Predicting Microenvironment in CXCR4- and FAP-Positive Solid Tumors—A Pan-Cancer Machine Learning Workflow for Theranostic Target Structures
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Acquisition
2.2. Machine Learning Model
2.3. Bioinformatical Analyses
2.4. Literature Search Regarding MicroRNA Functions
3. Results
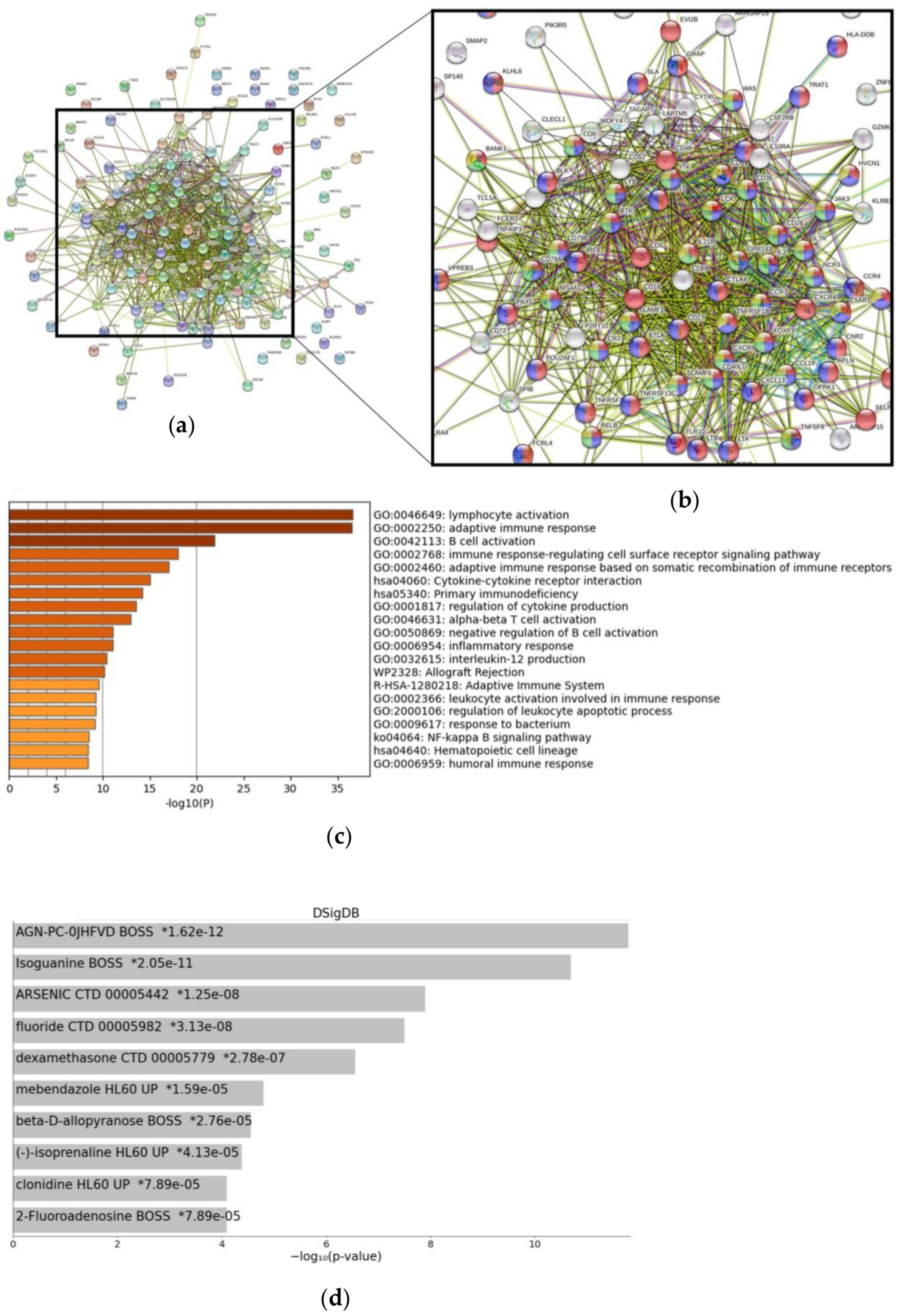
3.1. Signaling Pathways and Drug-Specific Signatures Related to CXCR4 Overexpression
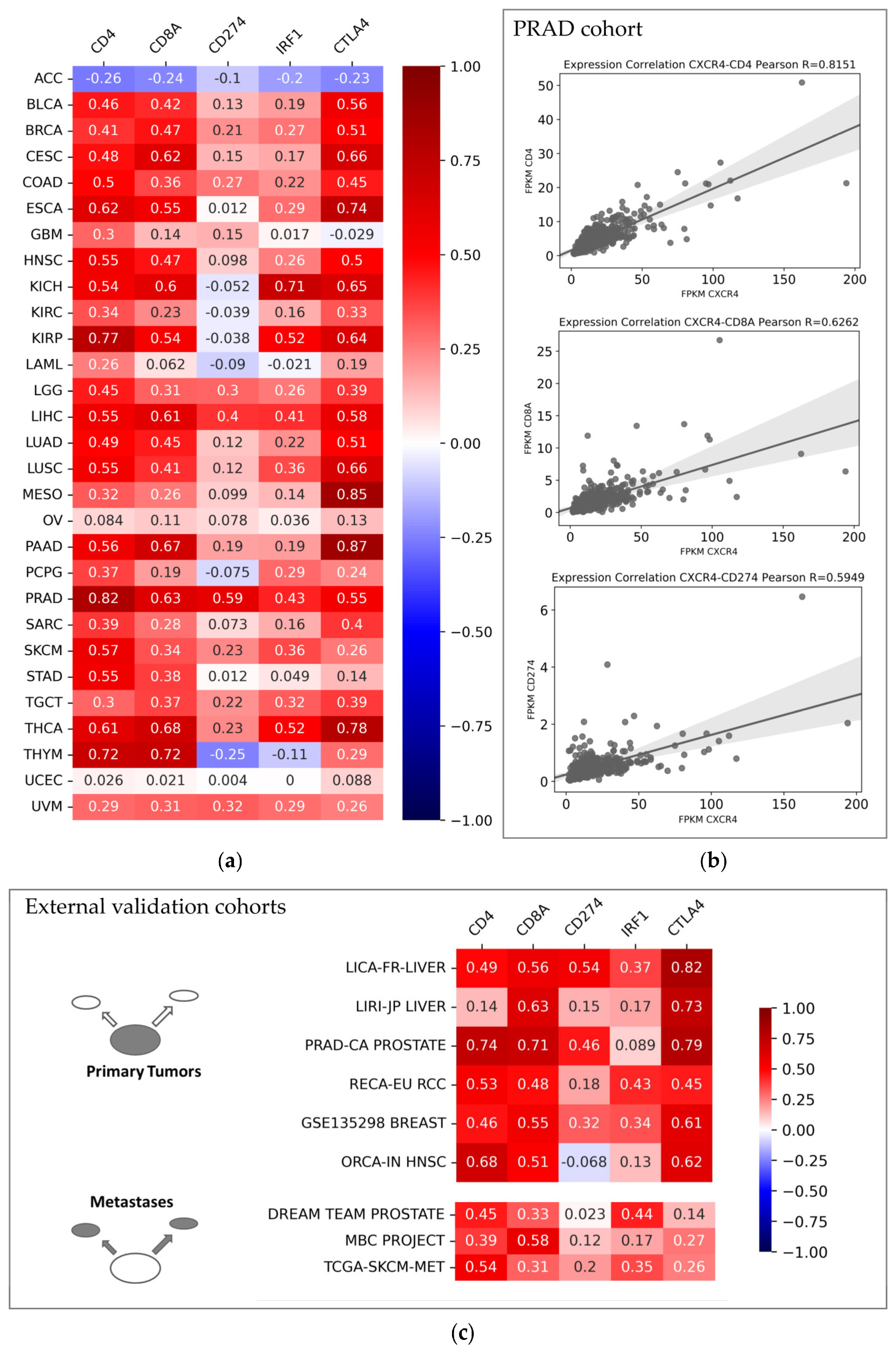
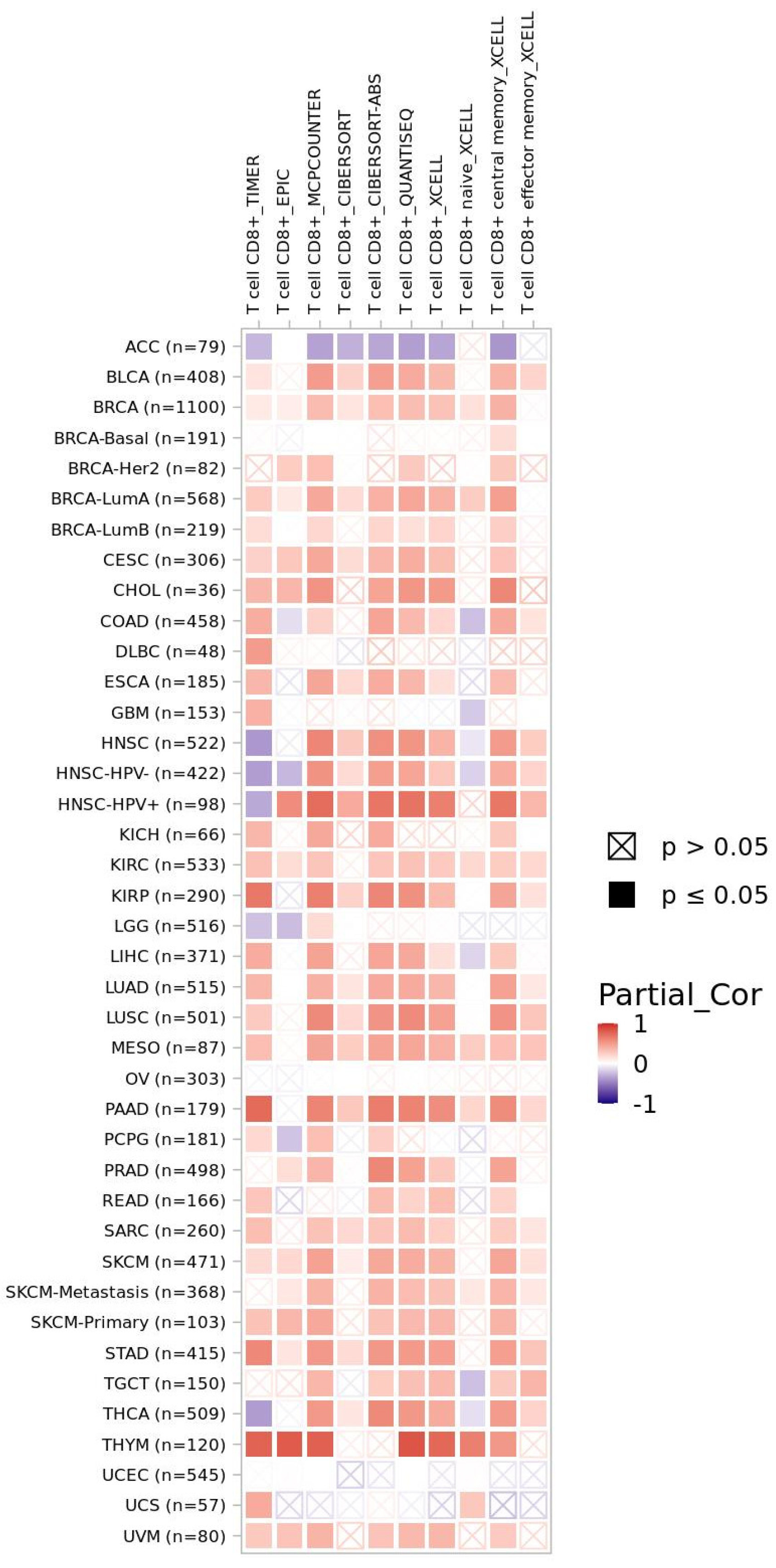
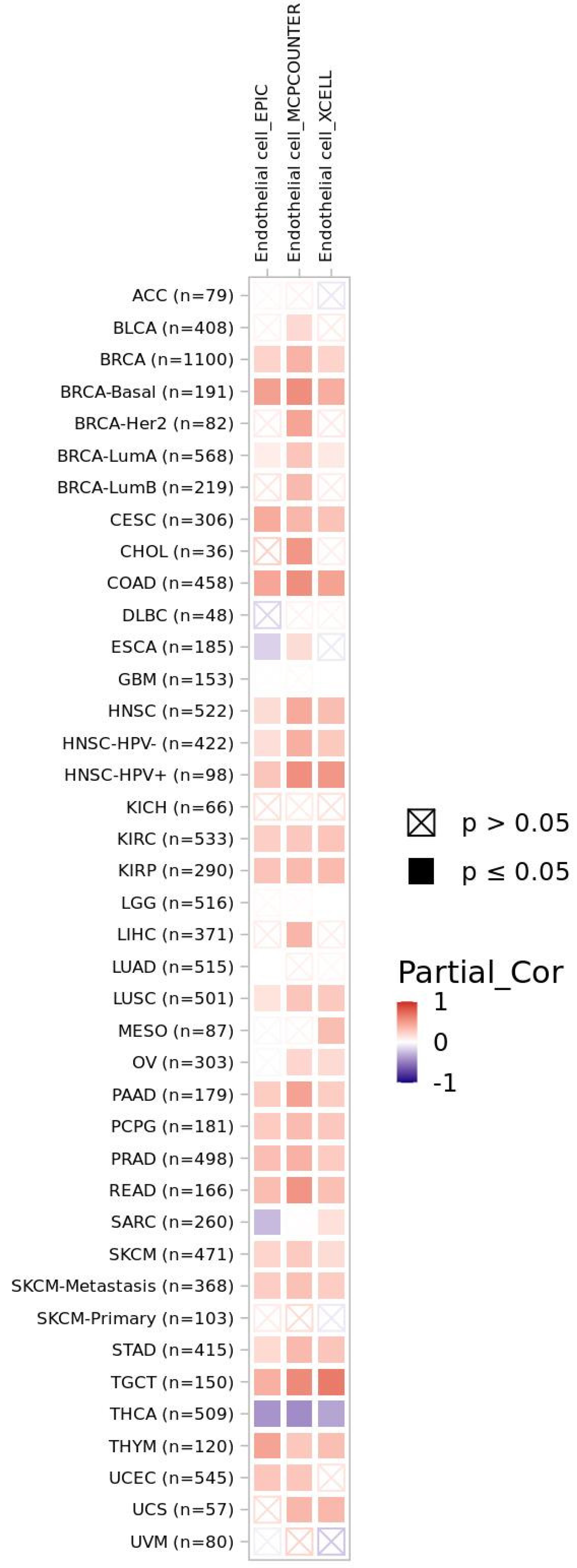
3.2. CXCR4-Associated Tumor Microenvironment from a Pan-Cancer Perspective
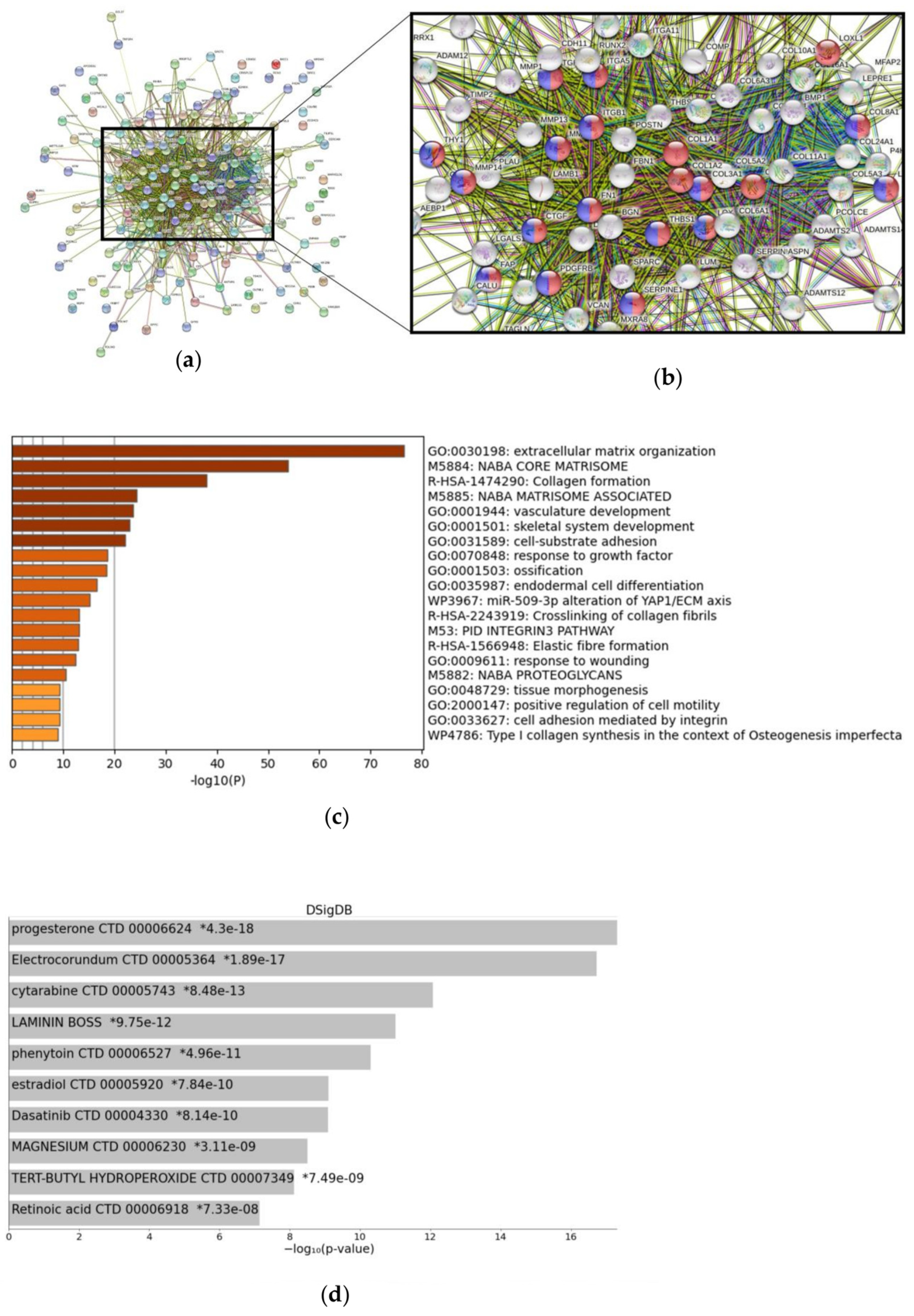
3.3. FAP-Associated Signaling and Tumor Microenvironment
3.4. MicroRNAs Characterizing CXCR4 and FAP Overexpression
3.5. Transferability of Transcriptomic Results to Protein Expression and Theranostics
4. Discussion
4.1. CXCR4 as Immune-Related Biomarker in Solid Tumors
4.2. FAP as Potential Biomarker for Anti-Angiogenic Therapy Stratification
4.3. Limitations and Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Solnes, L.B.; Werner, R.A.; Jones, K.M.; Sadaghiani, M.S.; Bailey, C.R.; Lapa, C.; Pomper, M.G.; Rowe, S.P. Theranostics: Leveraging Molecular Imaging and Therapy to Impact Patient Management and Secure the Future of Nuclear Medicine. J. Nucl. Med. 2020, 61, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Sartor, O.; de Bono, J.; Chi, K.N.; Fizazi, K.; Herrmann, K.; Rahbar, K.; Tagawa, S.T.; Nordquist, L.T.; Vaishampayan, N.; El-Haddad, G.; et al. Lutetium-177–PSMA-617 for Metastatic Castration-Resistant Prostate Cancer. N. Engl. J. Med. 2021, 385, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Strosberg, J.; El-Haddad, G.; Wolin, E.; Hendifar, A.; Yao, J.; Chasen, B.; Mittra, E.; Kunz, P.L.; Kulke, M.H.; Jacene, H.; et al. Phase 3 Trial of 177Lu-Dotatate for Midgut Neuroendocrine Tumors. N. Engl. J. Med. 2017, 376, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Kircher, M.; Herhaus, P.; Schottelius, M.; Buck, A.K.; Werner, R.A.; Wester, H.-J.; Keller, U.; Lapa, C. CXCR4-Directed Theranostics in Oncology and Inflammation. Ann. Nucl. Med. 2018, 32, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Kesch, C.; Yirga, L.; Dendl, K.; Handke, A.; Darr, C.; Krafft, U.; Radtke, J.P.; Tschirdewahn, S.; Szarvas, T.; Fazli, L.; et al. High Fibroblast-Activation-Protein Expression in Castration-Resistant Prostate Cancer Supports the Use of FAPI-Molecular Theranostics. Eur. J. Nucl. Med. Mol. Imaging 2021, 49, 385–389. [Google Scholar] [CrossRef]
- Chatterjee, S.; Behnam Azad, B.; Nimmagadda, S. The Intricate Role of CXCR4 in Cancer. Adv. Cancer Res. 2014, 124, 31–82. [Google Scholar] [CrossRef]
- Werner, R.A.; Kircher, S.; Higuchi, T.; Kircher, M.; Schirbel, A.; Wester, H.-J.; Buck, A.K.; Pomper, M.G.; Rowe, S.P.; Lapa, C. CXCR4-Directed Imaging in Solid Tumors. Front. Oncol. 2019, 9, 770. [Google Scholar] [CrossRef]
- Kalluri, R. The Biology and Function of Fibroblasts in Cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef]
- Fitzgerald, A.A.; Weiner, L.M. The Role of Fibroblast Activation Protein in Health and Malignancy. Cancer Metastasis Rev. 2020, 39, 783–803. [Google Scholar] [CrossRef]
- Mona, C.; Benz, M.R.; Hikmat, F.; Grogan, T.; Lueckerath, K.; Razmaria, A.A.; Riahi, R.; Slavik, R.; Grigis, M.D.; Carlucci, G.; et al. Validation of FAPi PET Biodistribution by Immunohistochemistry in Patients with Solid Cancers: A Prospective Exploratory Study. JCO 2021, 39, 3066. [Google Scholar] [CrossRef]
- Lapa, C.; Hänscheid, H.; Kircher, M.; Schirbel, A.; Wunderlich, G.; Werner, R.A.; Samnick, S.; Kotzerke, J.; Einsele, H.; Buck, A.K.; et al. Feasibility of CXCR4-Directed Radioligand Therapy in Advanced Diffuse Large B-Cell Lymphoma. J. Nucl. Med. 2019, 60, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Giesel, F.L.; Kratochwil, C.; Lindner, T.; Marschalek, M.M.; Loktev, A.; Lehnert, W.; Debus, J.; Jäger, D.; Flechsig, P.; Altmann, A.; et al. 68Ga-FAPI PET/CT: Biodistribution and Preliminary Dosimetry Estimate of 2 DOTA-Containing FAP-Targeting Agents in Patients with Various Cancers. J. Nucl. Med. 2019, 60, 386–392. [Google Scholar] [CrossRef] [PubMed]
- Kratochwil, C.; Flechsig, P.; Lindner, T.; Abderrahim, L.; Altmann, A.; Mier, W.; Adeberg, S.; Rathke, H.; Röhrich, M.; Winter, H.; et al. 68Ga-FAPI PET/CT: Tracer Uptake in 28 Different Kinds of Cancer. J. Nucl. Med. 2019, 60, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data: Figure 1. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Tekpli, X.; Lien, T.; Røssevold, A.H.; Nebdal, D.; Borgen, E.; Ohnstad, H.O.; Kyte, J.A.; Vallon-Christersson, J.; Fongaard, M.; Due, E.U.; et al. An Independent Poor-Prognosis Subtype of Breast Cancer Defined by a Distinct Tumor Immune Microenvironment. Nat. Commun. 2019, 10, 5499. [Google Scholar] [CrossRef]
- Abida, W.; Cyrta, J.; Heller, G.; Prandi, D.; Armenia, J.; Coleman, I.; Cieslik, M.; Benelli, M.; Robinson, D.; Van Allen, E.M.; et al. Genomic Correlates of Clinical Outcome in Advanced Prostate Cancer. Proc. Natl. Acad. Sci. USA 2019, 116, 11428–11436. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef]
- Li, J.; Lu, Y.; Akbani, R.; Ju, Z.; Roebuck, P.L.; Liu, W.; Yang, J.-Y.; Broom, B.M.; Verhaak, R.G.W.; Kane, D.W.; et al. TCPA: A Resource for Cancer Functional Proteomics Data. Nat. Methods 2013, 10, 1046–1047. [Google Scholar] [CrossRef]
- Li, J.; Akbani, R.; Zhao, W.; Lu, Y.; Weinstein, J.N.; Mills, G.B.; Liang, H. Explore, Visualize, and Analyze Functional Cancer Proteomic Data Using the Cancer Proteome Atlas. Cancer Res. 2017, 77, e51–e54. [Google Scholar] [CrossRef] [PubMed]
- SciPy 1. 0 Contributors; Virtanen, P.; Gommers, R.; Oliphant, T.E.; Haberland, M.; Reddy, T.; Cournapeau, D.; Burovski, E.; Peterson, P.; Weckesser, W.; Bright, J. et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 2020, 17, 261–272. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Müller, A.; Nothman, J.; Louppe, G.; et al. Scikit-Learn: Machine Learning in Python. arXiv 2018, arXiv:1201.0490. [Google Scholar]
- Marquardt, A.; Solimando, A.G.; Kerscher, A.; Bittrich, M.; Kalogirou, C.; Kübler, H.; Rosenwald, A.; Bargou, R.; Kollmannsberger, P.; Schilling, B.; et al. Subgroup-Independent Mapping of Renal Cell Carcinoma—Machine Learning Reveals Prognostic Mitochondrial Gene Signature Beyond Histopathologic Boundaries. Front. Oncol. 2021, 11, 621278. [Google Scholar] [CrossRef] [PubMed]
- Boughorbel, S.; Jarray, F.; El-Anbari, M. Optimal Classifier for Imbalanced Data Using Matthews Correlation Coefficient Metric. PLoS ONE 2017, 12, e0177678. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein–Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.-F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1alpha-Responsive Genes Involved in Oxidative Phosphorylation Are Coordinately Downregulated in Human Diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Severson, E.; Pignon, J.-C.; Zhao, H.; Li, T.; Novak, J.; Jiang, P.; Shen, H.; Aster, J.C.; Rodig, S.; et al. Comprehensive Analyses of Tumor Immunity: Implications for Cancer Immunotherapy. Genome Biol. 2016, 17, 174. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for Analysis of Tumor-Infiltrating Immune Cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef] [PubMed]
- Tsitsiou, E.; Lindsay, M.A. MicroRNAs and the Immune Response. Curr. Opin. Pharmacol. 2009, 9, 514–520. [Google Scholar] [CrossRef]
- Eichmüller, S.B.; Osen, W.; Mandelboim, O.; Seliger, B. Immune Modulatory MicroRNAs Involved in Tumor Attack and Tumor Immune Escape. JNCI J. Natl. Cancer Inst. 2017, 109, djx034. [Google Scholar] [CrossRef]
- Wang, S.; Olson, E.N. AngiomiRs--Key Regulators of Angiogenesis. Curr. Opin. Genet. Dev. 2009, 19, 205–211. [Google Scholar] [CrossRef]
- Salinas-Vera, Y.M.; Marchat, L.A.; Gallardo-Rincón, D.; Ruiz-García, E.; Astudillo-De La Vega, H.; Echavarría-Zepeda, R.; López-Camarillo, C. AngiomiRs: MicroRNAs Driving Angiogenesis in Cancer (Review). Int. J. Mol. Med. 2019, 43, 657–670. [Google Scholar] [CrossRef]
- Yoo, M.; Shin, J.; Kim, J.; Ryall, K.A.; Lee, K.; Lee, S.; Jeon, M.; Kang, J.; Tan, A.C. DSigDB: Drug Signatures Database for Gene Set Analysis. Bioinformatics 2015, 31, 3069–3071. [Google Scholar] [CrossRef]
- Bezman, N.A.; Chakraborty, T.; Bender, T.; Lanier, L.L. MiR-150 Regulates the Development of NK and INKT Cells. J. Exp. Med. 2011, 208, 2717–2731. [Google Scholar] [CrossRef]
- Sang, W.; Wang, Y.; Zhang, C.; Zhang, D.; Sun, C.; Niu, M.; Zhang, Z.; Wei, X.; Pan, B.; Chen, W.; et al. MiR-150 Impairs Inflammatory Cytokine Production by Targeting ARRB-2 after Blocking CD28/B7 Costimulatory Pathway. Immunol. Lett. 2016, 172, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Han, F.; Chen, G.; Guo, Y.; Li, B.; Sun, Y.; Qi, X.; Tian, H.; Zhao, X.; Zhang, H. MicroRNA-4491 Enhances Cell Proliferation and Inhibits Cell Apoptosis in Non-small Cell Lung Cancer via Targeting TRIM7. Oncol. Lett. 2021, 22, 591. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, H.-W.; Lu, M.-H.; He, X.-H.; Li, Y.; Gu, H.; Liu, M.-F.; Wang, E.-D. MicroRNA-155 Functions as an OncomiR in Breast Cancer by Targeting the Suppressor of Cytokine Signaling 1 Gene. Cancer Res. 2010, 70, 3119–3127. [Google Scholar] [CrossRef]
- Cardoso, A.L.; Guedes, J.R.; Pereira de Almeida, L.; Pedroso de Lima, M.C. MiR-155 Modulates Microglia-Mediated Immune Response by down-Regulating SOCS-1 and Promoting Cytokine and Nitric Oxide Production: MiR-155 Role during Microglia Activation. Immunology 2012, 135, 73–88. [Google Scholar] [CrossRef]
- Jia, L.; Xi, Q.; Wang, H.; Zhang, Z.; Liu, H.; Cheng, Y.; Guo, X.; Zhang, J.; Zhang, Q.; Zhang, L.; et al. MiR-142-5p Regulates Tumor Cell PD-L1 Expression and Enhances Anti-Tumor Immunity. Biochem. Biophys. Res. Commun. 2017, 488, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Noman, M.Z.; Buart, S.; Romero, P.; Ketari, S.; Janji, B.; Mari, B.; Mami-Chouaib, F.; Chouaib, S. Hypoxia-Inducible MiR-210 Regulates the Susceptibility of Tumor Cells to Lysis by Cytotoxic T Cells. Cancer Res. 2012, 72, 4629–4641. [Google Scholar] [CrossRef] [PubMed]
- Nygren, M.K.; Tekle, C.; Ingebrigtsen, V.A.; Mäkelä, R.; Krohn, M.; Aure, M.R.; Nunes-Xavier, C.E.; Perälä, M.; Tramm, T.; Alsner, J.; et al. Identifying MicroRNAs Regulating B7-H3 in Breast Cancer: The Clinical Impact of MicroRNA-29c. Br. J. Cancer 2014, 110, 2072–2080. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-M.; Lian, G.-Y.; Song, Y.; Huang, Y.-F.; Gong, Y. LncRNA MALAT1 Promotes Tumorigenesis and Immune Escape of Diffuse Large B Cell Lymphoma by Sponging MiR-195. Life Sci. 2019, 231, 116335. [Google Scholar] [CrossRef]
- Yang, L.; Cai, Y.; Zhang, D.; Sun, J.; Xu, C.; Zhao, W.; Jiang, W.; Pan, C. MiR-195/MiR-497 Regulate CD274 Expression of Immune Regulatory Ligands in Triple-Negative Breast Cancer. J. Breast Cancer 2018, 21, 371. [Google Scholar] [CrossRef]
- He, B.; Yan, F.; Wu, C. Overexpressed MiR-195 Attenuated Immune Escape of Diffuse Large B-Cell Lymphoma by Targeting PD-L1. Biomed. Pharmacother. 2018, 98, 95–101. [Google Scholar] [CrossRef]
- Saba, R.; Sorensen, D.L.; Booth, S.A. MicroRNA-146a: A Dominant, Negative Regulator of the Innate Immune Response. Front. Immunol. 2014, 5, 578. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Wang, S.; Liu, X.; He, T.; Lin, X.; Wu, S.; Wang, D.; Li, J.; Huang, W.; Yang, H. MiR-1307-5p Targeting TRAF3 Upregulates the MAPK/NF-ΚB Pathway and Promotes Lung Adenocarcinoma Proliferation. Cancer Cell Int. 2020, 20, 502. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Hong, L.; Sun, L.; Sang, H.; Qian, A.; Li, W.; Zhuang, H.; Liang, H.; Song, D.; Li, C.; et al. MiR-21 Induces Endothelial Progenitor Cells Proliferation and Angiogenesis via Targeting FASLG and Is a Potential Prognostic Marker in Deep Venous Thrombosis. J. Transl. Med. 2019, 17, 270. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Ye, A.; Ye, W.; Liao, X.; Qin, G.; Xu, Y.; Yin, Y.; Luo, H.; Yi, M.; Xian, L.; et al. Cancer-Secreted Exosomal MiR-21-5p Induces Angiogenesis and Vascular Permeability by Targeting KRIT1. Cell Death. Dis. 2021, 12, 576. [Google Scholar] [CrossRef] [PubMed]
- van Mil, A.; Grundmann, S.; Goumans, M.-J.; Lei, Z.; Oerlemans, M.I.; Jaksani, S.; Doevendans, P.A.; Sluijter, J.P.G. MicroRNA-214 Inhibits Angiogenesis by Targeting Quaking and Reducing Angiogenic Growth Factor Release. Cardiovasc. Res. 2012, 93, 655–665. [Google Scholar] [CrossRef]
- Jin, Y.; Yang, C.-J.; Xu, X.; Cao, J.-N.; Feng, Q.-T.; Yang, J. MiR-214 Regulates the Pathogenesis of Patients with Coronary Artery Disease by Targeting VEGF. Mol. Cell. Biochem. 2015, 402, 111–122. [Google Scholar] [CrossRef]
- Li, Q.; He, Q.; Baral, S.; Mao, L.; Li, Y.; Jin, H.; Chen, S.; An, T.; Xia, Y.; Hu, B. MicroRNA-493 Regulates Angiogenesis in a Rat Model of Ischemic Stroke by Targeting MIF. FEBS J. 2016, 283, 1720–1733. [Google Scholar] [CrossRef]
- Zhao, F.; Xing, Y.; Jiang, P.; Hu, L.; Deng, S. LncRNA MEG3 Inhibits the Proliferation of Neural Stem Cells after Ischemic Stroke via the MiR-493–5P/MIF Axis. Biochem. Biophys. Res. Commun. 2021, 568, 186–192. [Google Scholar] [CrossRef]
- Zhai, Z.; Chen, W.; Hu, Q.; Wang, X.; Zhao, Q.; Tuerxunyiming, M. High Glucose Inhibits Osteogenic Differentiation of Bone Marrow Mesenchymal Stem Cells via Regulating MiR-493-5p/ZEB2 Signalling. J. Biochem. 2020, 167, 613–621. [Google Scholar] [CrossRef]
- Deng, F.; Zhou, R.; Lin, C.; Yang, S.; Wang, H.; Li, W.; Zheng, K.; Lin, W.; Li, X.; Yao, X.; et al. Tumor-Secreted Dickkopf2 Accelerates Aerobic Glycolysis and Promotes Angiogenesis in Colorectal Cancer. Theranostics 2019, 9, 1001–1014. [Google Scholar] [CrossRef]
- Hu, J.; Cheng, Y.; Li, Y.; Jin, Z.; Pan, Y.; Liu, G.; Fu, S.; Zhang, Y.; Feng, K.; Feng, Y. MicroRNA-128 Plays a Critical Role in Human Non-Small Cell Lung Cancer Tumourigenesis, Angiogenesis and Lymphangiogenesis by Directly Targeting Vascular Endothelial Growth Factor-C. Eur. J. Cancer 2014, 50, 2336–2350. [Google Scholar] [CrossRef] [PubMed]
- Tao, T.; Li, G.; Dong, Q.; Liu, D.; Liu, C.; Han, D.; Huang, Y.; Chen, S.; Xu, B.; Chen, M. Loss of SNAIL Inhibits Cellular Growth and Metabolism through the MiR-128-Mediated RPS6KB1/HIF-1α/PKM2 Signaling Pathway in Prostate Cancer Cells. Tumor Biol. 2014, 35, 8543–8550. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Dasgupta, D.; Ghosh, A.; Roychoudhury, S.; Kumar, D.; Gorain, M.; Butti, R.; Datta, S.; Agarwal, S.; Gupta, S.; et al. MiRNA199a-3p Suppresses Tumor Growth, Migration, Invasion and Angiogenesis in Hepatocellular Carcinoma by Targeting VEGFA, VEGFR1, VEGFR2, HGF and MMP2. Cell Death. Dis. 2017, 8, e2706. [Google Scholar] [CrossRef]
- Pencheva, N.; Tran, H.; Buss, C.; Huh, D.; Drobnjak, M.; Busam, K.; Tavazoie, S.F. Convergent Multi-MiRNA Targeting of ApoE Drives LRP1/LRP8-Dependent Melanoma Metastasis and Angiogenesis. Cell 2012, 151, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Hsu, C.-Y.; Hsieh, T.-H.; Tsai, C.-F.; Tsai, H.-P.; Chen, H.-S.; Chang, Y.; Chuang, H.-Y.; Lee, J.-N.; Hsu, Y.-L.; Tsai, E.-M. MiRNA-199a-5p Regulates VEGFA in Endometrial Mesenchymal Stem Cells and Contributes to the Pathogenesis of Endometriosis: MiRNA99a-5p in Endometriosis. J. Pathol. 2014, 232, 330–343. [Google Scholar] [CrossRef]
- Gao, P.; Wang, D.; Liu, M.; Chen, S.; Yang, Z.; Zhang, J.; Wang, H.; Niu, Y.; Wang, W.; Yang, J.; et al. DNA Methylation-Mediated Repression of Exosomal MiR-652-5p Expression Promotes Oesophageal Squamous Cell Carcinoma Aggressiveness by Targeting PARG and VEGF Pathways. PLoS Genet. 2020, 16, e1008592. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Kang, C.W.; Hun Oh, J.; Kim, J.; Park, J.-P.; Moon, J.H.; Kim, E.H.; Lee, S.; Kim, S.H.; Ku, C.R.; et al. Downregulation of MiR-216a-5p and MiR-652-3p Is Associated with Growth and Invasion by Targeting JAK2 and PRRX1 in GH-Producing Pituitary Tumours. J. Mol. Endocrinol. 2022, 68, 51–62. [Google Scholar] [CrossRef]
- Li, Y.-Z.; Wen, L.; Wei, X.; Wang, Q.-R.; Xu, L.-W.; Zhang, H.-M.; Liu, W.-C. Inhibition of MiR-7 Promotes Angiogenesis in Human Umbilical Vein Endothelial Cells by Upregulating VEGF via KLF4. Oncol. Rep. 2016, 36, 1569–1575. [Google Scholar] [CrossRef]
- Lin, J.; Liu, Z.; Liao, S.; Li, E.; Wu, X.; Zeng, W. Elevated MicroRNA-7 Inhibits Proliferation and Tumor Angiogenesis and Promotes Apoptosis of Gastric Cancer Cells via Repression of Raf-1. Cell Cycle 2020, 19, 2496–2508. [Google Scholar] [CrossRef]
- Puram, S.V.; Tirosh, I.; Parikh, A.S.; Patel, A.P.; Yizhak, K.; Gillespie, S.; Rodman, C.; Luo, C.L.; Mroz, E.A.; Emerick, K.S.; et al. Single-Cell Transcriptomic Analysis of Primary and Metastatic Tumor Ecosystems in Head and Neck Cancer. Cell 2017, 171, 1611–1624.e24. [Google Scholar] [CrossRef]
- Akashi, T.; Koizumi, K.; Tsuneyama, K.; Saiki, I.; Takano, Y.; Fuse, H. Chemokine Receptor CXCR4 Expression and Prognosis in Patients with Metastatic Prostate Cancer. Cancer Sci. 2008, 99, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Shen, H.; Liu, L.; Shu, Y. The Impact of Chemokine Receptor CXCR4 on Breast Cancer Prognosis: A Meta-Analysis. Cancer Epidemiol. 2013, 37, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Yang, Y.; Kwon, S.; Han, M.; Zhao, F.; Kang, H.; Dai, C.; Wang, R. The Association of CXCR4 Expression with Prognosis and Clinicopathological Indicators in Colorectal Carcinoma Patients: A Meta-Analysis. Histopathology 2014, 64, 701–712. [Google Scholar] [CrossRef]
- Lapa, C.; Herrmann, K.; Schirbel, A.; Hänscheid, H.; Lückerath, K.; Schottelius, M.; Kircher, M.; Werner, R.A.; Schreder, M.; Samnick, S.; et al. CXCR4-Directed Endoradiotherapy Induces High Response Rates in Extramedullary Relapsed Multiple Myeloma. Theranostics 2017, 7, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Duell, J.; Krummenast, F.; Schirbel, A.; Klassen, P.; Samnick, S.; Rauert-Wunderlich, H.; Rasche, L.; Buck, A.K.; Wester, H.-J.; Rosenwald, A.; et al. Improved Primary Staging of Marginal Zone Lymphoma by Addition of CXCR4-Directed PET/CT. J. Nucl. Med. 2021, 62, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Bouter, C.; Meller, B.; Sahlmann, C.O.; Staab, W.; Wester, H.J.; Kropf, S.; Meller, J. 68Ga-Pentixafor PET/CT Imaging of Chemokine Receptor CXCR4 in Chronic Infection of the Bone: First Insights. J. Nucl. Med. 2018, 59, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Derlin, T.; Gueler, F.; Bräsen, J.H.; Schmitz, J.; Hartung, D.; Herrmann, T.R.; Ross, T.L.; Wacker, F.; Wester, H.-J.; Hiss, M.; et al. Integrating MRI and Chemokine Receptor CXCR4-Targeted PET for Detection of Leukocyte Infiltration in Complicated Urinary Tract Infections After Kidney Transplantation. J. Nucl. Med. 2017, 58, 1831–1837. [Google Scholar] [CrossRef]
- Marik, J.; Junutula, J.R. Emerging Role of ImmunoPET in Receptor Targeted Cancer Therapy. Curr. Drug. Deliv. 2011, 8, 70–78. [Google Scholar] [CrossRef]
- Wei, W.; Rosenkrans, Z.T.; Liu, J.; Huang, G.; Luo, Q.-Y.; Cai, W. ImmunoPET: Concept, Design, and Applications. Chem. Rev. 2020, 120, 3787–3851. [Google Scholar] [CrossRef]
- Luker, G.D.; Yang, J.; Richmond, A.; Scala, S.; Festuccia, C.; Schottelius, M.; Wester, H.; Zimmermann, J. At the Bench: Pre-clinical Evidence for Multiple Functions of CXCR4 in Cancer. J. Leukoc. Biol. 2021, 109, 969–989. [Google Scholar] [CrossRef]
- Biasci, D.; Smoragiewicz, M.; Connell, C.M.; Wang, Z.; Gao, Y.; Thaventhiran, J.E.D.; Basu, B.; Magiera, L.; Johnson, T.I.; Bax, L.; et al. CXCR4 Inhibition in Human Pancreatic and Colorectal Cancers Induces an Integrated Immune Response. Proc. Natl. Acad. Sci. USA 2020, 117, 28960–28970. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Shen, Y.; Qian, C.; Oupicky, D.; Sun, M. Targeting Pulmonary Tumor Microenvironment with CXCR4-Inhibiting Nanocomplex to Enhance Anti–PD-L1 Immunotherapy. Sci. Adv. 2020, 6, eaaz9240. [Google Scholar] [CrossRef] [PubMed]
- Buck, A.K.; Haug, A.; Dreher, N.; Lambertini, A.; Higuchi, T.; Lapa, C.; Weich, A.; Pomper, M.G.; Wester, H.-J.; Zehnder, A.; et al. Imaging of C-X-C Motif Chemokine Receptor 4 Expression in 690 Patients with Solid or Hematologic Neoplasms Using 68Ga-PentixaFor PET. J. Nucl. Med. 2022, 63, 1687–1692. [Google Scholar] [CrossRef] [PubMed]
- Le Tourneau, C.; Hoimes, C.; Zarwan, C.; Wong, D.J.; Bauer, S.; Claus, R.; Wermke, M.; Hariharan, S.; von Heydebreck, A.; Kasturi, V.; et al. Avelumab in Patients with Previously Treated Metastatic Adrenocortical Carcinoma: Phase 1b Results from the JAVELIN Solid Tumor Trial. J. Immunother. Cancer 2018, 6, 111. [Google Scholar] [CrossRef] [PubMed]
- Habra, M.A.; Stephen, B.; Campbell, M.; Hess, K.; Tapia, C.; Xu, M.; Rodon Ahnert, J.; Jimenez, C.; Lee, J.E.; Perrier, N.D.; et al. Phase II Clinical Trial of Pembrolizumab Efficacy and Safety in Advanced Adrenocortical Carcinoma. J. Immunother. Cancer 2019, 7, 253. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, B.A.; Konda, B.; Costa, R.B.; Costa, R.L.B.; Sagar, V.; Gursel, D.B.; Kirschner, L.S.; Chae, Y.K.; Abdulkadir, S.A.; Rademaker, A.; et al. Nivolumab in Metastatic Adrenocortical Carcinoma: Results of a Phase 2 Trial. J. Clin. Endocrinol. Metab. 2019, 104, 6193–6200. [Google Scholar] [CrossRef]
- Altieri, B.; Ronchi, C.L.; Kroiss, M.; Fassnacht, M. Next-Generation Therapies for Adrenocortical Carcinoma. Best Pract. Res. Clin. Endocrinol. Metab. 2020, 34, 101434. [Google Scholar] [CrossRef]
- Raj, N.; Zheng, Y.; Kelly, V.; Katz, S.S.; Chou, J.; Do, R.K.G.; Capanu, M.; Zamarin, D.; Saltz, L.B.; Ariyan, C.E.; et al. PD-1 Blockade in Advanced Adrenocortical Carcinoma. J Clin Oncol 2020, 38, 71–80. [Google Scholar] [CrossRef]
- Mhawech-Fauceglia, P.; Yan, L.; Sharifian, M.; Ren, X.; Liu, S.; Kim, G.; Gayther, S.A.; Pejovic, T.; Lawrenson, K. Stromal Expression of Fibroblast Activation Protein Alpha (FAP) Predicts Platinum Resistance and Shorter Recurrence in Patients with Epithelial Ovarian Cancer. Cancer Microenviron. 2015, 8, 23–31. [Google Scholar] [CrossRef]
- Moreno-Ruiz, P.; Corvigno, S.; te Grootenhuis, N.C.; La Fleur, L.; Backman, M.; Strell, C.; Mezheyeuski, A.; Hoelzlwimmer, G.; Klein, C.; Botling, J.; et al. Stromal FAP Is an Independent Poor Prognosis Marker in Non-Small Cell Lung Adenocarcinoma and Associated with P53 Mutation. Lung. Cancer 2021, 155, 10–19. [Google Scholar] [CrossRef]
- Wikberg, M.L.; Edin, S.; Lundberg, I.V.; Van Guelpen, B.; Dahlin, A.M.; Rutegård, J.; Stenling, R.; Öberg, Å.; Palmqvist, R. High Intratumoral Expression of Fibroblast Activation Protein (FAP) in Colon Cancer Is Associated with Poorer Patient Prognosis. Tumor Biol. 2013, 34, 1013–1020. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Park, H.J.; Seol, J.W.; Jang, J.Y.; Cho, Y.; Kim, K.R.; Choi, Y.; Lydon, J.P.; DeMayo, F.J.; Shibuya, M.; et al. VEGF-A Regulated by Progesterone Governs Uterine Angiogenesis and Vascular Remodelling during Pregnancy. EMBO Mol. Med. 2013, 5, 1415–1430. [Google Scholar] [CrossRef] [PubMed]
- Losordo, D.W.; Isner, J.M. Estrogen and Angiogenesis: A Review. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Coto-Llerena, M.; Ercan, C.; Kancherla, V.; Taha-Mehlitz, S.; Eppenberger-Castori, S.; Soysal, S.D.; Ng, C.K.Y.; Bolli, M.; von Flüe, M.; Nicolas, G.P.; et al. High Expression of FAP in Colorectal Cancer Is Associated With Angiogenesis and Immunoregulation Processes. Front. Oncol. 2020, 10, 979. [Google Scholar] [CrossRef]
- Solimando, A.G.; Kalogirou, C.; Krebs, M. Angiogenesis as Therapeutic Target in Metastatic Prostate Cancer—Narrowing the Gap Between Bench and Bedside. Front. Immunol. 2022, 13, 842038. [Google Scholar] [CrossRef]
- Mona, C.E.; Benz, M.R.; Hikmat, F.; Grogan, T.R.; Lückerath, K.; Razmaria, A.; Riahi, R.; Slavik, R.; Girgis, M.D.; Carlucci, G.; et al. Correlation of 68Ga-FAPi-46 PET Biodistribution with FAP Expression by Immunohistochemistry in Patients with Solid Cancers: A Prospective Translational Exploratory Study. J. Nucl. Med. 2021, 63, 1021–1026. [Google Scholar] [CrossRef]
- Cheng, J.; Zhang, J.; Wu, Z.; Sun, X. Inferring Microenvironmental Regulation of Gene Expression from Single-Cell RNA Sequencing Data Using ScMLnet with an Application to COVID-19. Brief. Bioinform 2021, 22, 988–1005. [Google Scholar] [CrossRef]
- Zhang, J.; Guan, M.; Wang, Q.; Zhang, J.; Zhou, T.; Sun, X. Single-Cell Transcriptome-Based Multilayer Network Biomarker for Predicting Prognosis and Therapeutic Response of Gliomas. Brief. Bioinform. 2020, 21, 1080–1097. [Google Scholar] [CrossRef]

| Rank | miR Candidate | Mean High (+/−Std High) | Mean Low (+/−Std Low) | p Value | Immune-Related Target Genes/In Vitro (Selection) | Covered in Review Articles |
|---|---|---|---|---|---|---|
| 1 | miR-150 | 2204.46 (+/−3768.76) | 734.17 (+/−1402.81) | 1.66 × 10−24 | c-Myb [40], ARBB2 [41] | [35,36] |
| 2 | miR-4491 | 1.24 (+/−2.97) | 0.50 (+/−3.06) | 1.87 × 10−15 | TRIM7 [42] | - |
| 3 | miR-155 | 992.76 (+/−1386.74) | 515.66 (+/−1180.80) | 2.17 × 10−25 | SOCS1 [43,44] | [35,36] |
| 4 | miR-5586 | 5.08 (+/−4.77) | 3.16 (+/−3.74) | 3.29 × 10−17 | - | - |
| 5 | miR-142 | 7184.13 (+/−18,169.54) | 4084.70 (+/−14,505.49) | 1.56 × 10−19 | PD-L1 [45] | [35,36] |
| 6 | miR-210 | 994.59 (+/−1418.87) | 1051.44 (+/−1527.88) | 0.166973729 | PTPN, HOXA1, TP53I11 [46] | - |
| 7 | miR-29c | 2861.48 (+/−2863.31) | 2195.79 (+/−2074.10) | 0.000214287 | B7-H3 [47] | [36] |
| 8 | miR-195 | 51.30 (+/−41.85) | 40.86 (+/−36.79) | 4.56 × 10−7 | PD-L1 [48,49,50] | - |
| 9 | miR-146a | 544.21 (+/−2503.58) | 354.89 (+/−1622.84) | 3.78 × 10−13 | IRAK1, TRAF6 [51] | [35,36] |
| 10 | miR-1307 | 1652.20 (+/−1937.24) | 1745.57 (+/−1999.00) | 0.016754954 | TRAF3 [52] | - |
| Rank | miR Candidate | Mean High (+/−Std High) | Mean Low (+/−Std Low) | p Value | Angiogenesis-Related Target Genes/In Vitro (Selection) | Covered in Review Articles |
|---|---|---|---|---|---|---|
| 1 | miR-21 | 305,318.37 (+/−139,932.32) | 226,877.88 (+/−143,668.68) | 1.81 × 10−27 | FASLG [53], KRIT1 [54] | [37,38] |
| 2 | miR-1245a | 3.78 (+/−4.81) | 1.68 (+/−3.34) | 2.39 × 10−44 | - | - |
| 3 | miR-214 | 48.98 (+/−49.57) | 28.67 (+/−63.28) | 1.89 × 10−48 | QKI [55], VEGFA [56] | - |
| 4 | miR-493 | 41.20 (+/−119.60) | 28.61 (+/−110.12) | 1.87 × 10−42 | MIF [57,58], ZEB2 [59], DKK2 [60] | - |
| 5 | miR-128-2 | 73.34 (+/−93.89) | 134.96 (+/−364.17) | 5.27 × 10−11 | VEGFC [61], RPS6KB1 [62] | [38] |
| 6 | miR-199a-1 | 1910.74 (+/−1744.58) | 1190.78 (+/−2028.65) | 1.26 × 10−36 | VEGFA, VEGFR1, VEGFR2, HGF, MMP2 [63], APOE [64] | [38] |
| 7 | miR-199a-2 | 3107.70 (+/−2798.14) | 1940.26 (+/−3134.63) | 2.47 × 10−35 | VEGFA [65], APOE [64] | [38] |
| 8 | miR-652 | 30.18 (+/−36.47) | 36.92 (+/−41.11) | 7.14 × 10−10 | VEGFA [66], PRRX1 [67] | - |
| 9 | miR-337 | 61.72 (+/−130.58) | 51.09 (+/−152.74) | 1.69 × 10−37 | - | - |
| 10 | miR-7-1 | 25.56 (+/−31.47) | 40.47 (+/−65.27) | 6.98 × 10−10 | KLF4 [68], RAF1 [69] | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marquardt, A.; Hartrampf, P.; Kollmannsberger, P.; Solimando, A.G.; Meierjohann, S.; Kübler, H.; Bargou, R.; Schilling, B.; Serfling, S.E.; Buck, A.; et al. Predicting Microenvironment in CXCR4- and FAP-Positive Solid Tumors—A Pan-Cancer Machine Learning Workflow for Theranostic Target Structures. Cancers 2023, 15, 392. https://doi.org/10.3390/cancers15020392
Marquardt A, Hartrampf P, Kollmannsberger P, Solimando AG, Meierjohann S, Kübler H, Bargou R, Schilling B, Serfling SE, Buck A, et al. Predicting Microenvironment in CXCR4- and FAP-Positive Solid Tumors—A Pan-Cancer Machine Learning Workflow for Theranostic Target Structures. Cancers. 2023; 15(2):392. https://doi.org/10.3390/cancers15020392
Chicago/Turabian StyleMarquardt, André, Philipp Hartrampf, Philip Kollmannsberger, Antonio G. Solimando, Svenja Meierjohann, Hubert Kübler, Ralf Bargou, Bastian Schilling, Sebastian E. Serfling, Andreas Buck, and et al. 2023. "Predicting Microenvironment in CXCR4- and FAP-Positive Solid Tumors—A Pan-Cancer Machine Learning Workflow for Theranostic Target Structures" Cancers 15, no. 2: 392. https://doi.org/10.3390/cancers15020392
APA StyleMarquardt, A., Hartrampf, P., Kollmannsberger, P., Solimando, A. G., Meierjohann, S., Kübler, H., Bargou, R., Schilling, B., Serfling, S. E., Buck, A., Werner, R. A., Lapa, C., & Krebs, M. (2023). Predicting Microenvironment in CXCR4- and FAP-Positive Solid Tumors—A Pan-Cancer Machine Learning Workflow for Theranostic Target Structures. Cancers, 15(2), 392. https://doi.org/10.3390/cancers15020392








