Molecular Characterization of Primary Mediastinal Large B-Cell Lymphomas
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection of Cases
2.2. Clinicopathological Characteristics
2.3. Immunochemistry and Study
2.4. Fluorescence In Situ Hybridization (FISH)
2.5. Targeted Next-Generation Sequencing Analysis (TNGS)
2.6. Statistical Analysis
3. Results
3.1. Epidemiological Characteristics
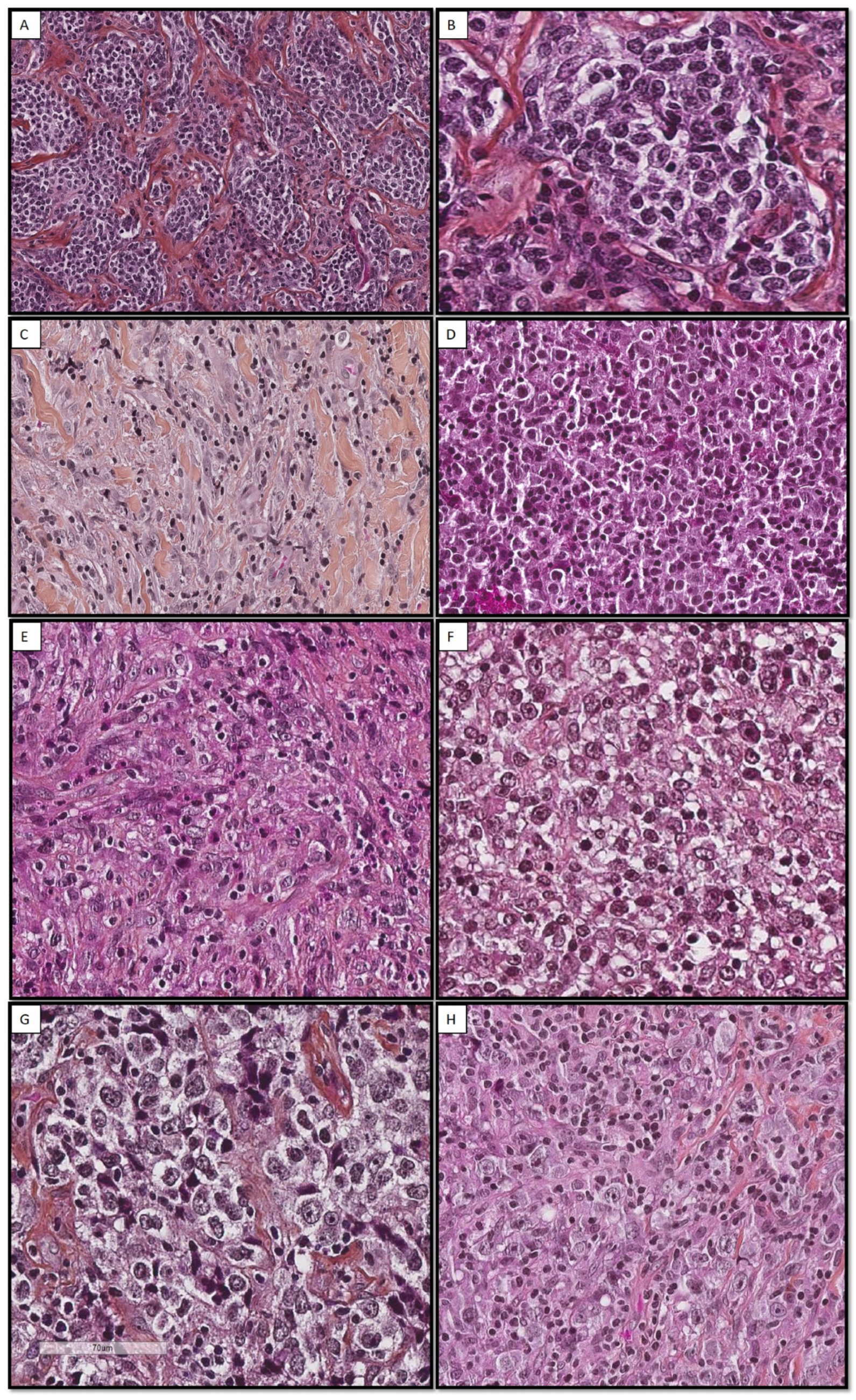
3.2. Histological Aspect
3.3. Immunohistochemistry Results
3.4. Fluorescence In Situ Hybridization (FISH) Results
3.5. Mutational Characteristics by Targeted Sequencing
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Laurent, C.; Baron, M.; Amara, N.; Haioun, C.; Dandoit, M.; Maynadié, M.; Parrens, M.; Vergier, B.; Copie-Bergman, C.; Fabiani, B.; et al. Impact of Expert Pathologic Review of Lymphoma Diagnosis: Study of Patients From the French Lymphopath Network. J. Clin. Oncol. 2017, 35, 2008–2017. [Google Scholar] [CrossRef] [PubMed]
- Sukswai, N.; Lyapichev, K.; Khoury, J.D.; Medeiros, L.J. Diffuse Large B-Cell Lymphoma Variants: An Update. Pathology 2020, 52, 53–67. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.; Afridi, S.S.; Shahid, Z.; Zamani, Z.; Rehman, S.; Aiman, W.; Khan, M.; Mir, M.A.; Awan, F.T.; Anwer, F.; et al. Primary Mediastinal B-Cell Lymphoma: A 2021 Update on Genetics, Diagnosis, and Novel Therapeutics. Clin. Lymphoma Myeloma Leuk. 2021, 21, e865–e875. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 Revision of the World Health Organization Classification of Lymphoid Neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Csernus, B.; Timár, B.; Fülöp, Z.; Bognár, Á.; Szepesi, Á.; László, T.; Jáksó, P.; Warnke, R.; Kopper, L.; Matolcsy, A. Mutational Analysis of IgV H and BCL-6 Genes Suggests Thymic B-Cells Origin of Mediastinal (Thymic) B-Cell Lymphoma. Leuk. Lymphoma 2004, 45, 2105–2110. [Google Scholar] [CrossRef]
- Salama, M.E.; Rajan Mariappan, M.; Inamdar, K.; Tripp, S.R.; Perkins, S.L. The Value of CD23 Expression as an Additional Marker in Distinguishing Mediastinal (Thymic) Large B-Cell Lymphoma from Hodgkin Lymphoma. Int. J. Surg. Pathol. 2010, 18, 121–128. [Google Scholar] [CrossRef]
- Liu, H.Y.; Zhong, R.Q.; Zhang, K.P.; Zhu, T.N.; Zhong, D.R. Clinicopathological features and prognosis of primary mediastinal large B-cell lymphoma: A series of sixty cases. Cancer Manag. Res. 2021, 50, 1139–1144. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, H.-Y. Primary Mediastinal Large B-Cell Lymphoma: Diagnostic Challenges and Recent Advances. J. Clin. Transl. Pathol. 2021, 1, 21–27. [Google Scholar] [CrossRef]
- Mangasarova, Y.K.; Sidorova, Y.V.; Magomedova, A.U.; Biderman, B.V.; Nikulina, E.E.; Sudarikov, A.B.; Kovrigina, A.M.; Kravchenko, S.K. Rearrangements of Immunoglobulin Genes in Tumor Cells of Patients with Primary Mediastinal (Thymic) Large B-Cell Lymphoma. Clin. Oncohematology 2019, 12, 271–277. [Google Scholar] [CrossRef]
- Bobée, V.; Ruminy, P.; Marchand, V.; Viailly, P.-J.; Sater, A.A.; Veresezan, L.; Drieux, F.; Bérard, C.; Bohers, E.; Mareschal, S.; et al. Determination of Molecular Subtypes of Diffuse Large B-Cell Lymphoma Using a Reverse Transcriptase Multiplex Ligation-Dependent Probe Amplification Classifier: A CALYM Study. J. Mol. Diagn. 2017, 19, 892–904. [Google Scholar] [CrossRef]
- Mottok, A.; Hung, S.S.; Chavez, E.A.; Woolcock, B.; Telenius, A.; Chong, L.C.; Meissner, B.; Nakamura, H.; Rushton, C.; Viganò, E.; et al. Integrative Genomic Analysis Identifies Key Pathogenic Mechanisms in Primary Mediastinal Large B-Cell Lymphoma. Blood 2019, 134, 802–813. [Google Scholar] [CrossRef]
- Dunleavy, K.; Wilson, W.H. Primary Mediastinal B-Cell Lymphoma and Mediastinal Gray Zone Lymphoma: Do They Require a Unique Therapeutic Approach? Blood 2015, 125, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Savage, K.J.; Monti, S.; Kutok, J.L.; Cattoretti, G.; Neuberg, D.; De Leval, L.; Kurtin, P.; Dal Cin, P.; Ladd, C.; Feuerhake, F.; et al. The Molecular Signature of Mediastinal Large B-Cell Lymphoma Differs from That of Other Diffuse Large B-Cell Lymphomas and Shares Features with Classical Hodgkin Lymphoma. Blood 2003, 102, 3871–3879. [Google Scholar] [CrossRef] [PubMed]
- Rosenwald, A.; Wright, G.; Leroy, K.; Yu, X.; Gaulard, P.; Gascoyne, R.D.; Chan, W.C.; Zhao, T.; Haioun, C.; Greiner, T.C.; et al. Molecular Diagnosis of Primary Mediastinal B Cell Lymphoma Identifies a Clinically Favorable Subgroup of Diffuse Large B Cell Lymphoma Related to Hodgkin Lymphoma. J. Exp. Med. 2003, 198, 851–862. [Google Scholar] [CrossRef] [PubMed]
- Campo, E. Holes in SOCS in Primary Mediastinal Large B-Cell Lymphoma. Blood 2005, 105, 2244–2245. [Google Scholar] [CrossRef][Green Version]
- Sarkozy, C.; Hung, S.S.; Chavez, E.A.; Duns, G.; Takata, K.; Chong, L.C.; Aoki, T.; Jiang, A.; Miyata-Takata, T.; Telenius, A.; et al. Mutational Landscape of Gray Zone Lymphoma. Blood 2021, 137, 1765–1776. [Google Scholar] [CrossRef]
- Chen, H.; Pan, T.; He, Y.; Zeng, R.; Li, Y.; Yi, L.; Zang, H.; Chen, S.; Duan, Q.; Xiao, L.; et al. Primary Mediastinal B-Cell Lymphoma: Novel Precision Therapies and Future Directions. Front. Oncol. 2021, 11, 654854. [Google Scholar] [CrossRef]
- Fakhri, B.; Ai, W. Current and Emerging Treatment Options in Primary Mediastinal B-Cell Lymphoma. Ther. Adv. Hematol. 2021, 12, 204062072110489. [Google Scholar] [CrossRef]
- Hatic, H.; Sampat, D.; Goyal, G. Immune Checkpoint Inhibitors in Lymphoma: Challenges and Opportunities. Ann. Transl. Med. 2021, 9, 1037. [Google Scholar] [CrossRef]
- Nann, D.; Ramis-Zaldivar, J.E.; Müller, I.; Gonzalez-Farre, B.; Schmidt, J.; Egan, C.; Salmeron-Villalobos, J.; Clot, G.; Mattern, S.; Otto, F.; et al. Follicular Lymphoma t(14;18)-Negative Is Genetically a Heterogeneous Disease. Blood Adv. 2020, 4, 5652–5665. [Google Scholar] [CrossRef]
- Dorfman, D.M.; Shahsafaei, A.; Alonso, M.A. Utility of CD200 Immunostaining in the Diagnosis of Primary Mediastinal Large B Cell Lymphoma: Comparison with MAL, CD23, and Other Markers. Mod. Pathol. 2012, 25, 1637–1643. [Google Scholar] [CrossRef] [PubMed]
- Salas, M.Q.; Climent, F.; Domingo Domenech, E.; Mercadal, S.; Paredes, V.; Oliveira, A.C.; Aguilera, C.; de la Banda, E.; Lucas, A.; Garcia, N.; et al. CD30 Expression in Diffuse Large B-Cell Lymphoma (DLBCL) Correlates with Non-GCB Subtype but Does Not Have Prognostic Impact in Patients Treated with First Line R-CHOP/R-CHOP-Like. Blood 2016, 128, 4209. [Google Scholar] [CrossRef]
- Almeida, R.; Abrantes, C.; Gigliano, D.; Oliveira, R.C.; Teixeira, P.; Viegas, M.; Rodrigues, Â.; Julião, M.J. Clinical and Pathological Features of Double-Hit and Triple-Hit High-Grade B-Cell Lymphomas: A Retrospective Study from Three Portuguese Tertiary Centers. Int. J. Hematol. Oncol. Stem Cell Res. 2022, 16, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, A.; Kato, S.; Okamoto, A.; Inaguma, Y.; Satou, A.; Tsuzuki, T.; Emi, N.; Okamoto, M.; Nakamura, S. Reappraisal of Epstein-Barr Virus (EBV) in Diffuse Large B-Cell Lymphoma (DLBCL): Comparative Analysis between EBV-Positive and EBV-Negative DLBCL with EBV-Positive Bystander Cells. Histopathology 2017, 71, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Bourbon, E.; Maucort-Boulch, D.; Fontaine, J.; Mauduit, C.; Sesques, P.; Safar, V.; Ferrant, E.; Golfier, C.; Ghergus, D.; Karlin, L.; et al. Clinicopathological Features and Survival in EBV-Positive Diffuse Large B-Cell Lymphoma Not Otherwise Specified. Blood Adv. 2021, 5, 3227–3239. [Google Scholar] [CrossRef]
- Donzel, M.; Bonjour, M.; Combes, J.-D.; Broussais, F.; Sesques, P.; Traverse-Glehen, A.; De Martel, C. Lymphomas Associated with Epstein-Barr Virus Infection in 2020: Results from a Large, Unselected Case Series in France. eClinicalMedicine 2022, 54, 101674. [Google Scholar] [CrossRef]
- Trecourt, A.; Mauduit, C.; Szablewski, V.; Fontaine, J.; Balme, B.; Donzel, M.; Laurent, C.; Sesques, P.; Ghesquières, H.; Bachy, E.; et al. Plasticity of Mature B Cells Between Follicular and Classic Hodgkin Lymphomas: A Series of 22 Cases Expanding the Spectrum of Transdifferentiation. Am. J. Surg. Pathol. 2022, 46, 58–70. [Google Scholar] [CrossRef]
- Bommier, C.; Mauduit, C.; Fontaine, J.; Bourbon, E.; Sujobert, P.; Huet, S.; Baseggio, L.; Hayette, S.; Laurent, C.; Bachy, E.; et al. Real-life Targeted Next-generation Sequencing for Lymphoma Diagnosis over 1 Year from the French Lymphoma Network. Br. J. Haematol. 2021, 193, 1110–1122. [Google Scholar] [CrossRef]
- Sujobert, P.; Le Bris, Y.; de Leval, L.; Gros, A.; Merlio, J.P.; Pastoret, C.; Huet, S.; Sarkozy, C.; Davi, F.; Callanan, M.; et al. The Need for a Consensus Next-Generation Sequencing Panel for Mature Lymphoid Malignancies. HemaSphere 2019, 3, e169. [Google Scholar] [CrossRef]
- Dubois, S.; Viailly, P.-J.; Mareschal, S.; Bohers, E.; Bertrand, P.; Ruminy, P.; Maingonnat, C.; Jais, J.-P.; Peyrouze, P.; Figeac, M.; et al. Next-Generation Sequencing in Diffuse Large B-Cell Lymphoma Highlights Molecular Divergence and Therapeutic Opportunities: A LYSA Study. Clin. Cancer Res. 2016, 22, 2919–2928. [Google Scholar] [CrossRef]
- Huet, S.; Paubelle, E.; Lours, C.; Grange, B.; Courtois, L.; Chabane, K.; Charlot, C.; Mosnier, I.; Simonet, T.; Hayette, S.; et al. Validation of the Prognostic Value of the Knowledge Bank Approach to Determine AML Prognosis in Real Life. Blood 2018, 132, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.J.; Arcila, M.E.; Corless, C.; Kamel-Reid, S.; Lubin, I.M.; Pfeifer, J.; Temple-Smolkin, R.L.; Voelkerding, K.V.; Nikiforova, M.N. Guidelines for Validation of Next-Generation Sequencing–Based Oncology Panels. J. Mol. Diagn. 2017, 19, 341–365. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Lakatos, E.; Bakir, I.A.; Curtius, K.; Graham, T.A.; Mustonen, V. The Mutational Signatures of Formalin Fixation on the Human Genome. Nat. Commun. 2022, 13, 4487. [Google Scholar] [CrossRef] [PubMed]
- Yildiz, M.; Li, H.; Bernard, D.; Amin, N.A.; Ouillette, P.; Jones, S.; Saiya-Cork, K.; Parkin, B.; Jacobi, K.; Shedden, K.; et al. Activating STAT6 Mutations in Follicular Lymphoma. Blood 2015, 125, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Camus, V.; Rossi, C.; Sesques, P.; Lequesne, J.; Tonnelet, D.; Haioun, C.; Durot, E.; Willaume, A.; Gauthier, M.; Moles-Moreau, M.-P.; et al. Outcomes after First-Line Immunochemotherapy for Primary Mediastinal B-Cell Lymphoma: A LYSA Study. Blood Adv. 2021, 5, 3862–3872. [Google Scholar] [CrossRef] [PubMed]
- Wästerlid, T.; Hasselblom, S.; Joelsson, J.; Weibull, C.E.; Rassidakis, G.; Sander, B.; Smedby, K.E. Real-World Data on Treatment and Outcomes of Patients with Primary Mediastinal Large B-Cell Lymphoma: A Swedish Lymphoma Register Study. Blood Cancer J. 2021, 11, 100. [Google Scholar] [CrossRef]
- De Mello, C.A.L.; De Andrade, V.P.; De Lima, V.C.C.; Carvalho, A.L.; Soares, F.A. Prognostic Impact of MUM1 Expression by Immunohistochemistry on Primary Mediastinal Large B-Cell Lymphoma. Leuk. Lymphoma 2011, 52, 1495–1503. [Google Scholar] [CrossRef]
- Maracaja, D.L.V.; Puthenpura, V.; Pels, S.G.; O’Malley, D.P.; Sklar, J.L.; Finberg, K.E.; Xu, M.L. EBV-Positive Primary Large B-Cell Lymphoma: The Role of Immunohistochemistry and XPO1 in the Diagnosis of Mediastinal Lymphomas. Appl. Immunohistochem. Mol. Morphol. 2020, 28, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Cazals-Hatem, D.; Lepage, E.; Brice, P.; Ferrant, A.; d’Agay, M.F.; Baumelou, E.; Brière, J.; Blanc, M.; Gaulard, P.; Biron, P.; et al. Primary Mediastinal Large B-Cell Lymphoma. A Clinicopathologic Study of 141 Cases Compared with 916 Nonmediastinal Large B-Cell Lymphomas, a GELA (“Groupe d’Etude Des Lymphomes de l’Adulte”) Study. Am. J. Surg. Pathol. 1996, 20, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Steidl, C.; Shah, S.P.; Woolcock, B.W.; Rui, L.; Kawahara, M.; Farinha, P.; Johnson, N.A.; Zhao, Y.; Telenius, A.; Neriah, S.B.; et al. MHC Class II Transactivator CIITA Is a Recurrent Gene Fusion Partner in Lymphoid Cancers. Nature 2011, 471, 377–381. [Google Scholar] [CrossRef]
- Ondrejka, S.L.; Ott, G. How I Diagnose Primary Mediastinal (Thymic) Large B-Cell Lymphoma. Am. J. Clin. Pathol. 2021, 156, 497–512. [Google Scholar] [CrossRef] [PubMed]
- Pileri, S.A.; Gaidano, G.; Zinzani, P.L.; Falini, B.; Gaulard, P.; Zucca, E.; Pieri, F.; Berra, E.; Sabattini, E.; Ascani, S.; et al. Primary Mediastinal B-Cell Lymphoma: High Frequency of BCL-6 Mutations and Consistent Expression of the Transcription Factors OCT-2, BOB.1, and PU.1 in the Absence of Immunoglobulins. Am. J. Pathol. 2003, 162, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Evidence of C-Myc Gene Abnormalities in Mediastinal Large B-Cell Lymphoma of Young Adult Age—PubMed. Available online: https://pubmed.ncbi.nlm.nih.gov/1713514/ (accessed on 10 February 2023).
- Calvo, K.R.; Traverse-Glehen, A.; Pittaluga, S.; Jaffe, E.S. Molecular Profiling Provides Evidence of Primary Mediastinal Large B-Cell Lymphoma as a Distinct Entity Related to Classic Hodgkin Lymphoma: Implications for Mediastinal Gray Zone Lymphomas as an Intermediate Form of B-Cell Lymphoma. Adv. Anat. Pathol. 2004, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Campuzano-Zuluaga, G.; Ortiz, D.; Peng, J.-H.; Francis Ikpatt, O.; Fan, Y.-S.; Barredo, J.C.; Vega, F.; Chapman, J.R. Primary Mediastinal Large B-Cell Lymphoma with Translocations Involving BCL6 and MYC (Double-Hit Lymphoma). Am. J. Clin. Pathol. 2016, 145, 710–716. [Google Scholar] [CrossRef]
- Ocal, J.L.; Feldman, A.L.; Greipp, P.T.; Rimsza, L.M. Mediastinal B-Cell Lymphoma with MYC, BCL2, and BCL6 Rearrangements. J. Hematopathol. 2022, 15, 151–155. [Google Scholar] [CrossRef]
- Caranfil, E.; Isnard, P.; Bruneau, J.; Brière, J.; Molina, T.J. Lymphome à grandes cellules B primitif du médiastin. Rev. Francoph. Lab. 2021, 2021, 57–63. [Google Scholar] [CrossRef]
- Gunawardana, J.; Chan, F.C.; Telenius, A.; Woolcock, B.; Kridel, R.; Tan, K.L.; Ben-Neriah, S.; Mottok, A.; Lim, R.S.; Boyle, M.; et al. Recurrent Somatic Mutations of PTPN1 in Primary Mediastinal B Cell Lymphoma and Hodgkin Lymphoma. Nat. Genet. 2014, 46, 329–335. [Google Scholar] [CrossRef]
- Mottok, A.; Wright, G.; Rosenwald, A.; Ott, G.; Ramsower, C.; Campo, E.; Braziel, R.M.; Delabie, J.; Weisenburger, D.D.; Song, J.Y.; et al. Molecular Classification of Primary Mediastinal Large B-Cell Lymphoma Using Routinely Available Tissue Specimens. Blood 2018, 132, 2401–2405. [Google Scholar] [CrossRef]
- Ritz, O.; Guiter, C.; Castellano, F.; Dorsch, K.; Melzner, J.; Jais, J.-P.; Dubois, G.; Gaulard, P.; Möller, P.; Leroy, K. Recurrent Mutations of the STAT6 DNA Binding Domain in Primary Mediastinal B-Cell Lymphoma. Blood 2009, 114, 1236–1242. [Google Scholar] [CrossRef]
- STAT6 Activity Is Regulated by SOCS-1 and Modulates BCL-XL Expression in Primary Mediastinal B-Cell Lymphoma|Leukemia. Available online: https://www.nature.com/articles/leu200885 (accessed on 20 December 2021).
- Mellert, K.; Martin, M.; Lennerz, J.K.; Lüdeke, M.; Staiger, A.M.; Kreuz, M.; Löffler, M.; Schmitz, N.; Trümper, L.; Feller, A.C.; et al. The Impact of SOCS1 Mutations in Diffuse Large B-cell Lymphoma. Br. J. Haematol. 2019, 187, 627–637. [Google Scholar] [CrossRef]
- Lennerz, J.K.; Hoffmann, K.; Bubolz, A.-M.; Lessel, D.; Welke, C.; Rüther, N.; Viardot, A.; Möller, P. Suppressor of Cytokine Signaling 1 Gene Mutation Status as a Prognostic Biomarker in Classical Hodgkin Lymphoma. Oncotarget 2015, 6, 29097–29110. [Google Scholar] [CrossRef]
- Karpathiou, G.; Ferrand, E.; Papoudou-Bai, A.; Camy, F.; Honeyman, F.; Dumollard, J.M.; Peoc’h, M. STAT6 and Phosphorylated STAT6 Are Differentially Expressed in Lymphomas. Pathol. Res. Pract. 2022, 229, 153697. [Google Scholar] [CrossRef]
- Weniger, M.A.; Melzner, I.; Menz, C.K.; Wegener, S.; Bucur, A.J.; Dorsch, K.; Mattfeldt, T.; Barth, T.F.E.; Möller, P. Mutations of the Tumor Suppressor Gene SOCS-1 in Classical Hodgkin Lymphoma Are Frequent and Associated with Nuclear Phospho-STAT5 Accumulation. Oncogene 2006, 25, 2679–2684. [Google Scholar] [CrossRef] [PubMed]
- Van Slambrouck, C.; Huh, J.; Suh, C.; Song, J.Y.; Menon, M.P.; Sohani, A.R.; Duffield, A.S.; Goldberg, R.C.; Dama, P.; Kiyotani, K.; et al. Diagnostic Utility of STAT6YE361 Expression in Classical Hodgkin Lymphoma and Related Entities. Mod. Pathol. 2020, 33, 834–845. [Google Scholar] [CrossRef] [PubMed]
- Xian, R.R.; Xie, Y.; Haley, L.M.; Yonescu, R.; Pallavajjala, A.; Pittaluga, S.; Jaffe, E.S.; Duffield, A.S.; McCall, C.M.; Gheith, S.M.F.; et al. CREBBP and STAT6 Co-Mutation and 16p13 and 1p36 Loss Define the t(14;18)-Negative Diffuse Variant of Follicular Lymphoma. Blood Cancer J. 2020, 10, 69. [Google Scholar] [CrossRef] [PubMed]
- Tiacci, E.; Ladewig, E.; Schiavoni, G.; Penson, A.; Fortini, E.; Pettirossi, V.; Wang, Y.; Rosseto, A.; Venanzi, A.; Vlasevska, S.; et al. Pervasive Mutations of JAK-STAT Pathway Genes in Classical Hodgkin Lymphoma. Blood 2018, 131, 2454–2465. [Google Scholar] [CrossRef] [PubMed]
- Brune, M.M.; Juskevicius, D.; Haslbauer, J.; Dirnhofer, S.; Tzankov, A. Genomic Landscape of Hodgkin Lymphoma. Cancers 2021, 13, 682. [Google Scholar] [CrossRef]
- Jiang, Y.; Mo, W.; Miao, Y.; Liang, Y.; Li, Y.; Zhang, R. Primary Mediastinal Large B Cell Lymphoma with Coexisting Aberrations of C-MYC and BCL-2: A Case Report and Literature Review. Med. Mol. Morphol. 2020, 53, 124–129. [Google Scholar] [CrossRef]
- Genomic Alterations in CIITA Are Frequent in Primary Mediastinal Large B Cell Lymphoma and are Associated with Diminished MHC Class II Expression|Elsevier Enhanced Reader. Available online: https://reader.elsevier.com/reader/sd/pii/S2211124715011626?token=1200B3842BB667553B83788369E37909682B53B29E8570BBECFF23B05A628792D05E9990B7DCA9B5BE33FB4AC535EB4B&originRegion=eu-west-1&originCreation=20221027153320 (accessed on 27 October 2022).
- Sarkozy, C.; Chong, L.; Takata, K.; Chavez, E.A.; Miyata-Takata, T.; Duns, G.; Telenius, A.; Boyle, M.; Slack, G.W.; Laurent, C.; et al. Gene Expression Profiling of Gray Zone Lymphoma. Blood Adv. 2020, 4, 2523–2535. [Google Scholar] [CrossRef]
- Guiter, C.; Dusanter-Fourt, I.; Copie-Bergman, C.; Boulland, M.-L.; Le Gouvello, S.; Gaulard, P.; Leroy, K.; Castellano, F. Constitutive STAT6 Activation in Primary Mediastinal Large B-Cell Lymphoma. Blood 2004, 104, 543–549. [Google Scholar] [CrossRef]
- Ritz, O.; Guiter, C.; Dorsch, K.; Dusanter-Fourt, I.; Wegener, S.; Jouault, H.; Gaulard, P.; Castellano, F.; Möller, P.; Leroy, K. STAT6 Activity Is Regulated by SOCS-1 and Modulates BCL-XL Expression in Primary Mediastinal B-Cell Lymphoma. Leukemia 2008, 22, 2106–2110. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Krantsevich, A.; MacCarthy, T. Deep Learning Model of Somatic Hypermutation Reveals Importance of Sequence Context beyond Hotspot Targeting. iScience 2022, 25, 103668. [Google Scholar] [CrossRef] [PubMed]
- Duns, G.; Viganò, E.; Ennishi, D.; Sarkozy, C.; Hung, S.S.; Chavez, E.; Takata, K.; Rushton, C.; Jiang, A.; Ben-Neriah, S.; et al. Characterization of DLBCL with a PMBL Gene Expression Signature. Blood 2021, 138, 136–148. [Google Scholar] [CrossRef] [PubMed]
- Goenka, S.; Kaplan, M.H. Transcriptional Regulation by STAT6. Immunol. Res. 2011, 50, 87–96. [Google Scholar] [CrossRef]
- Kneitz, C.; Goller, M.; Seggewiss, R.; Yaman, A.; Serfling, E.; Tony, H.P. STAT6 and the Regulation of CD23 Expression in B-Chronic Lymphocytic Leukemia. Leuk. Res. 2000, 24, 331–337. [Google Scholar] [CrossRef]
- Romejko-Jarosinska, J.; Ostrowska, B.; Dabrowska-Iwanicka, A.; Domanska-Czyz, K.; Rymkiewicz, G.; Paszkiewicz-Kozik, E.; Konecki, R.; Borawska, A.; Druzd-Sitek, A.; Lampka, E.; et al. High Efficacy of Intensive Immunochemotherapy for Primary Mediastinal B-Cell Lymphoma with Prolonged Follow Up. Sci. Rep. 2022, 12, 10551. [Google Scholar] [CrossRef]



| Number (n) | Percentage (%) | Average | Ranges | ||
|---|---|---|---|---|---|
| Age (n = 49) | 38.9 | 21–83 | |||
| Sex (n = 49) | Male | 23 | 47 | ||
| Female | 26 | 53 | |||
| Symptoms (n = 33) | Cough | 10 | |||
| Dyspnea | 13 | ||||
| Chest pain | 13 | ||||
| Superior vena cava syndrome | 6 | ||||
| Elevated LDH (N < 250 UI/L) (n = 25) | 23 | 92 | 409 | 249–802 | |
| Tumor size (mm) (n = 25) | 101 | 52–150 | |||
| Ann Arbor (n = 33) | II | 22 | 67 | ||
| III-IV | 11 | 33 | |||
| Chemotherapy (n = 25) | R-CHOP | 2 | 8 | ||
| R-DA-EPOCH | 4 | 16 | |||
| R-ACVBP | 19 | 76 | |||
| Evolution (n = 25) | Complete remission | 21 | 84 | ||
| Relapse | 4 | 16 |
| Expression | CD20 | CD10 | BCL6 | CD30 | CD23 | CD15 | BCL2 | MUM1 | MYC | P63 | PDL1 | EBER-ISH |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffuse (n, (%)) | 49 (100%) | 39 (79%) | 10 (20%) | 11 (22%) | 29 (59%) | 29 (59%) | 15 (31%) | 2 (6%) | 2 (7%) | |||
| Partial (n, (%)) | 2 (4%) | 8 (16%) | 33 (67%) | 22 (45%) | 2 (4%) | 14 (28%) | 7 (23%) | 13 (45%) | 2 (4%) | |||
| Focal (n, (%)) | 1 (2%) | 4 (8%) | 3 (6%) | 34 (69%) | 7 (23%) | 12 (41%) | ||||||
| Negative (n, (%)) | 47 (96%) | 1 (2%) | 6 (12%) | 12 (24%) | 33 (100%) | 15 (31%) | 6 (12%) | 15 (48%) | 2 (7%) | 47 (96%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donzel, M.; Pesce, F.; Trecourt, A.; Groussel, R.; Bachy, E.; Ghesquières, H.; Fontaine, J.; Benzerdjeb, N.; Mauduit, C.; Traverse-Glehen, A. Molecular Characterization of Primary Mediastinal Large B-Cell Lymphomas. Cancers 2023, 15, 4866. https://doi.org/10.3390/cancers15194866
Donzel M, Pesce F, Trecourt A, Groussel R, Bachy E, Ghesquières H, Fontaine J, Benzerdjeb N, Mauduit C, Traverse-Glehen A. Molecular Characterization of Primary Mediastinal Large B-Cell Lymphomas. Cancers. 2023; 15(19):4866. https://doi.org/10.3390/cancers15194866
Chicago/Turabian StyleDonzel, Marie, Florian Pesce, Alexis Trecourt, Razika Groussel, Emmanuel Bachy, Hervé Ghesquières, Juliette Fontaine, Nazim Benzerdjeb, Claire Mauduit, and Alexandra Traverse-Glehen. 2023. "Molecular Characterization of Primary Mediastinal Large B-Cell Lymphomas" Cancers 15, no. 19: 4866. https://doi.org/10.3390/cancers15194866
APA StyleDonzel, M., Pesce, F., Trecourt, A., Groussel, R., Bachy, E., Ghesquières, H., Fontaine, J., Benzerdjeb, N., Mauduit, C., & Traverse-Glehen, A. (2023). Molecular Characterization of Primary Mediastinal Large B-Cell Lymphomas. Cancers, 15(19), 4866. https://doi.org/10.3390/cancers15194866







