Prognostication in Advanced Cancer by Combining Actigraphy-Derived Rest-Activity and Sleep Parameters with Routine Clinical Data: An Exploratory Machine Learning Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Setting
2.2. Study Participants
2.3. Routine Data Collection
2.4. Wrist Actigraphy and Consensus Sleep Diary
| Sleep Parameter | Definition |
|---|---|
| Actigraphy and sleep diary | |
| Bed-time (BT) (hh:mm) | Clock time attempted to fall asleep based on actigraphy event marker or sleep diary |
| Get-up time (GUT) (hh:mm) | Clock time attempted to rise from bed for the final time based on actigraphy event marker or sleep diary |
| Time in bed (TIB) (hh:mm) | Duration between reported BT and GUT (reported in hours and minutes) or as self-reported in sleep diary |
| Sleep onset latency (SOL) (min) | Duration between reported BT and actigraph scored sleep onset time or as self-reported in sleep diary |
| Total sleep time (TST) (hh:mm) | Duration of sleep during the major sleep period calculated by Actiware; |
| Sleep diary manual calculation: TIB minus (SOL plus WASO plus TWAK) | |
| Sleep efficiency (SE) (%) | Proportion of time the patient is asleep out of the total time in bed (reported as a percentage) calculated by Actiware; |
| Sleep diary manual calculation: TST divided by TIB × 100 | |
| Wake after sleep onset (WASO) (min) | Sum of wake times from sleep onset to the final awakening calculated by Actiware or as self-reported in sleep diary |
| Number of awake episodes (NA) | Number of continuous blocks of wake during the major sleep period calculated by Actiware or as self-reported in sleep diary |
| Sleep Diary | |
| Time tried to sleep (hh:mm) | Self-reported time participant began ‘trying’ to fall asleep |
| Time of final awakening (hh:mm) | Self-reported time participant last woke up in the morning |
| Terminal awakening (TWAK) (hh:mm) | GUT minus time of final awakening |
2.5. Follow-Up
3. Statistical Analyses
4. Machine Learning Methods and Data Analysis
4.1. Machine Learning Dataset
4.2. Regularised Regression Methods
4.3. Model Development
5. Results
5.1. Acceptability of Actigraphy and Sleep Diary Acceptability
5.2. Univariate Analyses of Actigraphy Parameters
5.2.1. Characteristics of the Dichotomy Index (I < O) and Correlation with Other Actigraphy and Sleep Parameters
5.2.2. I < O: Predictor of Survival and Correlation with ECOG-PS
5.2.3. Autocorrelation Coefficient at 24 h (r24)
5.2.4. Other Actigraphy Parameters
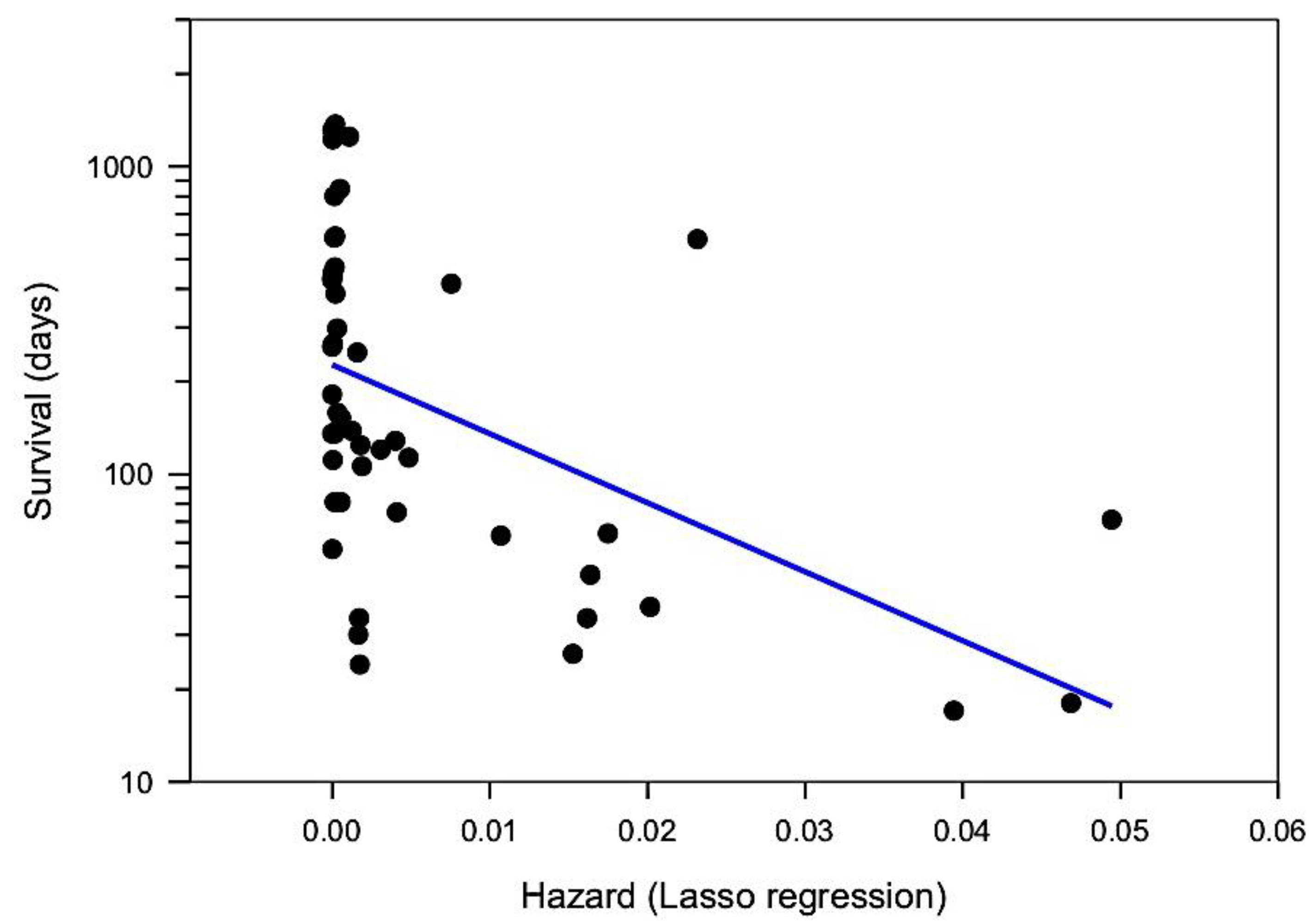
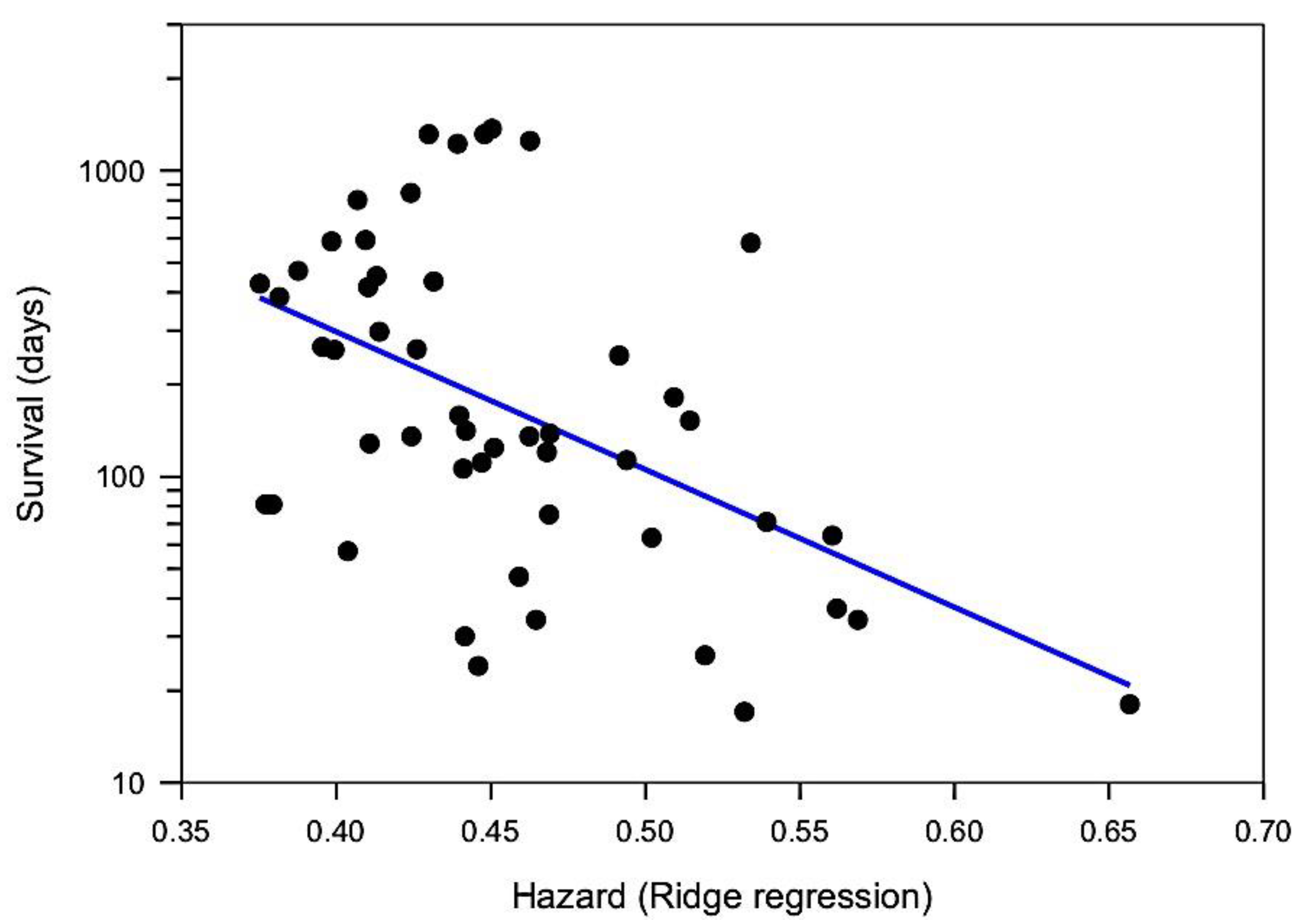
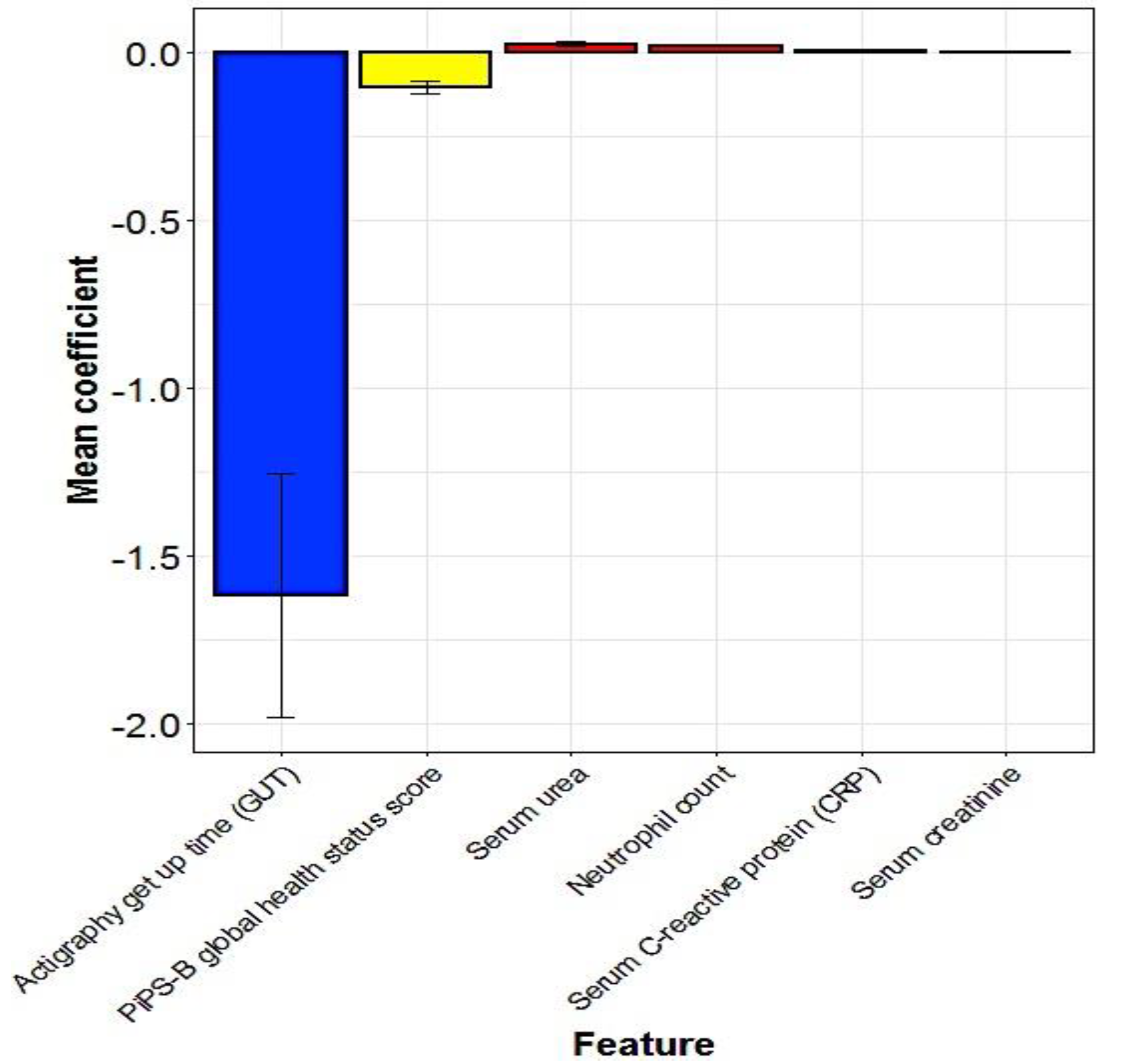
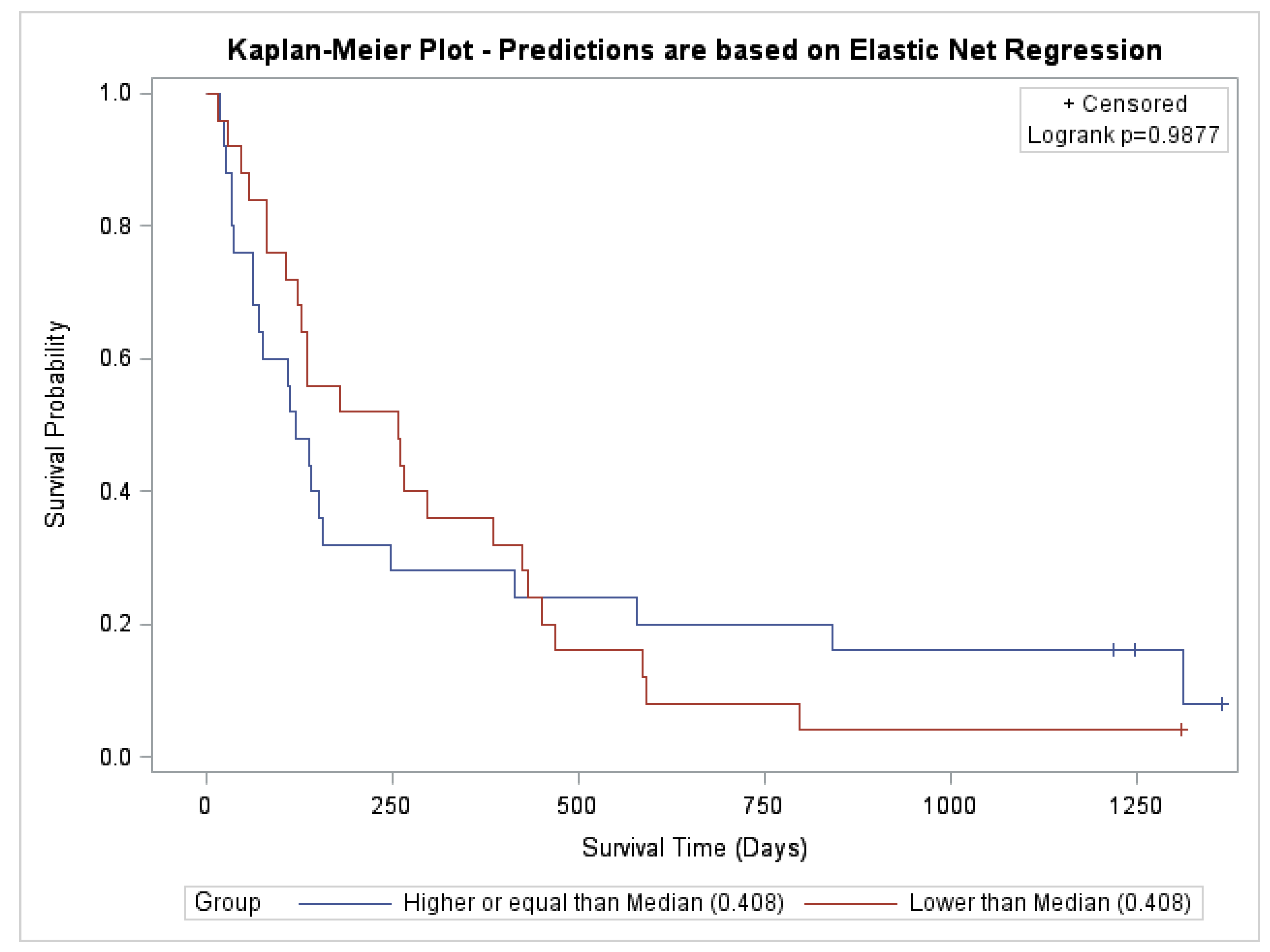
5.3. Multivariate Predictors of Survival: Machine Learning Results
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Prognostic Parameters for Machine Learning
| 1. Medication: Use of opioid analgesia |
| 2. ECOG-PS at baseline: Physician-assessed |
| 3. ECOG-PS at baseline: Patient-assessed |
| 4. ECOG-PS Day 8: Physician-assessed |
| 5. ECOG-PS Day 8: Patient-assessed |
| 6. MSAS-SF: Number of symptoms |
| 7. MSAS-SF: Physical symptom subscale score (MSASPHYS) |
| 8. MSAS-SF: Psychological symptom subscale score (MSASPSYCH) |
| 9. MSAS-SF: Total symptom distress score (TMSAS) |
| 10. MSAS-SF: Global Distress Index (GDI) |
| 11. PSQI: Usual Bedtime (BT) |
| 12. PSQI: Time to fall asleep (SOL) |
| 13. PSQI: Usual getting up time (GUT) |
| 14. PSQI: Hours of sleep per night (TST) |
| 15. PSQI: Sleep disturbance—Cannot get to sleep within 30 min |
| 16. PSQI: Sleep disturbance—Wake up in the middle of the night or early morning |
| 17. PSQI: Sleep disturbance—Have to get up to use bathroom |
| 18. PSQI Sleep disturbance—Cannot breathe comfortably |
| 19. PSQI Sleep disturbance—Cough or snore loudly |
| 20. PSQI Sleep disturbance—Feel too cold |
| 21. PSQI Sleep disturbance—Feel too hot |
| 22. PSQI: Sleep disturbance—Had bad dreams |
| 23. PSQI: Sleep disturbance—Have pain |
| 24. PSQI: Subjective sleep quality |
| 25. PSQI: Use of medication for sleep |
| 26. PSQI: Daytime dysfunction: Trouble staying awake |
| 27. PSQI: Keep up enough enthusiasm to get things done |
| 28. PSQI: Presence of bed partner or roommate |
| 29. PiPS-B algorithm: Abbreviated Mental Test Score (out of 10) |
| 30. PiPS-B algorithm: Patient’s pulse rate |
| 31. PiPS-B algorithm: Global Health Status Score (1 = extremely poor health; 7 = normal health) |
| 32. PiPS-B algorithm: Clinician’s estimate of survival (Days/Weeks/Months+) |
| 33. Modified Glasgow Prognostic Score (mGPS) |
| 34. Bloods: Haemoglobin (g/L) (130–180) |
| 35. Bloods: White Blood Count (109/L) (4–11) |
| 36. Bloods: Neutrophils (109/L) (2.0–7.5) |
| 37. Bloods: Lymphocytes (109/L) (1.0–4.0) |
| 38. Bloods: Platelets (109/L) (150–450) |
| 39. Bloods: Sodium (mmol/L) (133–146) |
| 40. Bloods: Potassium (mmol/L) (3.5–5.3) |
| 41. Bloods: Urea (mmol/L) (2.5–7.8) |
| 42. Bloods: Creatinine (µmol/L) (64–104) |
| 43. Bloods: ALP (IU/L) (30–130) |
| 44. Bloods: ALT (IU/L) (<50) |
| 45. Bloods: Albumin (g/L) (35–50) |
| 46. Bloods: C-reactive protein (CRP) (mg/L) (<10) |
| 47. Wrist actigraphy: Rest-activity parameter—Dichotomy Index (I < O) at least 72 h |
| 48. Wrist actigraphy: Rest-activity parameter—r24 (autocorrelation coefficient) at least 72 h |
| 49. Wrist actigraphy: Activity parameters—Mean activity during wakefulness at least 72 h |
| 50. Wrist actigraphy: Activity parameters—Mean daily activity (MDA) at least 72 h |
| 51. Wrist actigraphy: Sleep parameter—Bedtime (BT) |
| 52. Wrist actigraphy: Sleep parameter—Get up time (GUT) |
| 53. Wrist actigraphy: Sleep parameter—Time in bed (TIB) |
| 54. Wrist actigraphy: Sleep parameter—Total sleep time (TST) |
| 55. Wrist actigraphy: Sleep parameter—Sleep onset latency (SOL) |
| 56. Wrist actigraphy: Sleep parameter—Sleep Efficiency (%) |
| 57. Wrist actigraphy: Sleep parameter—Wake after sleep onset (WASO) |
| 58. Wrist actigraphy: Sleep parameter—Number of awake episodes (NA) |
| 59. Consensus Sleep Diary: Time in bed (BT) |
| 60. Consensus Sleep Diary: Time of final awakening |
| 61. Consensus Sleep Diary: Time out of bed (GUT) |
| 62. Consensus Sleep Diary: Time tried to go to sleep |
| 63. Consensus Sleep Diary: Time to fall asleep (SOL) |
| 64. Consensus Sleep Diary: Quality of Sleep |
| 65. Consensus Sleep Diary: Total amount of time awakenings lasted (WASO) |
| 66. Consensus Sleep Diary: Number of times awakened in the night (NA) |
| Numerical Prognostic Parameter (n = 42) | Mean | Standard Deviation |
|---|---|---|
| MSAS-SF: Number of symptoms | 11.9 | 5.2 |
| MSAS-SF: Physical Symptom Subscale Score (MSASPHYS) | 2.3 | 0.7 |
| MSAS-SF: Psychological Symptom Subscale Score (MSASPSYCH) | 1.9 | 0.8 |
| MSAS-SF: Total symptom distress score (TMSAS) | 2.2 | 0.6 |
| MSAS-SF: Global Distress Index (GDI) | 2.3 | 0.6 |
| PSQI: Usual Bedtime (BT) (hh:mm) | 22:28 | 1:13 |
| PSQI: Time to fall asleep (SOL) (min) | 28.3 | 38.3 |
| PSQI: Usual getting up time (GUT) (hh:mm) | 07:51 | 1:11 |
| PSQI: Hours of sleep per night (TST) (h) | 6.7 | 1.8 |
| PiPS-B algorithm: Patient’s pulse rate (beats per min) | 84 | 16 |
| Bloods: Haemoglobin (g/L) | 111.5 | 20.8 |
| Bloods: White Blood Count (109/L) | 7.8 | 4.3 |
| Bloods: Neutrophils (109/L) | 5.7 | 4.1 |
| Bloods: Lymphocytes (109/L) | 1.3 | 1.2 |
| Bloods: Platelets (109/L) | 315.3 | 166.7 |
| Bloods: Sodium (mmol/L) | 138.5 | 4.0 |
| Bloods: Potassium (mmol/L) | 4.3 | 0.6 |
| Bloods: Urea (mmol/L) | 6.2 | 2.7 |
| Bloods: Creatinine (µmol/L) | 71.4 | 26.1 |
| Bloods: ALP (IU/L) | 284.9 | 436.0 |
| Bloods: ALT (IU/L) | 59.4 | 149.3 |
| Bloods: Albumin (g/L) | 37.4 | 4.3 |
| Bloods: C-reactive protein (CRP) (mg/L) | 45.2 | 48.5 |
| Wrist actigraphy: (I < O) at least 72 h (%) | 89.0 | 6.5 |
| Wrist actigraphy: r24 at least 72 h (autocorrelation coefficient) | 0.17 | 0.1 |
| Wrist actigraphy: Mean activity during wakefulness at least 72 h (number of accelerations per min) | 143.7 | 62.1 |
| Wrist actigraphy: Mean daily activity (MDA) at least 72 h (number of accelerations per min) | 96.8 | 39.8 |
| Wrist actigraphy: Bedtime (BT) (hh:mm) | 22:41 | 1:07 |
| Wrist actigraphy: Get up time (GUT) (hh:mm) | 08:03 | 1:01 |
| Wrist actigraphy: Time in bed (TIB) (hh:mm) | 09:22 | 1:33 |
| Wrist actigraphy: Total sleep time (TST) (hh:mm) | 7:18 | 1:39 |
| Wrist actigraphy: Sleep onset latency (SOL) (min) | 21.7 | 21.6 |
| Wrist actigraphy: Sleep efficiency (SE) (%) | 78.2 | 12.0 |
| Wrist actigraphy: Wake after sleep onset (WASO) (min) | 68.4 | 31.6 |
| Wrist actigraphy: Number of awake episodes (NA) | 22.4 | 10.1 |
| Consensus Sleep Diary: Time in bed (BT) (hh:mm) | 22:35 | 1:06 |
| Consensus Sleep Diary: Time of final awakening (hh:mm) | 07:08 | 1:05 |
| Consensus Sleep Diary: Time out of bed (GUT) (hh:mm) | 08:03 | 1:01 |
| Consensus Sleep Diary: Time tried to go to sleep (hh:mm) | 22:58 | 1:02 |
| Consensus Sleep Diary: Time to fall asleep (SOL) (min) | 32.4 | 32.7 |
| Consensus Sleep Diary: Total amount of time awakenings lasted (WASO) (min) | 37.7 | 37.6 |
| Consensus Sleep Diary: Number of times awakened in the night (NA) | 2.5 | 1.3 |

References
- Maltoni, M.; Caraceni, A.; Brunelli, C.; Broeckaert, B.; Christakis, N.; Eychmueller, S.; Glare, P.; Nabal, M.; Vigano, A.; Larkin, P.; et al. Prognostic factors in advanced cancer patients: Evidence-based clinical recommendations--a study by the steering committee of the european association for palliative care. J. Clin. Oncol. 2005, 23, 6240–6248. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; White, N.; Stone, P. Prognostication in palliative care. Clin. Med. 2019, 19, 306–310. [Google Scholar] [CrossRef] [PubMed]
- White, N.; Reid, F.; Harris, A.; Harries, P.; Stone, P. A systematic review of predictions of survival in palliative care: How accurate are clinicians and who are the experts? PLoS ONE 2016, 11, e0161407. [Google Scholar] [CrossRef]
- Hui, D.; Paiva, C.E.; Del Fabbro, E.G.; Steer, C.; Naberhuis, J.; van de Wetering, M.; Fernández-Ortega, P.; Morita, T.; Suh, S.Y.; Bruera, E.; et al. Prognostication in advanced cancer: Update and directions for future research. Support. Care Cancer 2019, 27, 1973–1984. [Google Scholar] [CrossRef] [PubMed]
- Hui, D. Prognostication of survival in patients with advanced cancer: Predicting the unpredictable? Cancer Control 2015, 22, 489–497. [Google Scholar] [CrossRef]
- Allada, R.; Bass, J. Circadian mechanisms in medicine. N. Engl. J. Med. 2021, 384, 550–561. [Google Scholar] [CrossRef]
- Milanti, A.; Chan, D.N.S.; Li, C.; So, W.K.W. Actigraphy-measured rest-activity circadian rhythm disruption in patients with advanced cancer: A scoping review. Support. Care Cancer 2021, 29, 7145–7169. [Google Scholar] [CrossRef]
- Balachandran, D.D.; Miller, M.A.; Faiz, S.A.; Yennurajalingam, S.; Innominato, P.F. Evaluation and management of sleep and circadian rhythm disturbance in cancer. Curr. Treat. Options Oncol. 2021, 22, 81. [Google Scholar] [CrossRef]
- Medic, G.; Wille, M.; Hemels, M.E. Short- and long-term health consequences of sleep disruption. Nat. Sci. Sleep 2017, 9, 151–161. [Google Scholar] [CrossRef]
- Levi, F.; Dugue, P.A.; Innominato, P.; Karaboue, A.; Dispersyn, G.; Parganiha, A.; Giacchetti, S.; Moreau, T.; Focan, C.; Waterhouse, J.; et al. Wrist actimetry circadian rhythm as a robust predictor of colorectal cancer patients survival. Chronobiol. Int. 2014, 31, 891–900. [Google Scholar] [CrossRef]
- Innominato, P.F.; Komarzynski, S.; Palesh, O.G.; Dallmann, R.; Bjarnason, G.A.; Giacchetti, S.; Ulusakarya, A.; Bouchahda, M.; Haydar, M.; Ballesta, A.; et al. Circadian rest-activity rhythm as an objective biomarker of patient-reported outcomes in patients with advanced cancer. Cancer Med. 2018, 7, 4396–4405. [Google Scholar] [CrossRef] [PubMed]
- Mormont, M.C.; Waterhouse, J.; Bleuzen, P.; Giacchetti, S.; Jami, A.; Bogdan, A.; Lellouch, J.; Misset, J.L.; Touitou, Y.; Levi, F. Marked 24-h rest/activity rhythms are associated with better quality of life, better response, and longer survival in patients with metastatic colorectal cancer and good performance status. Clin. Cancer Res. 2000, 6, 3038–3045. [Google Scholar] [PubMed]
- Palesh, O.; Aldridge-Gerry, A.; Zeitzer, J.M.; Koopman, C.; Neri, E.; Giese-Davis, J.; Jo, B.; Kraemer, H.; Nouriani, B.; Spiegel, D. Actigraphy-measured sleep disruption as a predictor of survival among women with advanced breast cancer. Sleep 2014, 37, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Treatment and Care towards the End of Life: Good Practice in Decision-Making; General Medical Council: London, UK, 2010.
- Oken, M.M.; Creech, R.H.; Tormey, D.C.; Horton, J.; Davis, T.E.; McFadden, E.T.; Carbone, P.P. Toxicity and response criteria of the eastern cooperative oncology group. Am. J. Clin. Oncol. 1982, 5, 649–655. [Google Scholar] [CrossRef] [PubMed]
- Hodkinson, H.M. Evaluation of a mental test score for assessment of mental impairment in the elderly. Age Ageing 1972, 1, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Chang, V.T.; Hwang, S.S.; Feuerman, M.; Kasimis, B.S.; Thaler, H.T. The memorial symptom assessment scale short form (msas-sf). Cancer 2000, 89, 1162–1171. [Google Scholar] [CrossRef] [PubMed]
- Gwilliam, B.; Keeley, V.; Todd, C.; Gittins, M.; Roberts, C.; Kelly, L.; Barclay, S.; Stone, P.C. Development of prognosis in palliative care study (pips) predictor models to improve prognostication in advanced cancer: Prospective cohort study. BMJ 2011, 343, d4920. [Google Scholar] [CrossRef]
- Buysse, D.J.; Reynolds, C.F., 3rd; Monk, T.H.; Berman, S.R.; Kupfer, D.J. The pittsburgh sleep quality index: A new instrument for psychiatric practice and research. Psychiatry Res. 1989, 28, 193–213. [Google Scholar] [CrossRef]
- McMillan, D.C. The systemic inflammation-based glasgow prognostic score: A decade of experience in patients with cancer. Cancer Treat. Rev. 2013, 39, 534–540. [Google Scholar] [CrossRef]
- Philips Respironics. Professional Sleep and Activity Monitoring Solutions: Specifications for Actiwatch 2, Actiwatch Spectrum Plus, and Actiwatch Spectrum Pro; Koninklijke Philips Electronics N.V.: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Carney, C.E.; Buysse, D.J.; Ancoli-Israel, S.; Edinger, J.D.; Krystal, A.D.; Lichstein, K.L.; Morin, C.M. The consensus sleep diary: Standardizing prospective sleep self-monitoring. Sleep 2012, 35, 287–302. [Google Scholar] [CrossRef]
- Berger, A.M.; Wielgus, K.K.; Young-McCaughan, S.; Fischer, P.; Farr, L.; Lee, K.A. Methodological challenges when using actigraphy in research. J. Pain Symptom Manag. 2008, 36, 191–199. [Google Scholar] [CrossRef]
- Berger, A.M. Update on the state of the science: Sleep-wake disturbances in adult patients with cancer. Oncol. Nurs. Forum 2009, 36, E165–E177. [Google Scholar] [CrossRef] [PubMed]
- Innominato, P.F.; Focan, C.; Gorlia, T.; Moreau, T.; Garufi, C.; Waterhouse, J.; Giacchetti, S.; Coudert, B.; Iacobelli, S.; Genet, D.; et al. Circadian rhythm in rest and activity: A biological correlate of quality of life and a predictor of survival in patients with metastatic colorectal cancer. Cancer Res. 2009, 69, 4700–4707. [Google Scholar] [CrossRef]
- Ancoli-Israel, S.; Martin, J.L.; Blackwell, T.; Buenaver, L.; Liu, L.; Meltzer, L.J.; Sadeh, A.; Spira, A.P.; Taylor, D.J. The sbsm guide to actigraphy monitoring: Clinical and research applications. Behav. Sleep Med. 2015, 13, S4–S38. [Google Scholar] [CrossRef]
- Sadeh, A.; Hauri, P.J.; Kripke, D.F.; Lavie, P. The role of actigraphy in the evaluation of sleep disorders. Sleep 1995, 18, 288–302. [Google Scholar] [CrossRef] [PubMed]
- Schutte-Rodin, S.; Broch, L.; Buysse, D.; Dorsey, C.; Sateia, M. Clinical guideline for the evaluation and management of chronic insomnia in adults. J. Clin. Sleep Med. 2008, 4, 487–504. [Google Scholar] [CrossRef] [PubMed]
- Research Design Service London. Justify Sample Size for a Feasibility Study. Available online: https://www.rds-london.nihr.ac.uk/resources/justify-sample-size-for-a-feasibility-study/ (accessed on 14 June 2022).
- Mukaka, M.M. Statistics corner: A guide to appropriate use of correlation coefficient in medical research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference and Prediction, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning: With Applications in R; Springer: New York, NY, USA, 2013. [Google Scholar]
- Tibshirani, R. Regression shrinkage and selection via the lasso. J. R. Stat. Soc. Ser. B 1996, 58, 267–288. [Google Scholar] [CrossRef]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. Ser. B 2005, 67, 301–320. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Koo, T.K.; Li, M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J. Chiropr. Med. 2016, 15, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Storick, V.; O’Herlihy, A.; Abdelhafeez, S.; Ahmed, R.; May, P. Improving palliative and end-of-life care with machine learning and routine data: A rapid review. HRB Open Res. 2019, 2, 13. [Google Scholar] [CrossRef] [PubMed]
- Innominato, P.F.; Lim, A.S.; Palesh, O.; Clemons, M.; Trudeau, M.; Eisen, A.; Wang, C.; Kiss, A.; Pritchard, K.I.; Bjarnason, G.A. The effect of melatonin on sleep and quality of life in patients with advanced breast cancer. Support. Care Cancer 2016, 24, 1097–1105. [Google Scholar] [CrossRef]
- Innominato, P.F.; Giacchetti, S.; Bjarnason, G.A.; Focan, C.; Garufi, C.; Coudert, B.; Iacobelli, S.; Tampellini, M.; Durando, X.; Mormont, M.C.; et al. Prediction of overall survival through circadian rest-activity monitoring during chemotherapy for metastatic colorectal cancer. Int. J. Cancer 2012, 131, 2684–2692. [Google Scholar] [CrossRef]
- Collins, K.P.; Geller, D.A.; Antoni, M.; Donnell, D.M.; Tsung, A.; Marsh, J.W.; Burke, L.; Penedo, F.; Terhorst, L.; Kamarck, T.W.; et al. Sleep duration is associated with survival in advanced cancer patients. Sleep Med. 2017, 32, 208–212. [Google Scholar] [CrossRef] [PubMed]
- Laird, B.J.; Kaasa, S.; McMillan, D.C.; Fallon, M.T.; Hjermstad, M.J.; Fayers, P.; Klepstad, P. Prognostic factors in patients with advanced cancer: A comparison of clinicopathological factors and the development of an inflammation-based prognostic system. Clin. Cancer Res. 2013, 19, 5456–5464. [Google Scholar] [CrossRef]
- Ellershaw, J.; Ward, C. Care of the dying patient: The last hours or days of life. BMJ 2003, 326, 30–34. [Google Scholar] [CrossRef]
- Natale, V.; Innominato, P.F.; Boreggiani, M.; Tonetti, L.; Filardi, M.; Parganiha, A.; Fabbri, M.; Martoni, M.; Lévi, F. The difference between in bed and out of bed activity as a behavioral marker of cancer patients: A comparative actigraphic study. Chronobiol. Int. 2015, 32, 925–933. [Google Scholar] [CrossRef]
- Palesh, O.; Haitz, K.; Levi, F.; Bjarnason, G.A.; Deguzman, C.; Alizeh, I.; Ulusakarya, A.; Packer, M.M.; Innominato, P.F. Relationship between subjective and actigraphy-measured sleep in 237 patients with metastatic colorectal cancer. Qual. Life Res. 2017, 26, 2783–2791. [Google Scholar] [CrossRef]
- Innominato, P.F.; Roche, V.P.; Palesh, O.G.; Ulusakarya, A.; Spiegel, D.; Levi, F.A. The circadian timing system in clinical oncology. Ann. Med. 2014, 46, 191–207. [Google Scholar] [CrossRef]

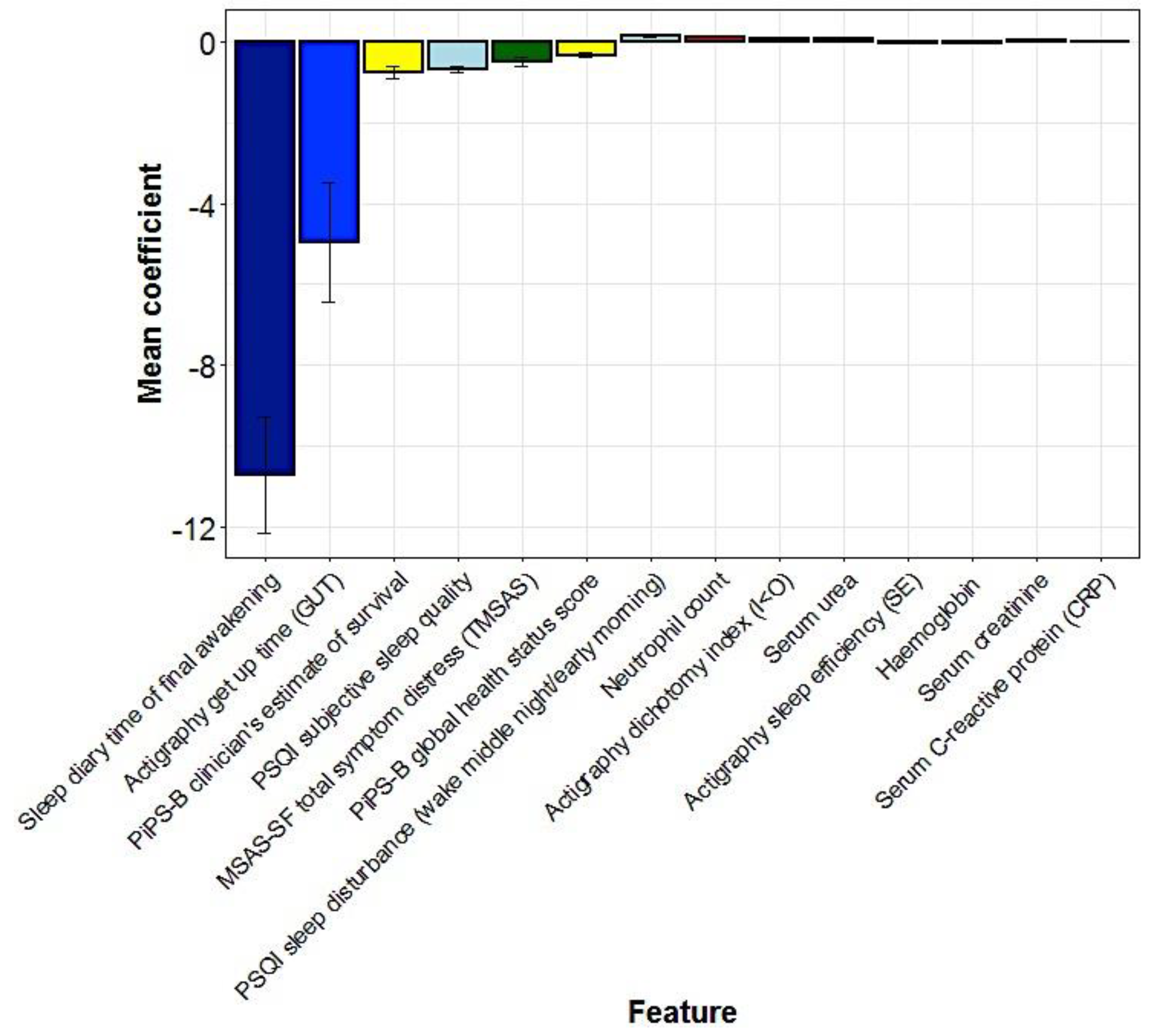
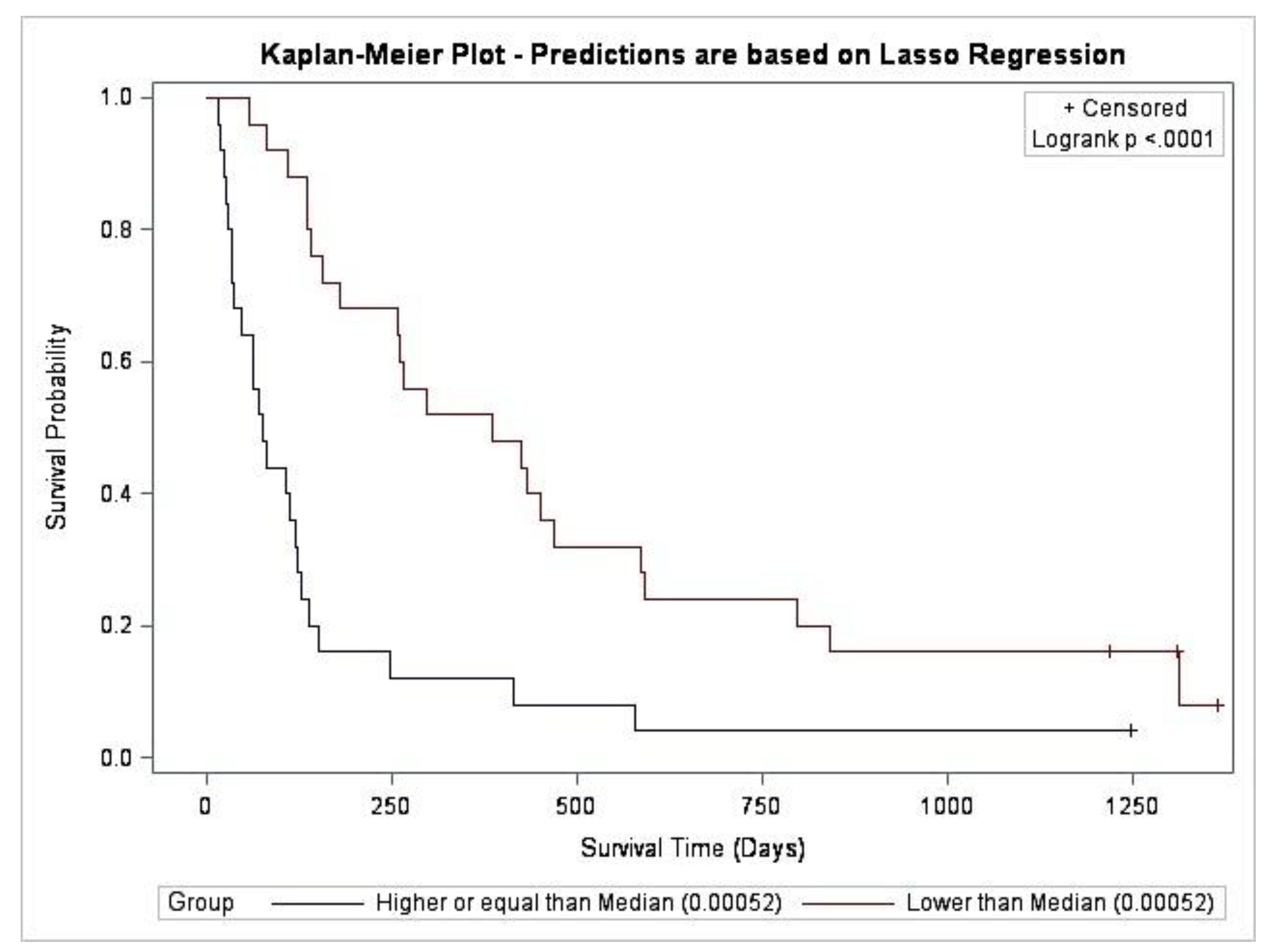
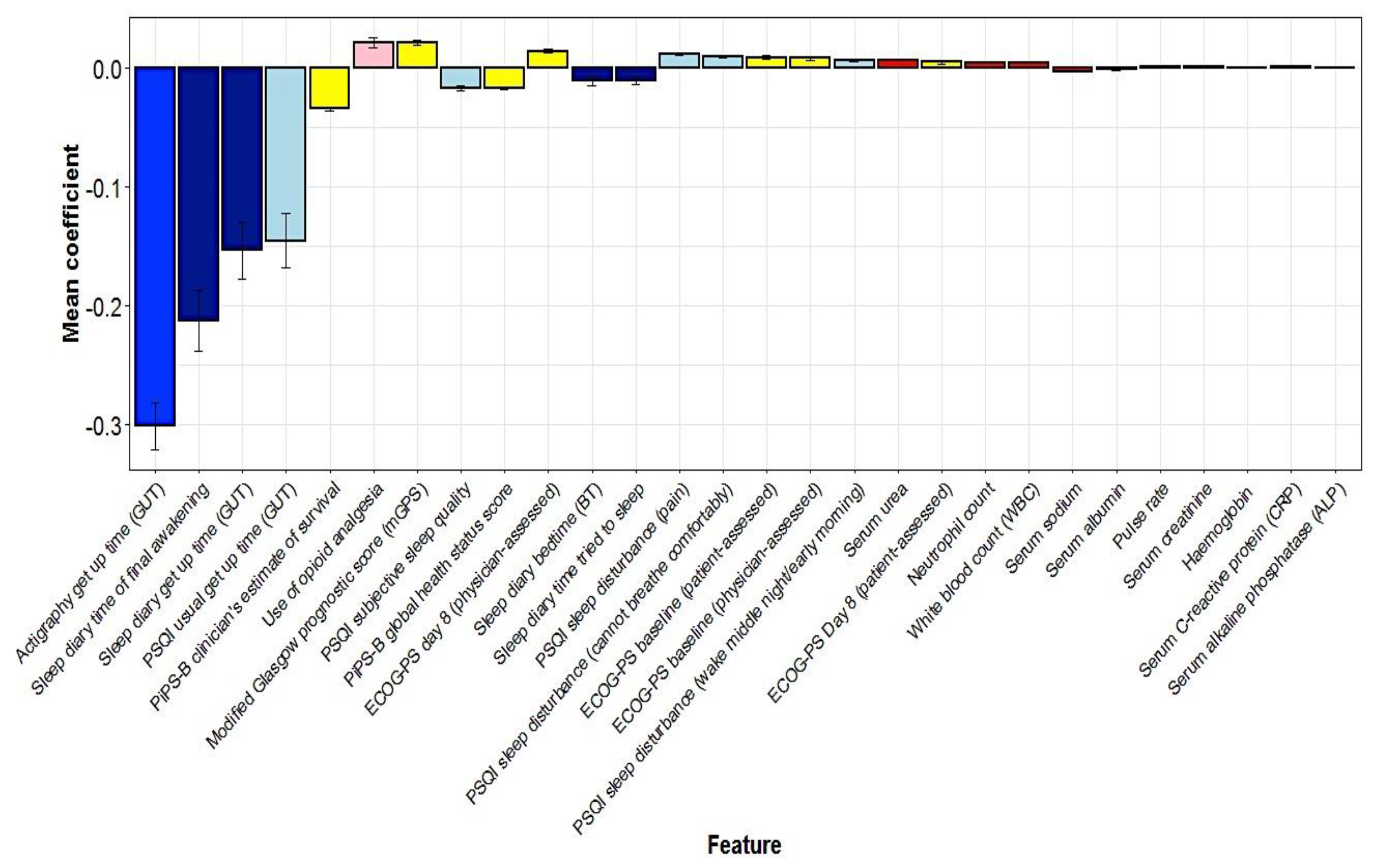

| Characteristic | All Participants (n = 50) | “Full Analysis Set” (n = 40) |
|---|---|---|
| Age | Median—63 yr | Median—66 yr |
| (range 40–81 yr) | (range 43–81 yr) | |
| Sex | Female—21 (42%) | Female–17 (39%) |
| Male—29 (58%) | Male—27 (61%) | |
| Cancer diagnosis | Breast—6 (12%) | Breast—6 (14%) |
| Endocrine—1 (2%) | Endocrine—1 (2%) | |
| Gastrointestinal—16 (32%) | Gastrointestinal—14 (32%) | |
| Gynaecological—6 (12%) | Gynaecological—4 (9%) | |
| Haematological—2 (4%) | Haematological—2 (5%) | |
| Head and Neck—3 (6%) | Head and Neck—2 (5%) | |
| Lung—6 (12%) | Lung—6 (14%) | |
| Skin—2 (4%) | Skin—2 (5%) | |
| Urological—8 (16%) | Urological—7 (16%) | |
| ECOG-PS | 0–0 (0%) | 0–0 (0%) |
| (Physician-assessed | 1–26 (52%) | 1–24 (55%) |
| at baseline) | 2–13 (26%) | 2–10 (23%) |
| 3–11 (22%) | 3–10 (23%) | |
| 4–0 (0%) | 4–0 (0%) |
| I < O Parameter | Full Analysis Set (n = 44) | Per Protocol Set (n = 37) |
|---|---|---|
| Mean | 88.90% | 89.90% |
| (+/− standard error) | (+/− 1.04) | (+/− 0.97) |
| Minimum | 70.90% | 70.90% |
| 25th Centile | 86.90% | 87.40% |
| Median | 90.40% | 90.80% |
| 75th Centile | 93.60% | 93.60% |
| Maximum | 98.10% | 97.60% |
| Distribution | Non-normal | Non-normal |
| (Shapiro-Wilk | (Shapiro-Wilk | |
| test: p = 0.001) | test: p = 0.001) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Patel, S.D.; Davies, A.; Laing, E.; Wu, H.; Mendis, J.; Dijk, D.-J. Prognostication in Advanced Cancer by Combining Actigraphy-Derived Rest-Activity and Sleep Parameters with Routine Clinical Data: An Exploratory Machine Learning Study. Cancers 2023, 15, 503. https://doi.org/10.3390/cancers15020503
Patel SD, Davies A, Laing E, Wu H, Mendis J, Dijk D-J. Prognostication in Advanced Cancer by Combining Actigraphy-Derived Rest-Activity and Sleep Parameters with Routine Clinical Data: An Exploratory Machine Learning Study. Cancers. 2023; 15(2):503. https://doi.org/10.3390/cancers15020503
Chicago/Turabian StylePatel, Shuchita Dhwiren, Andrew Davies, Emma Laing, Huihai Wu, Jeewaka Mendis, and Derk-Jan Dijk. 2023. "Prognostication in Advanced Cancer by Combining Actigraphy-Derived Rest-Activity and Sleep Parameters with Routine Clinical Data: An Exploratory Machine Learning Study" Cancers 15, no. 2: 503. https://doi.org/10.3390/cancers15020503
APA StylePatel, S. D., Davies, A., Laing, E., Wu, H., Mendis, J., & Dijk, D.-J. (2023). Prognostication in Advanced Cancer by Combining Actigraphy-Derived Rest-Activity and Sleep Parameters with Routine Clinical Data: An Exploratory Machine Learning Study. Cancers, 15(2), 503. https://doi.org/10.3390/cancers15020503









