Immune Pathways with Aging Characteristics Improve Immunotherapy Benefits and Drug Prediction in Human Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Identification of Dysregulated IRPs in 25 Cancer Types
- (1)
- Identification of differentially expressed IRPs in tumor samples
- (2)
- Identification of IRPs differentially expressed with age
2.3. Benchmark Genesets to Evaluate the Aging Characteristics of IRPs
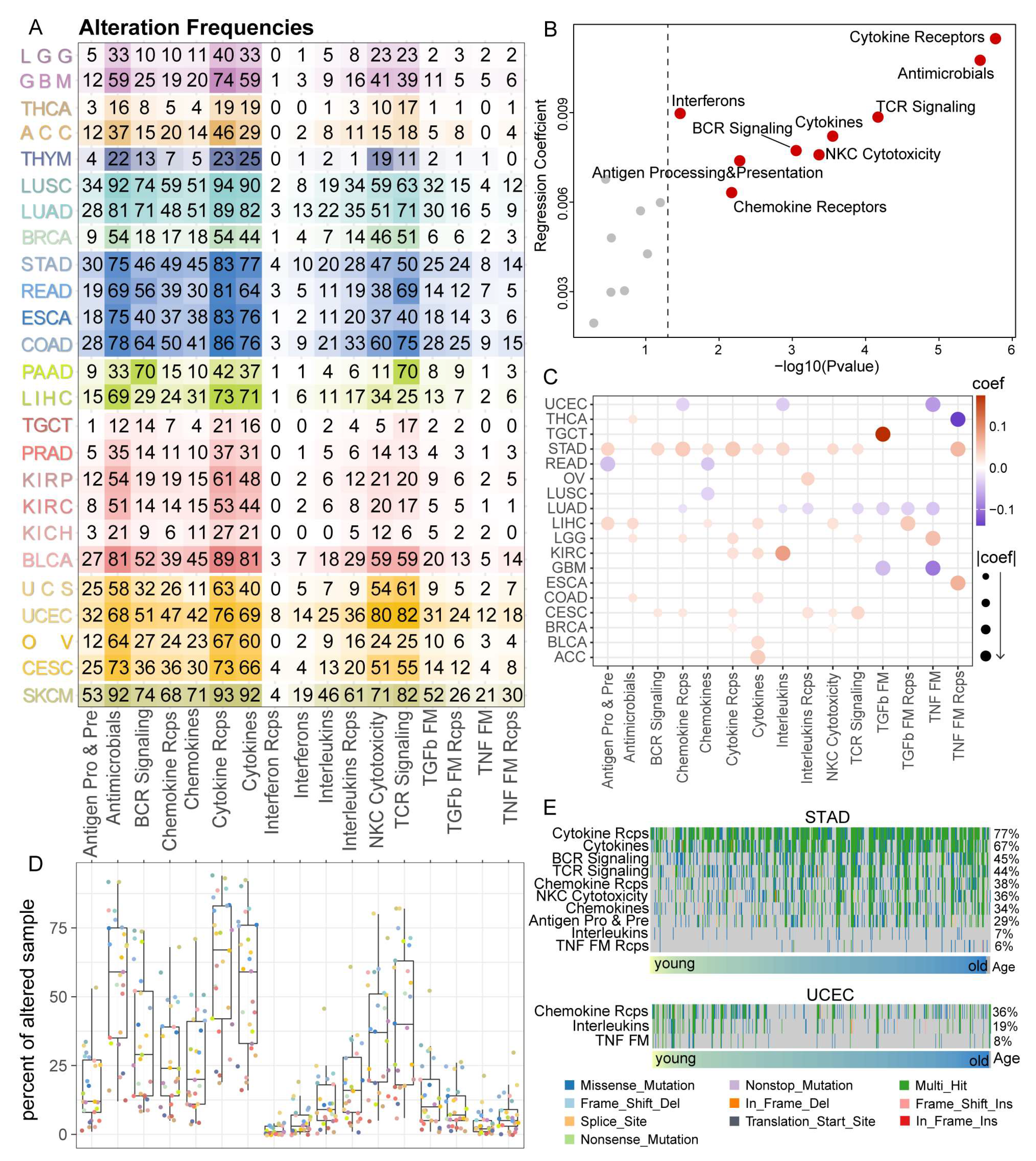
2.4. Mutation Analysis
2.5. Survival Analysis
2.6. Prediction of Immunotherapy Response of IRPs
2.7. Network-Based Method Identifying Immune-Related Genes as Drug Targets
2.8. Functional Enrichment Analysis
3. Results
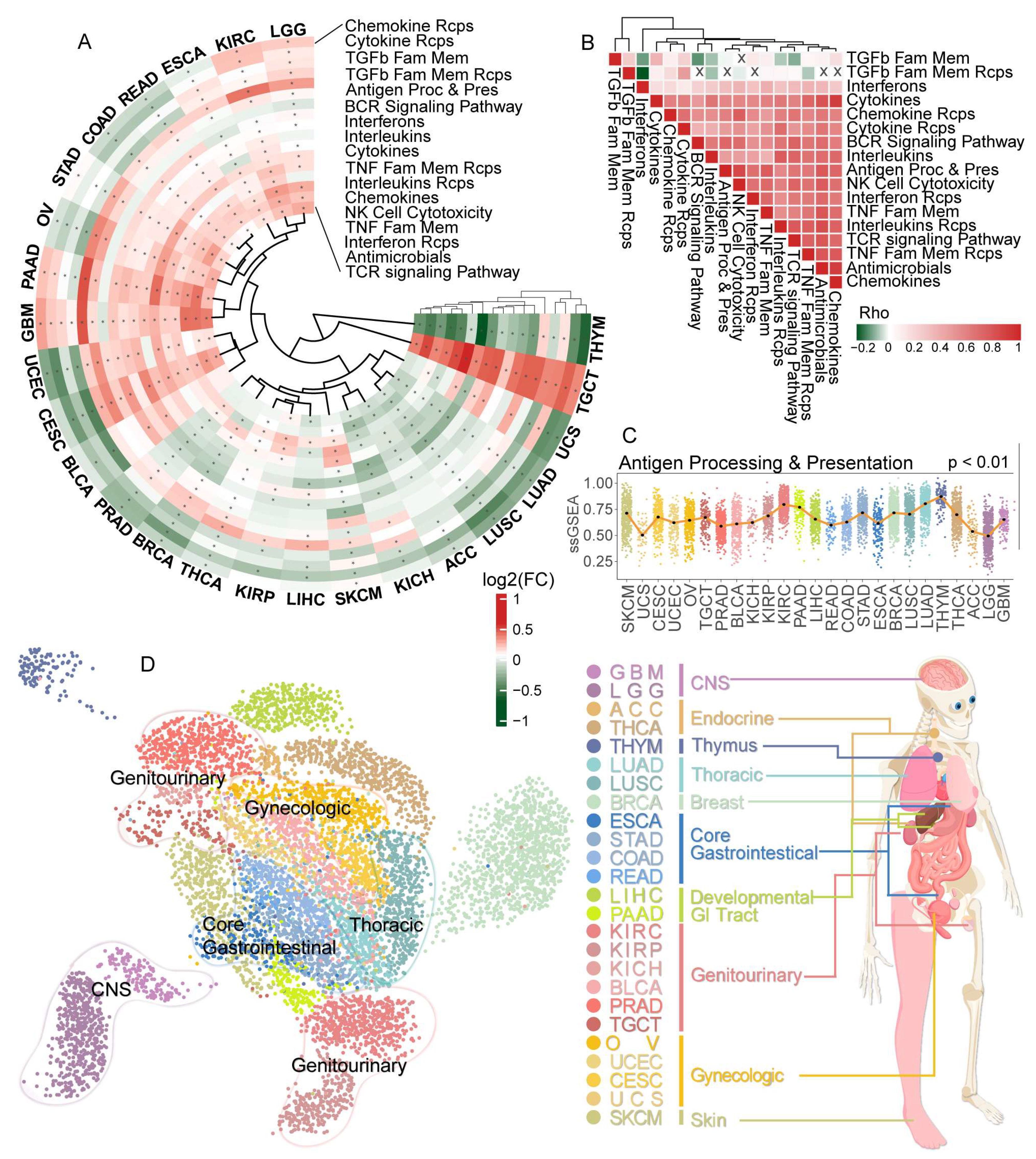
3.1. Disturbance of Immune-Related Pathways between Normal and Tumor Tissues among 25 Cancer Types
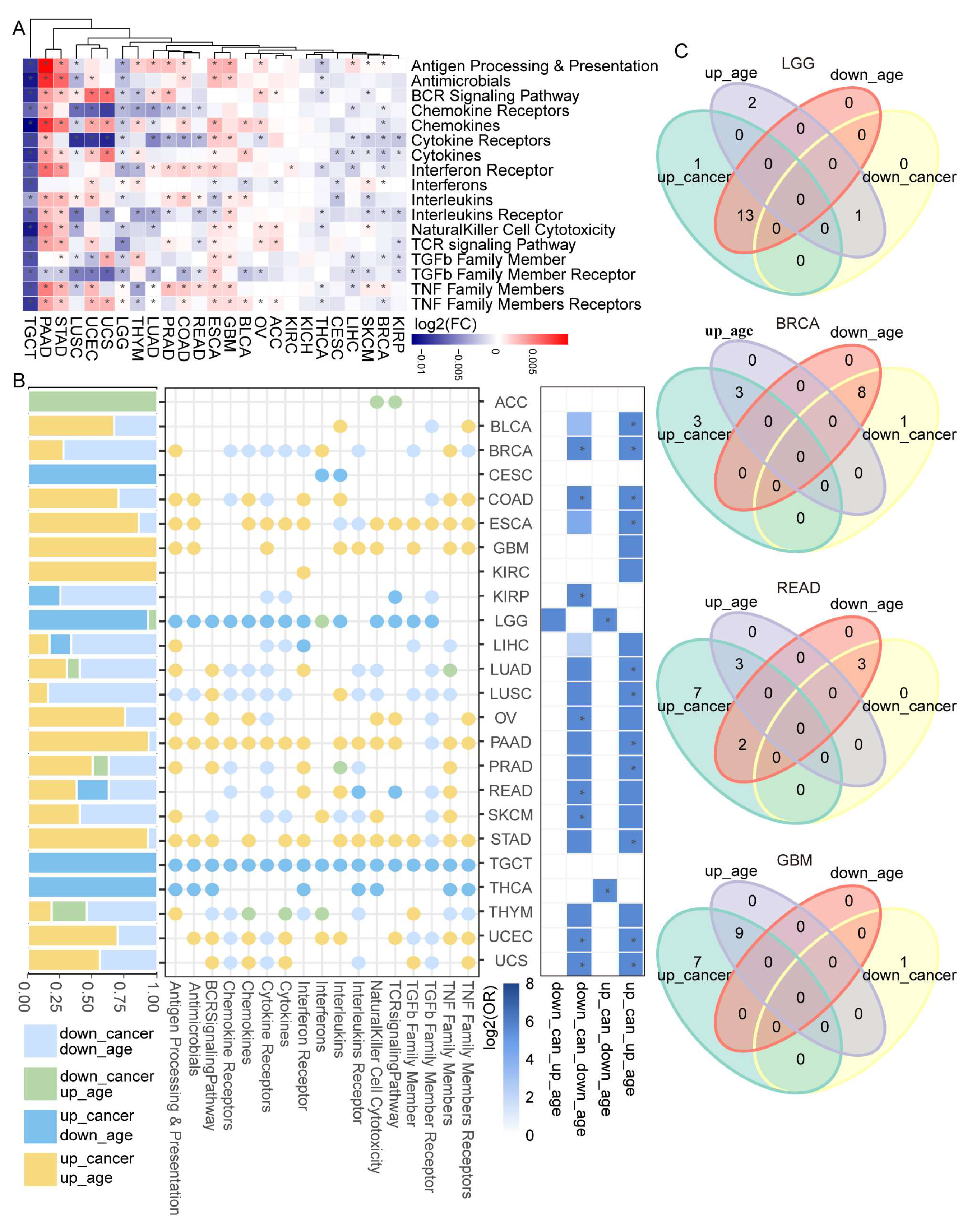
3.2. Dysregulated Immune-Related Pathways Showed Age-Associated Characteristics among Cancers
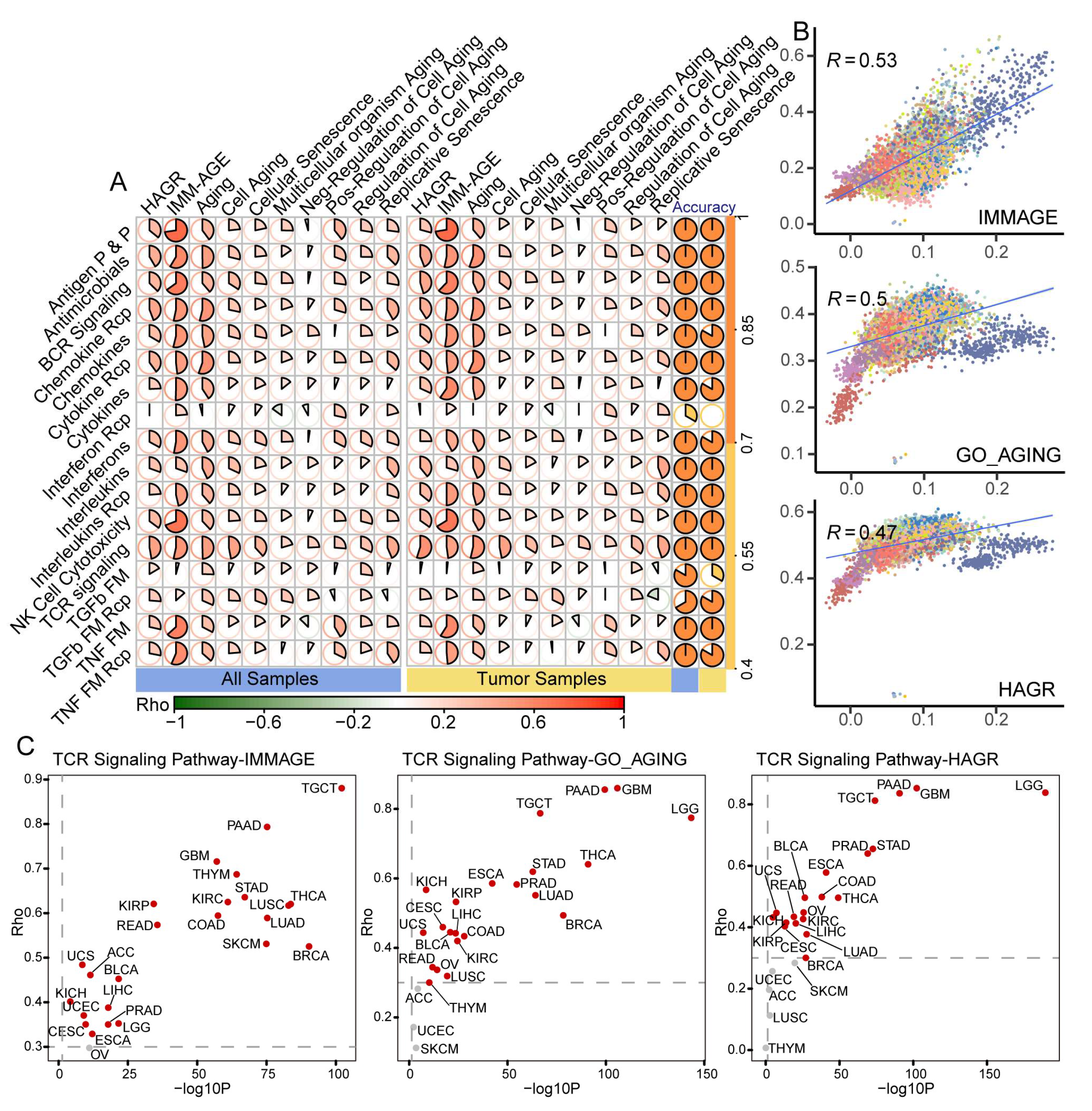
3.3. Benchmark Gene Sets Validated the Aging-Related Characteristics of IRPs
3.4. Cytokine Receptors Pathway Showed a High Alteration Frequency among 25 Cancer Types
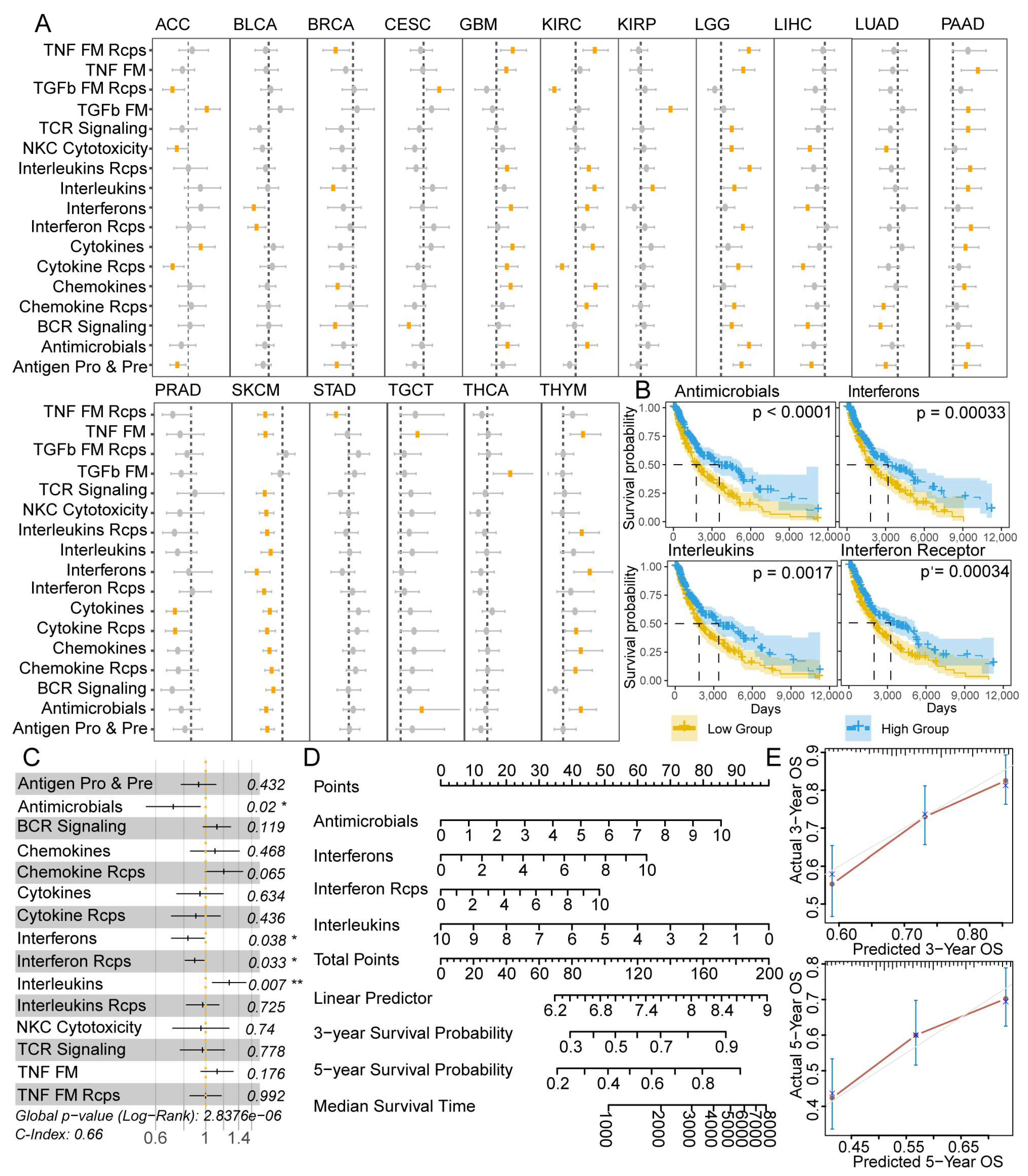
3.5. IRPs Displayed a Strong Clinical Relevance among 25 Cancer Types
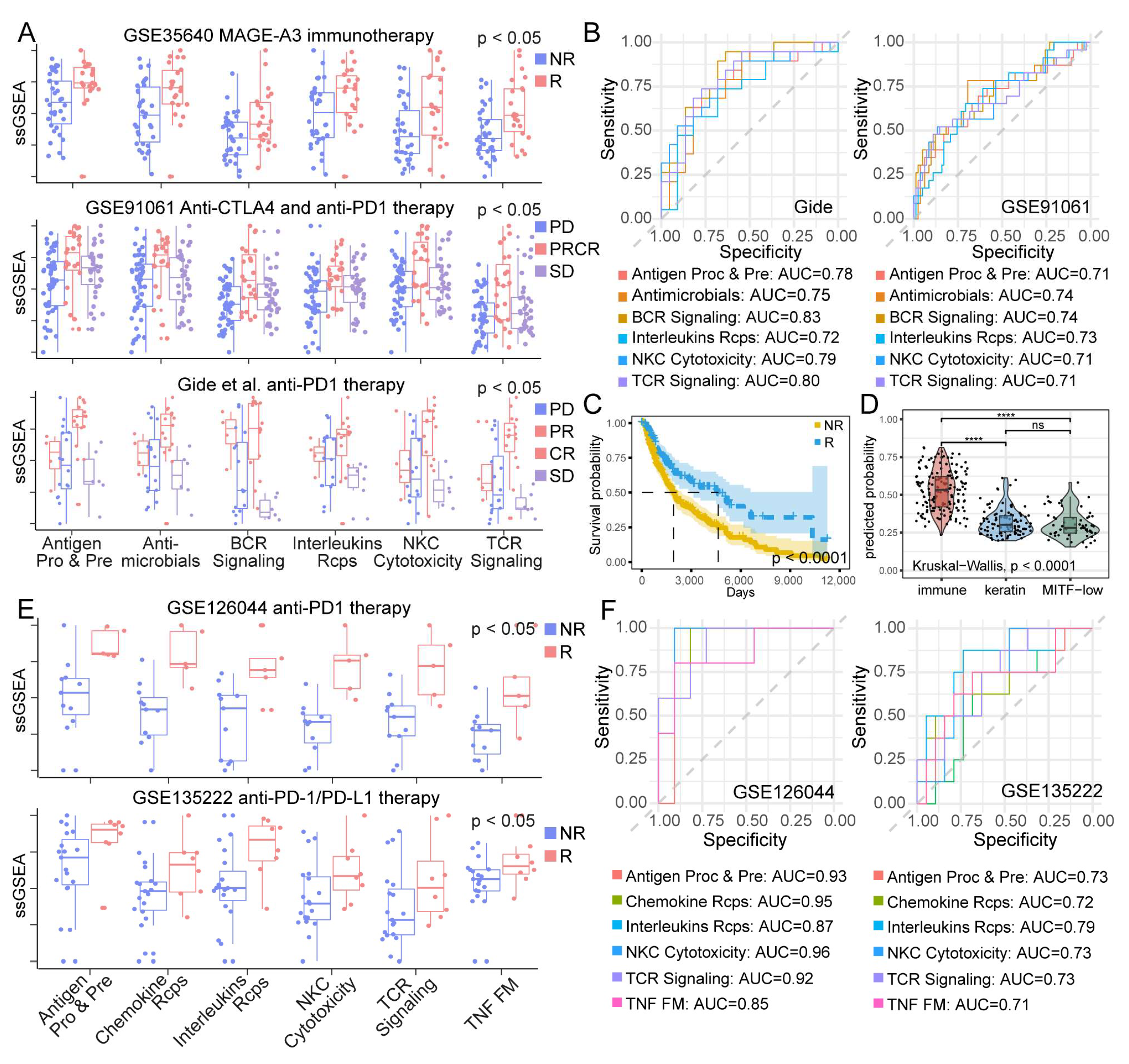
3.6. IRP Scores Were Associated with Immunotherapy Response of Patients
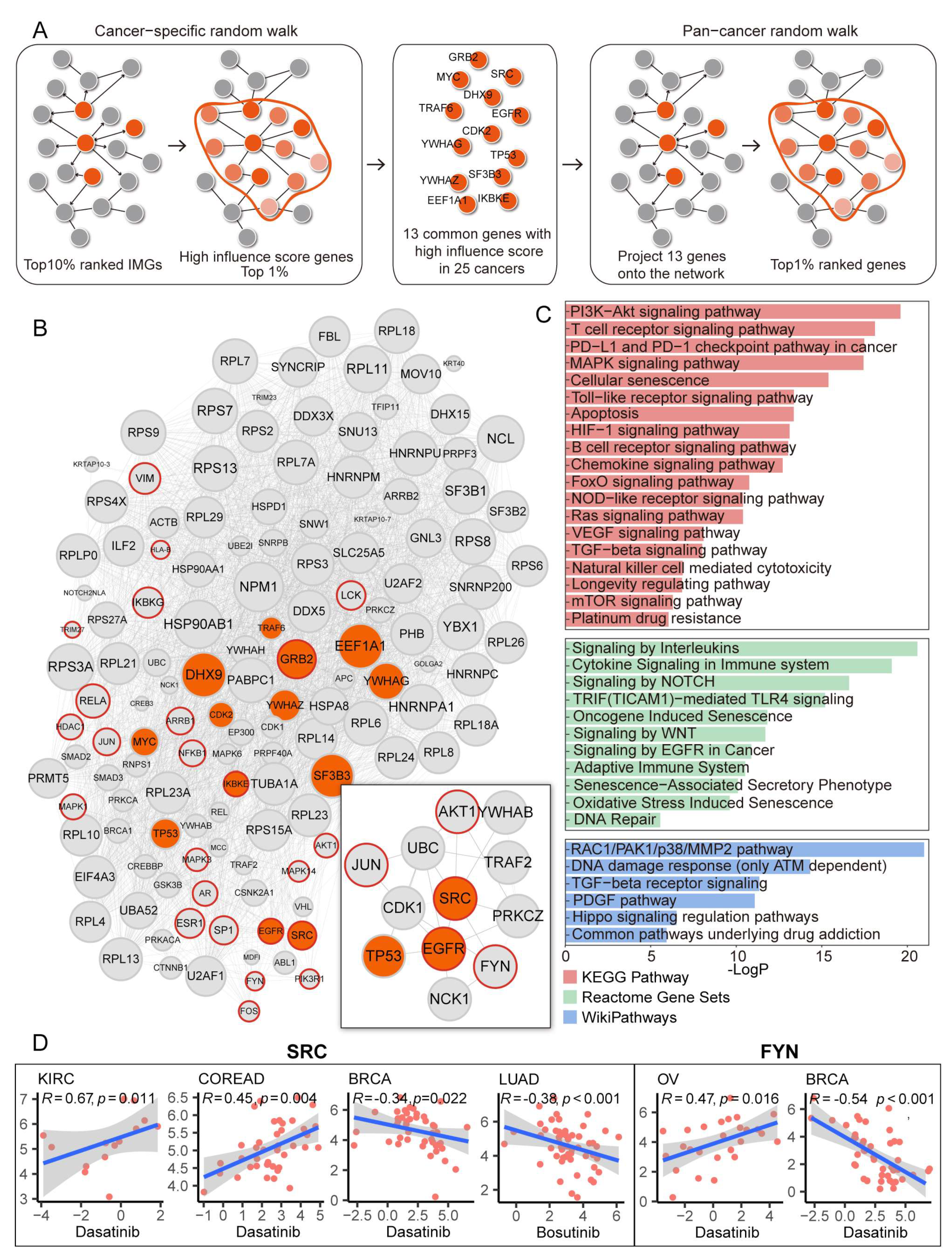
3.7. Identification of Potential Therapeutic Target Genes and Drugs Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fane, M.; Weeraratna, A.T. How the ageing microenvironment influences tumour progression. Nat. Rev. Cancer 2020, 20, 89–106. [Google Scholar] [CrossRef]
- De Magalhães, J.P. How ageing processes influence cancer. Nat. Rev. Cancer 2013, 13, 357–365. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Mancera, P.A.; Young, A.R.J.; Narita, M. Inside and out: The activities of senescence in cancer. Nat. Rev. Cancer 2014, 14, 547–558. [Google Scholar] [CrossRef] [PubMed]
- Maggiorani, D.; Beauséjour, C. Senescence and Aging: Does It Impact Cancer Immunotherapies? Cells 2021, 10, 1568. [Google Scholar] [CrossRef] [PubMed]
- Bruni, D.; Angell, H.K.; Galon, J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer 2020, 20, 662–680. [Google Scholar] [CrossRef]
- Lin, W.M.; Fisher, D.E. Signaling and Immune Regulation in Melanoma Development and Responses to Therapy. Annu. Rev. Pathol. Mech. Dis. 2017, 12, 75–102. [Google Scholar] [CrossRef]
- Pao, W.; Ooi, C.-H.; Birzele, F.; Ruefli-Brasse, A.; Cannarile, M.A.; Reis, B.; Scharf, S.H.; Schubert, D.A.; Hatje, K.; Pelletier, N.; et al. Tissue-Specific Immunoregulation: A Call for Better Understanding of the “Immunostat” in the Context of Cancer. Cancer Discov. 2018, 8, 395–402. [Google Scholar] [CrossRef]
- Kaiser, M.; Semeraro, M.; Herrmann, M.; Absenger, G.; Gerger, A.; Renner, W. Immune Aging and Immunotherapy in Cancer. Int. J. Mol. Sci. 2021, 22, 7016. [Google Scholar] [CrossRef]
- Mandrup-Poulsen, T. Metabolism and the inflammasome in health and ageing. Nat. Rev. Endocrinol. 2017, 14, 72–74. [Google Scholar] [CrossRef]
- Nikolich-Žugich, J. The twilight of immunity: Emerging concepts in aging of the immune system. Nat. Immunol. 2018, 19, 10–19. [Google Scholar] [CrossRef]
- Borgoni, S.; Kudryashova, K.S.; Burka, K.; De Magalhães, J.P. Targeting immune dysfunction in aging. Ageing Res. Rev. 2021, 70, 101410. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed]
- Hughes, P.E.; Caenepeel, S.; Wu, L.C. Targeted Therapy and Checkpoint Immunotherapy Combinations for the Treatment of Cancer. Trends Immunol. 2016, 37, 462–476. [Google Scholar] [CrossRef] [PubMed]
- Lesterhuis, W.J.; Bosco, A.; Millward, M.J.; Small, M.; Nowak, A.K.; Lake, R.A. Dynamic versus static biomarkers in cancer immune checkpoint blockade: Unravelling complexity. Nat. Rev. Drug Discov. 2017, 16, 264–272. [Google Scholar] [CrossRef]
- Leko, V.; Rosenberg, S.A. Identifying and Targeting Human Tumor Antigens for T Cell-Based Immunotherapy of Solid Tumors. Cancer Cell 2020, 38, 454–472. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hogg, G.D.; DeNardo, D.G. Rethinking immune checkpoint blockade: ‘Beyond the T cell’. J. Immunother. Cancer 2021, 9, e001460. [Google Scholar] [CrossRef]
- Jackaman, C.; Tomay, F.; Duong, L.; Razak, N.B.A.; Pixley, F.J.; Metharom, P.; Nelson, D.J. Aging and cancer: The role of macrophages and neutrophils. Ageing Res. Rev. 2017, 36, 105–116. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Sun, C.; Mezzadra, R.; Schumacher, T.N. Regulation and Function of the PD-L1 Checkpoint. Immunity 2018, 48, 434–452. [Google Scholar] [CrossRef]
- Zhao, X.; Subramanian, S. Oncogenic pathways that affect antitumor immune response and immune checkpoint blockade therapy. Pharmacol. Ther. 2018, 181, 76–84. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Vivian, J.; Rao, A.A.; Nothaft, F.A.; Ketchum, C.; Armstrong, J.; Novak, A.; Pfeil, J.; Narkizian, J.; Deran, A.D.; Musselman-Brown, A.; et al. Toil enables reproducible, open source, big biomedical data analyses. Nat. Biotechnol. 2017, 35, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2012, 41, D955–D961. [Google Scholar] [CrossRef]
- Bhattacharya, S.; Dunn, P.; Thomas, C.; Smith, B.; Schaefer, H.; Chen, J.; Hu, Z.; Zalocusky, K.A.; Shankar, R.D.; Shen-Orr, S.S.; et al. ImmPort, toward repurposing of open access immunological assay data for translational and clinical research. Sci. Data 2018, 5, 180015. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Belinda, P.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Chatsirisupachai, K.; Lesluyes, T.; Paraoan, L.; Van Loo, P.; De Magalhães, J.P. An integrative analysis of the age-associated multi-omic landscape across cancers. Nat. Commun. 2021, 12, 2345. [Google Scholar] [CrossRef] [PubMed]
- Chatsirisupachai, K.; Palmer, D.; Ferreira, S.; Magalhães, J.P. A human tissue-specific transcriptomic analysis reveals a complex relationship between aging, cancer, and cellular senescence. Aging Cell 2019, 18, e13041. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Tacutu, R.; Thornton, D.; Johnson, E.; Budovsky, A.; Barardo, D.; Craig, T.; Diana, E.; Lehmann, G.; Toren, D.; Wang, J.; et al. Human Ageing Genomic Resources: New and updated databases. Nucleic Acids Res. 2018, 46, D1083–D1090. [Google Scholar] [CrossRef]
- Alpert, A.; Pickman, Y.; Leipold, M.; Rosenberg-Hasson, Y.; Ji, X.; Gaujoux, R.; Rabani, H.; Starosvetsky, E.; Kveler, K.; Schaffert, S.; et al. A clinically meaningful metric of immune age derived from high-dimensional longitudinal monitoring. Nat. Med. 2019, 25, 487–495. [Google Scholar] [CrossRef]
- Tong, Y.; Sun, J.; Wong, C.F.; Kang, Q.; Ru, B.; Wong, C.N.; Chan, A.S.; Leung, S.Y.; Zhang, J. MICMIC: Identification of DNA methylation of distal regulatory regions with causal effects on tumorigenesis. Genome Biol. 2018, 19, 73. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Ng, P.K.-S.; Kucherlapati, M.; Chen, F.; Liu, Y.; Tsang, Y.H.; De Velasco, G.; Jeong, K.J.; Akbani, R.; Hadjipanayis, A.; et al. A Pan-Cancer Proteogenomic Atlas of PI3K/AKT/mTOR Pathway Alterations. Cancer Cell 2017, 31, 820–832.e3. [Google Scholar] [CrossRef] [PubMed]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Diaz, A.; Shin, D.S.; Moreno, B.H.; Saco, J.; Escuin-Ordinas, H.; Rodriguez, G.A.; Zaretsky, J.M.; Sun, L.; Hugo, W.; Wang, X.; et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017, 19, 1189–1201. [Google Scholar] [CrossRef]
- Gide, T.N.; Quek, C.; Menzies, A.M.; Tasker, A.T.; Shang, P.; Holst, J.; Madore, J.; Lim, S.Y.; Velickovic, R.; Wongchenko, M.; et al. Distinct Immune Cell Populations Define Response to Anti-PD-1 Monotherapy and Anti-PD-1/Anti-CTLA-4 Combined Therapy. Cancer Cell 2019, 35, 238–255.e6. [Google Scholar] [CrossRef]
- Riaz, N.; Havel, J.J.; Makarov, V.; Desrichard, A.; Urba, W.J.; Sims, J.S.; Hodi, F.S.; Martín-Algarra, S.; Mandal, R.; Sharfman, W.H.; et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell 2017, 171, 934–949.e16. [Google Scholar] [CrossRef] [PubMed]
- Ulloa-Montoya, F.; Louahed, J.; Dizier, B.; Gruselle, O.; Spiessens, B.; Lehmann, F.F.; Suciu, S.; Kruit, W.H.; Eggermont, A.M.; Vansteenkiste, J.; et al. Predictive Gene Signature in MAGE-A3 Antigen-Specific Cancer Immunotherapy. J. Clin. Oncol. 2013, 31, 2388–2395. [Google Scholar] [CrossRef]
- Van Allen, E.M.; Miao, D.; Schilling, B.; Shukla, S.A.; Blank, C.; Zimmer, L.; Sucker, A.; Hillen, U.; Foppen, M.H.G.; Goldinger, S.M.; et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 2015, 350, 207–211. [Google Scholar] [CrossRef]
- Cho, J.-W.; Hong, M.H.; Ha, S.-J.; Kim, Y.-J.; Cho, B.C.; Lee, I.; Kim, H.R. Genome-wide identification of differentially methylated promoters and enhancers associated with response to anti-PD-1 therapy in non-small cell lung cancer. Exp. Mol. Med. 2020, 52, 1550–1563. [Google Scholar] [CrossRef]
- Jung, H.; Kim, H.S.; Kim, J.Y.; Sun, J.-M.; Ahn, J.S.; Ahn, M.-J.; Park, K.; Esteller, M.; Lee, S.-H.; Choi, J.K. DNA methylation loss promotes immune evasion of tumours with high mutation and copy number load. Nat. Commun. 2019, 10, 4278. [Google Scholar] [CrossRef]
- Menche, J.; Sharma, A.; Kitsak, M.; Ghiassian, S.D.; Vidal, M.; Loscalzo, J.; Barabási, A.-L. Uncovering disease-disease relationships through the incomplete interactome. Science 2015, 347, 1257601. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Burgman, B.; McGrail, D.J.; Sun, M.; Qi, D.; Shukla, S.A.; Wu, E.; Capasso, A.; Lin, S.-Y.; Wu, C.J.; et al. Integrated Genomic Characterization of the Human Immunome in Cancer. Cancer Res. 2020, 80, 4854–4867. [Google Scholar] [CrossRef]
- Wu, C.-C.; Wang, Y.A.; Livingston, J.A.; Zhang, J.; Futreal, P.A. Prediction of biomarkers and therapeutic combinations for anti-PD-1 immunotherapy using the global gene network association. Nat. Commun. 2022, 13, 42. [Google Scholar] [CrossRef]
- Freshour, S.L.; Kiwala, S.; Cotto, K.C.; Coffman, A.C.; McMichael, J.F.; Song, J.J.; Griffith, M.; Griffith, O.L.; Wagner, A.H. Integration of the Drug–Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Res. 2021, 49, D1144–D1151. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Chan, T.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 2019, 30, 44–56. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell 2015, 161, 1681–1696. [Google Scholar] [CrossRef]
- Vanneman, M.; Dranoff, G. Combining immunotherapy and targeted therapies in cancer treatment. Nat. Rev. Cancer 2012, 12, 237–251. [Google Scholar] [CrossRef]
- Chalupny, N.J.; Aruffo, A.; Esselstyn, J.M.; Chan, P.-Y.; Bajorath, J.; Blake, J.; Gilliland, L.K.; Ledbetter, J.A.; Tepper, M.A. Specific binding of Fyn and phosphatidylinositol 3-kinase to the B cell surface glycoprotein CD19 through their src homology 2 domains. Eur. J. Immunol. 1995, 25, 2978–2984. [Google Scholar] [CrossRef]
- Zhang, X.; Huang, Z.; Guo, Y.; Xiao, T.; Tang, L.; Zhao, S.; Wu, L.; Su, J.; Zeng, W.; Huang, H.; et al. Correction to: The phosphorylation of CD147 by Fyn plays a critical role for melanoma cells growth and metastasis. Oncogene 2021, 40, 1925. [Google Scholar] [CrossRef]
- Elias, D.; Ditzel, H.J. Fyn is an important molecule in cancer pathogenesis and drug resistance. Pharmacol. Res. 2015, 100, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Kani, K.; Garri, C.; Tiemann, K.; Malihi, P.D.; Punj, V.; Nguyen, A.L.; Lee, J.; Hughes, L.D.; Alvarez, R.M.; Wood, D.M.; et al. JUN-Mediated Downregulation of EGFR Signaling Is Associated with Resistance to Gefitinib in EGFR-mutant NSCLC Cell Lines. Mol. Cancer Ther. 2017, 16, 1645–1657. [Google Scholar] [CrossRef]
- Lukey, M.J.; Greene, K.S.; Erickson, J.W.; Wilson, K.F.; Cerione, R.A. The oncogenic transcription factor c-Jun regulates glutaminase expression and sensitizes cells to glutaminase-targeted therapy. Nat. Commun. 2016, 7, 11321. [Google Scholar] [CrossRef] [PubMed]
- Blonska, M.; Zhu, Y.; Chuang, H.H.; You, M.J.; Kunkalla, K.; Vega, F.; Lin, X. Jun-regulated genes promote interaction of diffuse large B-cell lymphoma with the microenvironment. Blood 2015, 125, 981–991. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Chen, S.-Y.; Lerbs, T.; Lee, J.-W.; Domizi, P.; Gordon, S.; Kim, Y.-H.; Nolan, G.; Betancur, P.; Wernig, G. Activation of JUN in fibroblasts promotes pro-fibrotic programme and modulates protective immunity. Nat. Commun. 2020, 11, 2795. [Google Scholar] [CrossRef]
- Riesenberg, S.; Groetchen, A.; Siddaway, R.; Bald, T.; Reinhardt, J.; Smorra, D.; Kohlmeyer, J.; Renn, M.; Phung, B.; Aymans, P.; et al. MITF and c-Jun antagonism interconnects melanoma dedifferentiation with pro-inflammatory cytokine responsiveness and myeloid cell recruitment. Nat. Commun. 2015, 6, 8755. [Google Scholar] [CrossRef]
- Ye, F.; Li, J.; Xu, P.; Xie, Z.; Zheng, G.; Liu, W.; Ye, G.; Yu, W.; Lin, J.; Su, Z.; et al. Osteogenic differentiation of mesenchymal stem cells promotes c-Jun-dependent secretion of interleukin 8 and mediates the migration and differentiation of CD4+ T cells. Stem Cell Res. Ther. 2022, 13, 58. [Google Scholar] [CrossRef]
- Song, L.; Liu, Z.; Hu, H.-H.; Yang, Y.; Li, T.Y.; Lin, Z.-Z.; Ye, J.; Chen, J.; Huang, X.; Liu, D.-T.; et al. Proto-oncogene Src links lipogenesis via lipin-1 to breast cancer malignancy. Nat. Commun. 2020, 11, 5842. [Google Scholar] [CrossRef]
- Zhang, S.; Yang, Z.; Bao, W.; Liu, L.; You, Y.; Wang, X.; Shao, L.; Fu, W.; Kou, X.; Shen, W.; et al. SNX10 (sorting nexin 10) inhibits colorectal cancer initiation and progression by controlling autophagic degradation of SRC. Autophagy 2020, 16, 735–749. [Google Scholar] [CrossRef]
- Zhu, L.; Qin, C.; Li, T.; Ma, X.; Qiu, Y.; Lin, Y.; Ma, D.; Qin, Z.; Sun, C.; Shen, X.; et al. The E3 ubiquitin ligase TRIM7 suppressed hepatocellular carcinoma progression by directly targeting Src protein. Cell Death Differ. 2020, 27, 1819–1831. [Google Scholar] [CrossRef]
- Krishnan, H.; Miller, W.T.; Blanco, F.J.; Goldberg, G.S. Src and podoplanin forge a path to destruction. Drug Discov. Today 2019, 24, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Musumeci, F.; Schenone, S.; Brullo, C.; Botta, M. An update on dual Src/Abl inhibitors. Futur. Med. Chem. 2012, 4, 799–822. [Google Scholar] [CrossRef] [PubMed]
- Irizar, P.A.; Schäuble, S.; Esser, D.; Groth, M.; Frahm, C.; Priebe, S.; Baumgart, M.; Hartmann, N.; Marthandan, S.; Menzel, U.; et al. Publisher Correction: Transcriptomic alterations during ageing reflect the shift from cancer to degenerative diseases in the elderly. Nat. Commun. 2019, 10, 2459. [Google Scholar] [CrossRef]
- Gargini, R.; Segura-Collar, B.; Herránz, B.; García-Escudero, V.; Romero-Bravo, A.; Núñez, F.J.; García-Pérez, D.; Gutiérrez-Guamán, J.; Ayuso-Sacido, A.; Seoane, J.; et al. The IDH-TAU-EGFR triad defines the neovascular landscape of diffuse gliomas. Sci. Transl. Med. 2020, 12, 541326. [Google Scholar] [CrossRef] [PubMed]
- Rudd, B.D.; Venturi, V.; Li, G.; Samadder, P.; Ertelt, J.M.; Way, S.S.; Davenport, M.P.; Nikolich-Žugich, J. Nonrandom attrition of the naive CD8+ T-cell pool with aging governed by T-cell receptor:pMHC interactions. Proc. Natl. Acad. Sci. USA 2011, 108, 13694–13699. [Google Scholar] [CrossRef]
- Larbi, A.; Pawelec, G.; Wong, S.C.; Goldeck, D.; Tai, J.J.-Y.; Fulop, T. Impact of age on T cell signaling: A general defect or specific alterations? Ageing Res. Rev. 2011, 10, 370–378. [Google Scholar] [CrossRef]
- Goronzy, J.J.; Li, G.; Yu, M.; Weyand, C.M. Signaling pathways in aged T cells—A reflection of T cell differentiation, cell senescence and host environment. Semin. Immunol. 2012, 24, 365–372. [Google Scholar] [CrossRef] [PubMed]
- Laumont, C.M.; Wouters, M.C.; Smazynski, J.; Gierc, N.S.; Chavez, E.A.; Chong, L.C.; Thornton, S.; Milne, K.; Webb, J.R.; Steidl, C.; et al. Single-cell Profiles and Prognostic Impact of Tumor-Infiltrating Lymphocytes Coexpressing CD39, CD103, and PD-1 in Ovarian Cancer. Clin. Cancer Res. 2021, 27, 4089–4100. [Google Scholar] [CrossRef]
- Mazzaschi, G.; Madeddu, D.; Falco, A.; Bocchialini, G.; Goldoni, M.; Sogni, F.; Armani, G.; Lagrasta, C.A.; Lorusso, B.; Mangiaracina, C.; et al. Low PD-1 Expression in Cytotoxic CD8+ Tumor-Infiltrating Lymphocytes Confers an Immune-Privileged Tissue Microenvironment in NSCLC with a Prognostic and Predictive Value. Clin. Cancer Res. 2018, 24, 407–419. [Google Scholar] [CrossRef]
- Owen, D.; Chu, B.; Lehman, A.M.; Annamalai, L.; Yearley, J.H.; Shilo, K.; Otterson, G.A. Expression Patterns, Prognostic Value, and Intratumoral Heterogeneity of PD-L1 and PD-1 in Thymoma and Thymic Carcinoma. J. Thorac. Oncol. 2018, 13, 1204–1212. [Google Scholar] [CrossRef]
- Weiss, S.A.; Wolchok, J.D.; Sznol, M. Immunotherapy of Melanoma: Facts and Hopes. Clin. Cancer Res. 2019, 25, 5191–5201. [Google Scholar] [CrossRef]
- Yeong, J.; Lim, J.C.T.; Lee, B.; Li, H.; Ong, C.C.H.; Thike, A.A.; Yeap, W.H.; Yang, Y.; Lim, A.Y.H.; Tay, T.K.Y.; et al. Prognostic value of CD8 + PD-1+ immune infiltrates and PDCD1 gene expression in triple negative breast cancer. J. Immunother. Cancer 2019, 7, 34. [Google Scholar] [CrossRef]
- Kalaora, S.; Nagler, A.; Wargo, J.A.; Samuels, Y. Mechanisms of immune activation and regulation: Lessons from melanoma. Nat. Rev. Cancer 2022, 22, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Watson, R.A.; Tong, O.; Cooper, R.; Taylor, C.A.; Sharma, P.K.; Aires, A.V.D.L.; Mahé, E.A.; Ruffieux, H.; Nassiri, I.; Middleton, M.R.; et al. Immune checkpoint blockade sensitivity and progression-free survival associates with baseline CD8 + T cell clone size and cytotoxicity. Sci. Immunol. 2021, 6, eabj8825. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e310. [Google Scholar] [CrossRef] [PubMed]
- Dranoff, G. Cytokines in cancer pathogenesis and cancer therapy. Nat. Rev. Cancer 2004, 4, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Waldmann, T.A. Cytokines in Cancer Immunotherapy. Cold Spring Harb. Perspect. Biol. 2018, 10, a028472. [Google Scholar] [CrossRef]
- Liu, C.; Chu, D.; Kalantar-Zadeh, K.; George, J.; Young, H.A.; Liu, G. Cytokines: From Clinical Significance to Quantification. Adv. Sci. 2021, 8, e2004433. [Google Scholar] [CrossRef]
- Morad, G.; Helmink, B.A.; Sharma, P.; Wargo, J.A. Hallmarks of response, resistance, and toxicity to immune checkpoint blockade. Cell 2022, 185, 576. [Google Scholar] [CrossRef]
- Sade-Feldman, M.; Jiao, Y.J.; Chen, J.H.; Rooney, M.S.; Barzily-Rokni, M.; Eliane, J.-P.; Bjorgaard, S.L.; Hammond, M.R.; Vitzthum, H.; Blackmon, S.M.; et al. Resistance to checkpoint blockade therapy through inactivation of antigen presentation. Nat. Commun. 2017, 8, 1136. [Google Scholar] [CrossRef] [PubMed]
- Elliot, T.A.; Jennings, E.K.; Lecky, D.A.; Thawait, N.; Flores-Langarica, A.; Copland, A.; Maslowski, K.M.; Wraith, D.C.; Bending, D. Antigen and checkpoint receptor engagement recalibrates T cell receptor signal strength. Immunity 2021, 54, 2481–2496.e6. [Google Scholar] [CrossRef]
- Yuan, B.; A Clark, C.; Wu, B.; Yang, J.; Drerup, J.M.; Li, T.; Jin, V.X.; Hu, Y.; Curiel, T.J.; Li, R. Estrogen receptor beta signaling in CD8+ T cells boosts T cell receptor activation and antitumor immunity through a phosphotyrosine switch. J. Immunother. Cancer 2021, 9, e001932. [Google Scholar] [CrossRef] [PubMed]
- Hodi, F.S.; O’Day, S.J.; McDermott, D.F.; Weber, R.W.; Sosman, J.A.; Haanen, J.B.; Gonzalez, R.; Robert, C.; Schadendorf, D.; Hassel, J.C.; et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. N. Engl. J. Med. 2010, 363, 711–723. [Google Scholar] [CrossRef]
- Hughes, T.; Klairmont, M.; Sharfman, W.H.; Kaufman, H.L. Interleukin-2, Ipilimumab, and Anti-PD-1: Clinical management and the evolving role of immunotherapy for the treatment of patients with metastatic melanoma. Cancer Biol. Ther. 2021, 22, 513–526. [Google Scholar] [CrossRef]
- Robert, C.; Thomas, L.; Bondarenko, I.; O’Day, S.; Weber, J.; Garbe, C.; Lebbe, C.; Baurain, J.-F.; Testori, A.; Grob, J.-J.; et al. Ipilimumab plus Dacarbazine for Previously Untreated Metastatic Melanoma. N. Engl. J. Med. 2011, 364, 2517–2526. [Google Scholar] [CrossRef]
- Schadendorf, D.; Hodi, F.S.; Robert, C.; Weber, J.S.; Margolin, K.; Hamid, O.; Patt, D.; Chen, T.-T.; Berman, D.M.; Wolchok, J.D. Pooled Analysis of Long-Term Survival Data From Phase II and Phase III Trials of Ipilimumab in Unresectable or Metastatic Melanoma. J. Clin. Oncol. 2015, 33, 1889–1894. [Google Scholar] [CrossRef] [PubMed]
- Larkin, J.; Lao, C.D.; Urba, W.J.; McDermott, D.F.; Horak, C.; Jiang, J.; Wolchok, J.D. Efficacy and Safety of Nivolumab in Patients With BRAF V600 Mutant and BRAF Wild-Type Advanced Melanoma: A Pooled Analysis of 4 Clinical Trials. JAMA Oncol. 2015, 1, 433–440. [Google Scholar] [CrossRef]
- Larkin, J.; Chiarion-Sileni, V.; Gonzalez, R.; Grob, J.-J.; Cowey, C.L.; Lao, C.D.; Schadendorf, D.; Dummer, R.; Smylie, M.; Rutkowski, P.; et al. Combined Nivolumab and Ipilimumab or Monotherapy in Untreated Melanoma. N. Engl. J. Med. 2015, 373, 23–34. [Google Scholar] [CrossRef]
- Redin, E.; Garmendia, I.; Lozano, T.; Serrano, D.; Senent, Y.; Redrado, M.; Villalba, M.; E De Andrea, C.; Exposito, F.; Ajona, D.; et al. SRC family kinase (SFK) inhibitor dasatinib improves the antitumor activity of anti-PD-1 in NSCLC models by inhibiting Treg cell conversion and proliferation. J. Immunother. Cancer 2021, 9, e001496. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.M.; Jing, X.; Pike, L.A.; Zhou, Q.; Lim, D.-J.; Sams, S.B.; Lund, G.S.; Sharma, V.; Haugen, B.R.; Schweppe, R.E. Targeted Inhibition of Src Kinase with Dasatinib Blocks Thyroid Cancer Growth and Metastasis. Clin. Cancer Res. 2012, 18, 3580–3591. [Google Scholar] [CrossRef]
- Huang, W.-C.; Kuo, K.-T.; Wang, C.-H.; Yeh, C.-T.; Wang, Y. Cisplatin resistant lung cancer cells promoted M2 polarization of tumor-associated macrophages via the Src/CD155/MIF functional pathway. J. Exp. Clin. Cancer Res. 2019, 38, 180. [Google Scholar] [CrossRef]
- Parseghian, C.M.; Parikh, N.U.; Wu, J.Y.; Jiang, Z.-Q.; Henderson, L.; Tian, F.; Pastor, B.; Ychou, M.; Raghav, K.; Dasari, A.; et al. Dual Inhibition of EGFR and c-Src by Cetuximab and Dasatinib Combined with FOLFOX Chemotherapy in Patients with Metastatic Colorectal Cancer. Clin. Cancer Res. 2017, 23, 4146–4154. [Google Scholar] [CrossRef]
- Secord, A.A.; Teoh, D.K.; Barry, W.T.; Yu, M.; Broadwater, G.; Havrilesky, L.J.; Lee, P.S.; Berchuck, A.; Lancaster, J.; Wenham, R.M. A Phase I Trial of Dasatinib, an Src-Family Kinase Inhibitor, in Combination with Paclitaxel and Carboplatin in Patients with Advanced or Recurrent Ovarian Cancer. Clin. Cancer Res. 2012, 18, 5489–5498. [Google Scholar] [CrossRef]
- Zhang, H.; Hu, Y.; Shao, M.; Teng, X.; Jiang, P.; Wang, X.; Wang, H.; Cui, J.; Yu, J.; Liang, Z.; et al. Dasatinib enhances anti-leukemia efficacy of chimeric antigen receptor T cells by inhibiting cell differentiation and exhaustion. J. Hematol. Oncol. 2021, 14, 113. [Google Scholar] [CrossRef]
- Lee, D.; Park, Y.H.; Lee, J.E.; Kim, H.S.; Min, K.Y.; Jo, M.G.; Kim, H.S.; Choi, W.S.; Kim, Y.M. Dasatinib Inhibits Lyn and Fyn Src-Family Kinases in Mast Cells to Suppress Type I Hypersensitivity in Mice. Biomol. Ther. 2020, 28, 456–464. [Google Scholar] [CrossRef]
- Li, J.; Rix, U.; Fang, B.; Bai, Y.; Edwards, A.; Colinge, J.; Bennett, K.L.; Gao, J.; Song, L.; Eschrich, S.; et al. A chemical and phosphoproteomic characterization of dasatinib action in lung cancer. Nat. Chem. Biol. 2010, 6, 291–299. [Google Scholar] [CrossRef]
- De Wispelaere, M.; LaCroix, A.J.; Yang, P.L. The Small Molecules AZD0530 and Dasatinib Inhibit Dengue Virus RNA Replication via Fyn Kinase. J. Virol. 2013, 87, 7367–7381. [Google Scholar] [CrossRef]
- Halbrook, C.J.; Pontious, C.; Kovalenko, I.; Lapienyte, L.; Dreyer, S.; Lee, H.-J.; Thurston, G.; Zhang, Y.; Lazarus, J.; Sajjakulnukit, P.; et al. Macrophage-Released Pyrimidines Inhibit Gemcitabine Therapy in Pancreatic Cancer. Cell Metab. 2019, 29, 1390–1399.e6. [Google Scholar] [CrossRef]
- Carpenter, E.S.; Steele, N.G.; Di Magliano, M.P. Targeting the Microenvironment to Overcome Gemcitabine Resistance in Pancreatic Cancer. Cancer Res. 2020, 80, 3070–3071. [Google Scholar] [CrossRef]
- Ho, T.T.B.; Nasti, A.; Seki, A.; Komura, T.; Inui, H.; Kozaka, T.; Kitamura, Y.; Shiba, K.; Yamashita, T.; Yamashita, T.; et al. Combination of gemcitabine and anti-PD-1 antibody enhances the anticancer effect of M1 macrophages and the Th1 response in a murine model of pancreatic cancer liver metastasis. J. Immunother. Cancer 2020, 8, e001367. [Google Scholar] [CrossRef]
- Principe, D.R.; Narbutis, M.; Kumar, S.; Park, A.; Viswakarma, N.; Dorman, M.J.; Kamath, S.D.; Grippo, P.J.; Fishel, M.L.; Hwang, R.F.; et al. Long-Term Gemcitabine Treatment Reshapes the Pancreatic Tumor Microenvironment and Sensitizes Murine Carcinoma to Combination Immunotherapy. Cancer Res. 2020, 80, 3101–3115. [Google Scholar] [CrossRef] [PubMed]
| Symbol | Entrez ID | Rank in Our Prediction | Compound |
|---|---|---|---|
| AKT1 | 207 | 0.906% | TEMSIROLIMUS |
| TP53 | 7157 | 0.030% | BEVACIZUMAB |
| BORTEZOMIB | |||
| EGFR | 1956 | 0.097% | BEVACIZUMAB |
| IBRUTINIB | |||
| SUNITINIB | |||
| FYN | 2534 | 0.178% | DASATINIB |
| JUN | 3725 | 0.253% | COLCHICINE |
| SRC | 6714 | 0.059% | DASATINIB |
| BOSUTINIB | |||
| GEMCITABINE |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Guo, S.; Zhou, H.; Sun, Y.; Gan, J.; Zhang, Y.; Zheng, W.; Zhang, C.; Zhao, X.; Xiao, J.; et al. Immune Pathways with Aging Characteristics Improve Immunotherapy Benefits and Drug Prediction in Human Cancer. Cancers 2023, 15, 342. https://doi.org/10.3390/cancers15020342
Wang X, Guo S, Zhou H, Sun Y, Gan J, Zhang Y, Zheng W, Zhang C, Zhao X, Xiao J, et al. Immune Pathways with Aging Characteristics Improve Immunotherapy Benefits and Drug Prediction in Human Cancer. Cancers. 2023; 15(2):342. https://doi.org/10.3390/cancers15020342
Chicago/Turabian StyleWang, Xinyue, Shuang Guo, Hanxiao Zhou, Yue Sun, Jing Gan, Yakun Zhang, Wen Zheng, Caiyu Zhang, Xiaoxi Zhao, Jiebin Xiao, and et al. 2023. "Immune Pathways with Aging Characteristics Improve Immunotherapy Benefits and Drug Prediction in Human Cancer" Cancers 15, no. 2: 342. https://doi.org/10.3390/cancers15020342
APA StyleWang, X., Guo, S., Zhou, H., Sun, Y., Gan, J., Zhang, Y., Zheng, W., Zhang, C., Zhao, X., Xiao, J., Wang, L., Gao, Y., & Ning, S. (2023). Immune Pathways with Aging Characteristics Improve Immunotherapy Benefits and Drug Prediction in Human Cancer. Cancers, 15(2), 342. https://doi.org/10.3390/cancers15020342






