]
- Strahl, B.D.; Allis, C.D. The language of covalent histone modifications. Nature 2000, 403, 41–45. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.J.; Yang, L.X. Gamma-H2AX—A novel biomarker for DNA double-strand breaks. In Vivo 2008, 22, 305–309. [Google Scholar]
- Morozova, E.A.; Bazhulina, N.P.; Anufrieva, N.V.; Mamaeva, D.V.; Tkachev, Y.V.; Streltsov, S.A.; Timofeev, V.P.; Faleev, N.G.; Demidkina, T.V. Kinetic and Spectral Parameters of Interaction of Citrobacter Freundii Methionine γ-lyase with Amino Acids. Biochem. Mosc. 2010, 75, 1272–1280. [Google Scholar] [CrossRef]
- Garcia, B.A.; Mollah, S.; Ueberheide, B.M.; Busby, S.A.; Muratore, T.L.; Shabanowitz, J.; Hunt, D.F. Chemical Derivatization of Histones for Facilitated Analysis by Mass Spectrometry. Nat. Protoc. 2007, 2, 933–938. [Google Scholar] [CrossRef]
- Yuan, Z.F.; Sidoli, S.; Marchione, D.M.; Simithy, J.; Janssen, K.A.; Szurgot, M.R.; Garcia, B.A. EpiProfile 2.0: A Computational Platform for Processing Epi-Proteomics Mass Spectrometry Data. J. Proteome Res. 2018, 17, 2533–2541. [Google Scholar] [CrossRef] [PubMed]
- Sancho, M.; Diani, E.; Beato, M.; Jordan, A. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet. 2008, 4, e1000227. [Google Scholar] [CrossRef] [PubMed]
- Terme, J.M.; Sesé, B.; Millán-Ariño, L.; Mayor, R.; Belmonte, J.C.I.; Barrero, M.J.; Jordan, A. Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency. J. Biol. Chem. 2011, 286, 35347–35357. [Google Scholar] [CrossRef]
- Mayor, R.; Izquierdo-Bouldstridge, A.; Millán-Ariño, L.; Bustillos, A.; Sampaio, C.; Luque, N.; Jordan, A. Genome distribution of replication-independent histone H1 variants shows H1.0 associated with nucleolar domains and H1X associated with RNA polymerase II-enriched regions. J. Biol. Chem. 2015, 290, 7474–7491. [Google Scholar] [CrossRef] [PubMed]
- Redon, C.; Pilch, D.; Rogakou, E.; Sedelnikova, O.; Newrock, K.; Bonner, W. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 2002, 12, 162–169. [Google Scholar] [CrossRef]
- Shi, L.; Wen, H.; Shi, X. The Histone Variant H3.3 in Transcriptional Regulation and Human Disease. J. Mol. Biol. 2017, 429, 1934–1945. [Google Scholar] [CrossRef]
- Xu, H.; Wu, M.; Ma, X.; Huang, W.; Xu, Y. Function and Mechanism of Novel Histone Posttranslational Modifications in Health and Disease. Biomed. Res. Int. 2021, 2021, 6635225. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Krautkramer, K.A.; Feldman, J.L.; Denu, J.M. Metabolic regulation of histone post-translational modifications. ACS Chem. Biol. 2015, 10, 95–108. [Google Scholar] [CrossRef]
- Lu, C.; Coradin, M.; Porter, E.G.; Garcia, B.A. Accelerating the Field of Epigenetic Histone Modification Through Mass Spectrometry-Based Approaches. Mol. Cell Proteom. 2021, 20, 100006. [Google Scholar] [CrossRef]
- Méndez-Acuña, L.; Di Tomaso, M.V.; Palitti, F.; Martínez-López, W. Histone post-translational modifications in DNA damage response. Cytogenet. Genome Res. 2010, 128, 28–36. [Google Scholar] [CrossRef] [PubMed]
- Aricthota, S.; Rana, P.P.; Haldar, D. Histone acetylation dynamics in repair of DNA double-strand breaks. Front. Genet. 2022, 13, 926577. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343–357. [Google Scholar] [CrossRef]
- Kokkinakis, D.M.; Liu, X.; Chada, S.; Ahmed, M.M.; Shareef, M.M.; Singha, U.K.; Yang, S.; Luo, J. Modulation of gene expression in human central nervous system tumors under methionine deprivation-induced stress. Cancer Res. 2004, 64, 7513–7525. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M. Clinical Studies of Methionine-Restricted Diets for Cancer Patients. Methods Mol. Biol. 2019, 1866, 95–105. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.G.; Mandloi, T.; Kunte, P.; Natu, A.; Rashid, M.; Reddy, D.; Gadewal, N.; Gupta, S. HISTome2: A database of histone proteins, modifiers for multiple organisms and epidrugs. Epigenetics Chromatin 2020, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhao, J.; Wang, Y.; Wang, M.; Long, H.; Liang, D.; Huang, L.; Wen, Z.; Li, W.; Li, X.; et al. H3.3 actively marks enhancers and primes gene transcription via opening higher-ordered chromatin. Genes Dev. 2013, 27, 2109–2124. [Google Scholar] [CrossRef]
- Santenard, A.; Ziegler-Birling, C.; Koch, M.; Tora, L.; Bannister, A.J.; Torres-Padilla, M.E. Heterochromatin formation in the mouse embryo requires critical residues of the histone variant H3.3. Nat. Cell Biol. 2010, 12, 853–862. [Google Scholar] [CrossRef]
- Szenker, E.; Ray-Gallet, D.; Almouzni, G. The double face of the histone variant H3.3. Cell Res. 2011, 21, 421–434. [Google Scholar] [CrossRef]
- Kallappagoudar, S.; Yadav, R.K.; Lowe, B.R.; Partridge, J.F. Histone H3 mutations--a special role for H3.3 in tumorigenesis? Chromosoma 2015, 124, 177–189. [Google Scholar] [CrossRef]
| hPMTs | Mean | Standard Error | p-Value | ||
|---|---|---|---|---|---|
| Hs27 | HT29 | Hs27 | HT29 | ||
| H14_25_32 K25me3 | 0.50% | 1.62% | 0.24% | 0.12% | 7.15 |
| H2A1_4_11 unmod | 73.43% | 93.18% | 2.46% | 1.37% | 9.95 |
| H2A1_4_11 K5acK9ac | 0.01% | 0.22% | 0.01% | 0.05% | 7.29 |
| H2A1_4_11 K9me1 | 13.59% | 0.98% | 4.18% | 0.80% | 5.49 |
| H2A1_4_11 K5me1 | 10.70% | 2.60% | 1.74% | 0.39% | 7.61 |
| H2AJ_4_11 unmod | 6.33% | 19.01% | 4.15% | 0.51% | 4.00 |
| H2A1_1_11 unmod | 50.90% | 3.13% | 9.77% | 0.39% | 5.23 |
| H2A1_1_11 K5ac | 47.64% | 96.87% | 9.83% | 0.39% | 5.30 |
| H2AZ_1_19 K7ac | 4.95% | 2.68% | 0.80% | 0.44% | 4.72 |
| H2AZ_1_19 K15ac | 0.01% | 0.12% | 0.01% | 0.05% | 4.17 |
| H2A1_12_17 unmod | 56.29% | 48.09% | 0.44% | 2.58% | 5.75 |
| H2A1_12_17 K13ac | 0.00% | 0.02% | 0.00% | 0.01% | 4.88 |
| H2A1_12_17 K15ac | 4.10% | 18.99% | 1.29% | 6.49% | 4.29 |
| H2A1_12_17 K13me1 | 0.40% | 2.50% | 0.21% | 0.72% | 5.24 |
| H2A3_12_17 unmod | 5.73% | 19.49% | 1.62% | 4.88% | 5.03 |
| H2A3_12_17 K15me1 | 14.10% | 80.20% | 2.36% | 4.96% | 12.98 |
| H2A3_12_17 K13me1 | 80.18% | 0.31% | 3.97% | 0.18% | 9.27 |
| H2A_1_88 H2A14s.HLQLAIR | 44.43% | 63.99% | 8.29% | 2.25% | 4.34 |
| H2A_1_88 H2AZ.AGGKAGKDSGKAKTKAVSR | 53.41% | 34.15% | 7.96% | 2.02% | 4.46 |
| H2B_1_29 1C.PEPAKSAPAPKKGSKKAVTKAQKKDGKKR | 62.08% | 46.65% | 1.31% | 5.79% | 4.91 |
| H2B_1_29 1H.PDPAKSAPAPKKGSKKAVTKAQKKDGKKR | 11.11% | 5.20% | 0.84% | 1.20% | 7.01 |
| H33_27_40 K27me2 | 4.87% | 13.33% | 3.20% | 2.12% | 4.21 |
| H33_27_40 K27me3 | 0.74% | 5.17% | 0.91% | 0.88% | 6.28 |
| H33_27_40 K36me3 | 4.09% | 1.08% | 0.92% | 0.23% | 5.83 |
| H33_27_40 K27me1K36me2 | 13.23% | 5.41% | 2.30% | 0.92% | 5.79 |
| H33_27_40 K27me1K36me3 | 4.62% | 1.57% | 0.90% | 1.07% | 4.17 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montalbano, S.; Raboni, S.; Sidoli, S.; Mozzarelli, A.; Bettati, S.; Buschini, A. Post-Translational Modifications of Histone Variants in the Absence and Presence of a Methionine-Depleting Enzyme in Normal and Cancer Cells. Cancers 2023, 15, 527. https://doi.org/10.3390/cancers15020527
Montalbano S, Raboni S, Sidoli S, Mozzarelli A, Bettati S, Buschini A. Post-Translational Modifications of Histone Variants in the Absence and Presence of a Methionine-Depleting Enzyme in Normal and Cancer Cells. Cancers. 2023; 15(2):527. https://doi.org/10.3390/cancers15020527
Chicago/Turabian StyleMontalbano, Serena, Samanta Raboni, Simone Sidoli, Andrea Mozzarelli, Stefano Bettati, and Annamaria Buschini. 2023. "Post-Translational Modifications of Histone Variants in the Absence and Presence of a Methionine-Depleting Enzyme in Normal and Cancer Cells" Cancers 15, no. 2: 527. https://doi.org/10.3390/cancers15020527
APA StyleMontalbano, S., Raboni, S., Sidoli, S., Mozzarelli, A., Bettati, S., & Buschini, A. (2023). Post-Translational Modifications of Histone Variants in the Absence and Presence of a Methionine-Depleting Enzyme in Normal and Cancer Cells. Cancers, 15(2), 527. https://doi.org/10.3390/cancers15020527








