Integrated Multi-Omics Profiling of Young Breast Cancer Patients Reveals a Correlation between Galactose Metabolism Pathway and Poor Disease-Free Survival
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Study Cohorts and Statistical Analyses
2.2. scRNA-Seq Cohorts and Data Processing
2.3. Cell Lines and D-Galactose Treatment
2.4. RNA Extraction, RT, and Real-Time qPCR
2.5. Mammosphere Formation Assay
3. Results
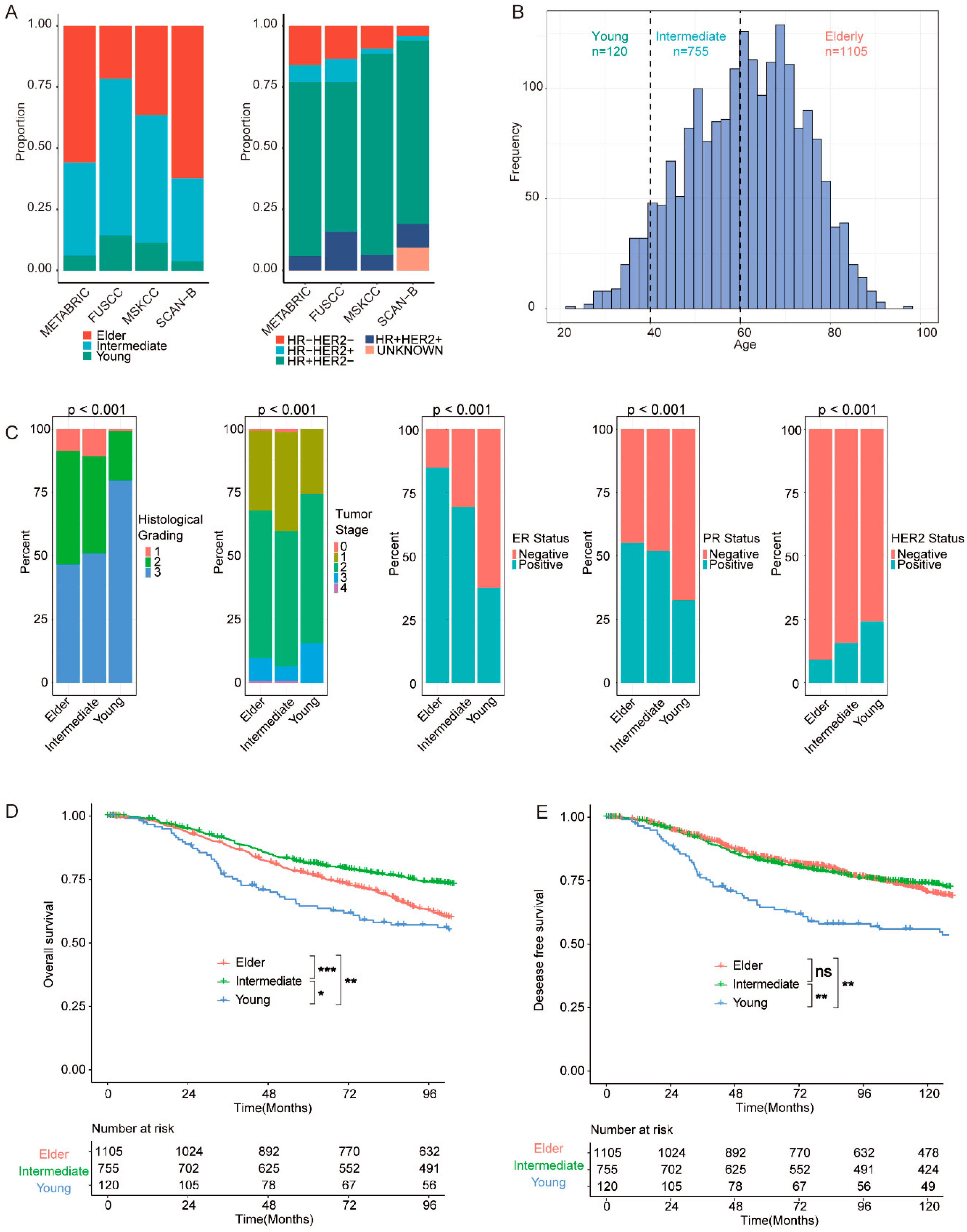
3.1. Clinicopathological and Survival Features of BC in Young Patients
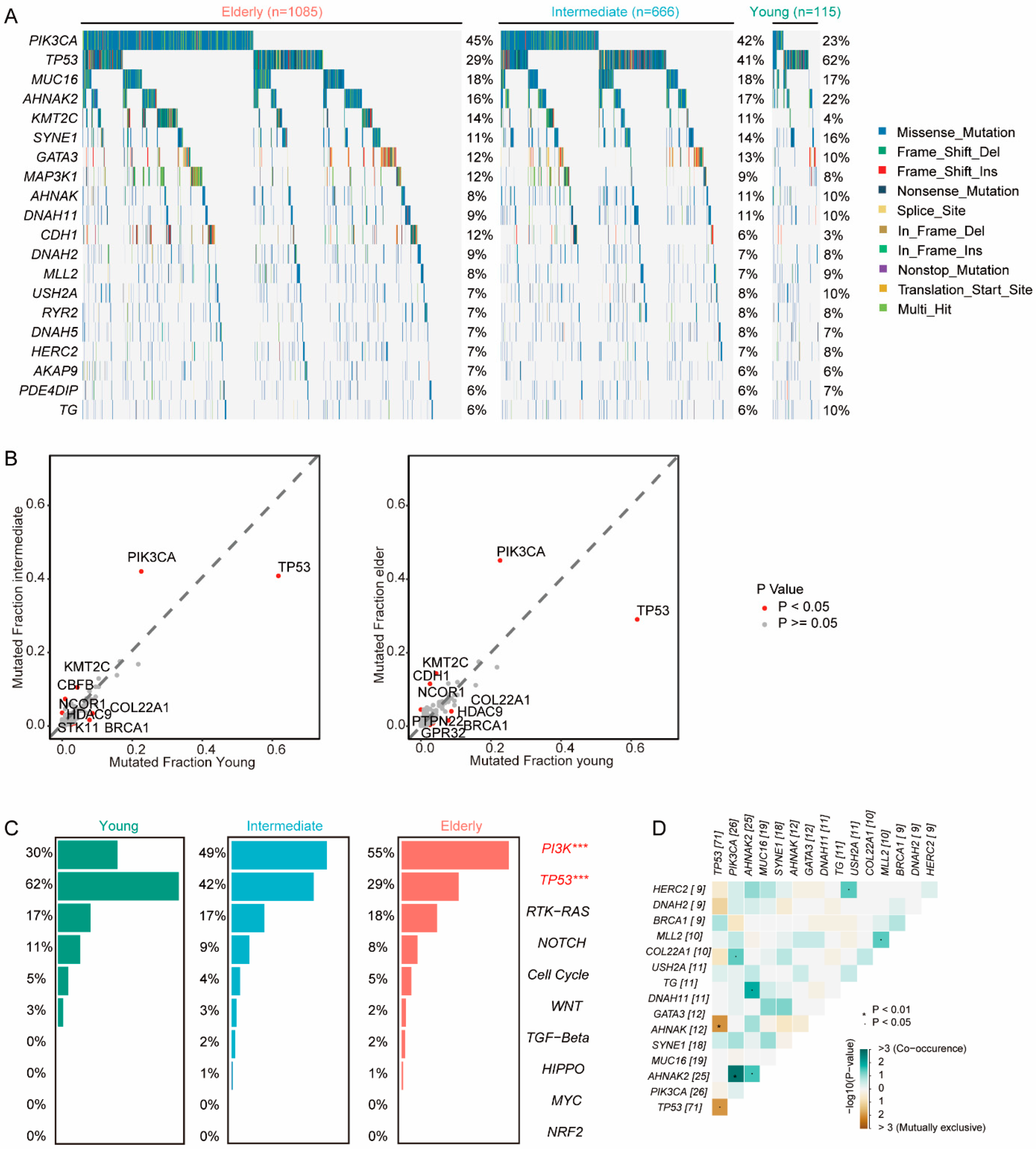
3.2. Genomics Features of BC in Young Patients
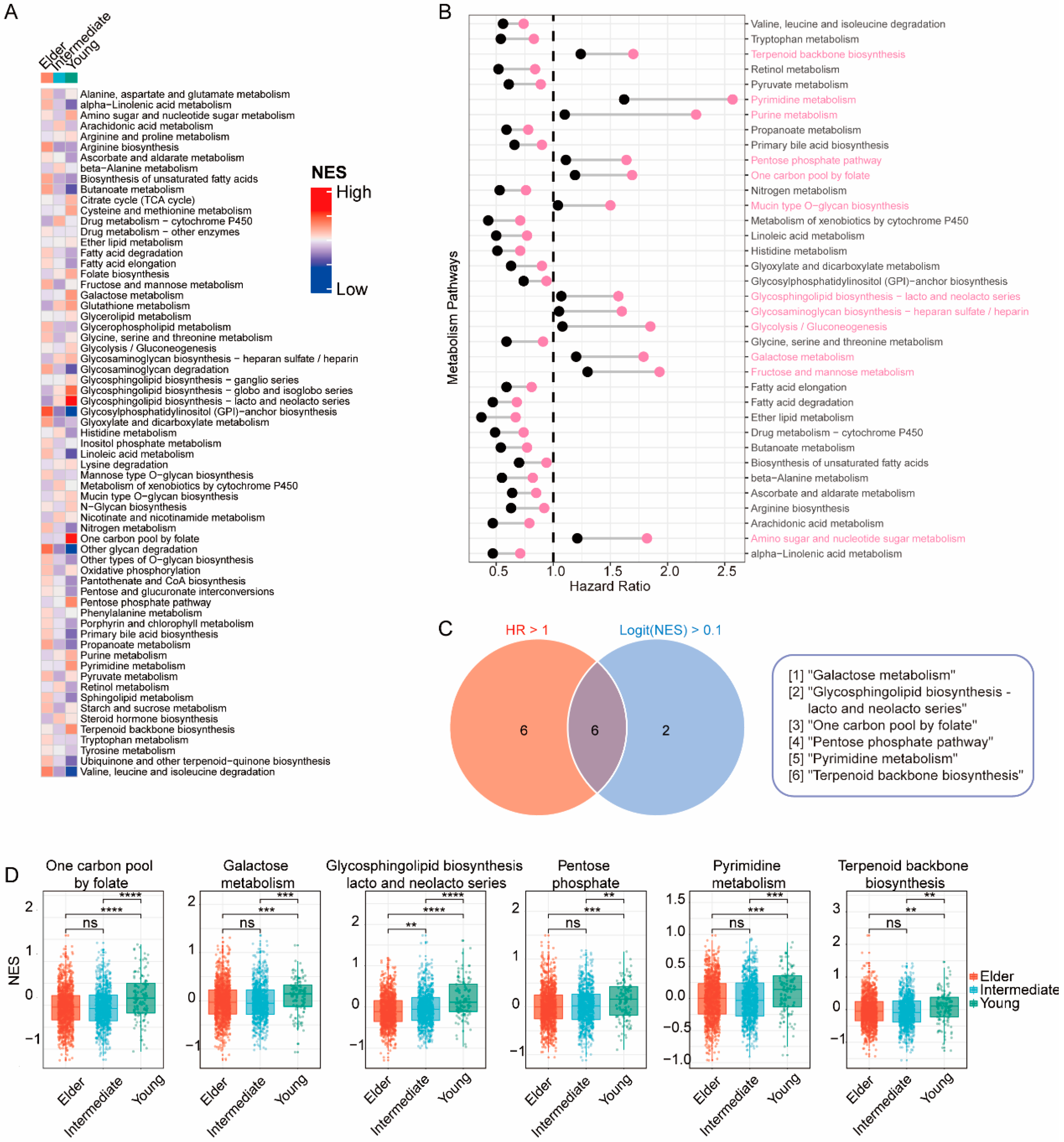
3.3. Transcriptomic Features of BC in Young Patients
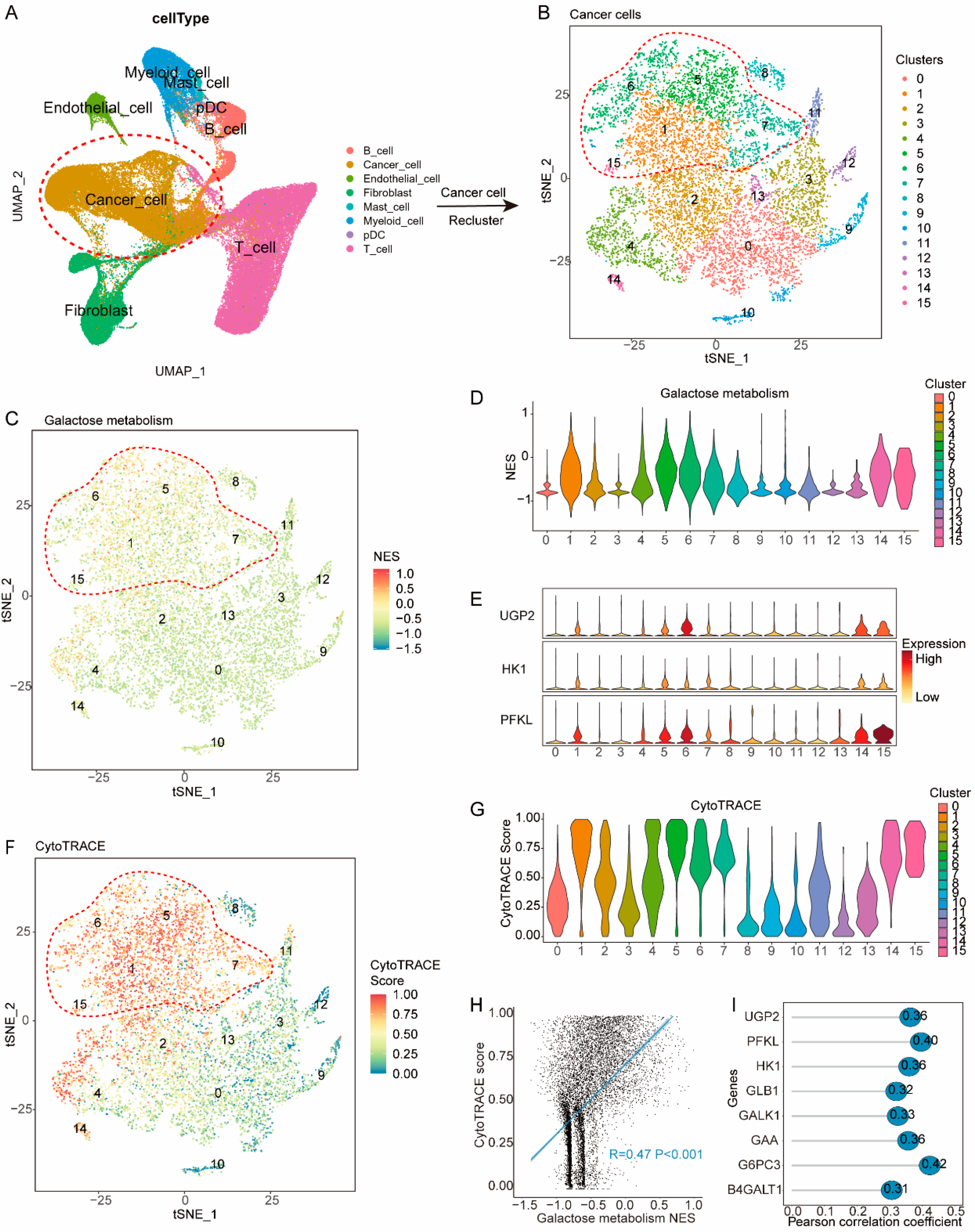
3.4. Galactose Metabolic Pathways Are Upregulated in Less-Differentiated Cancer Cells
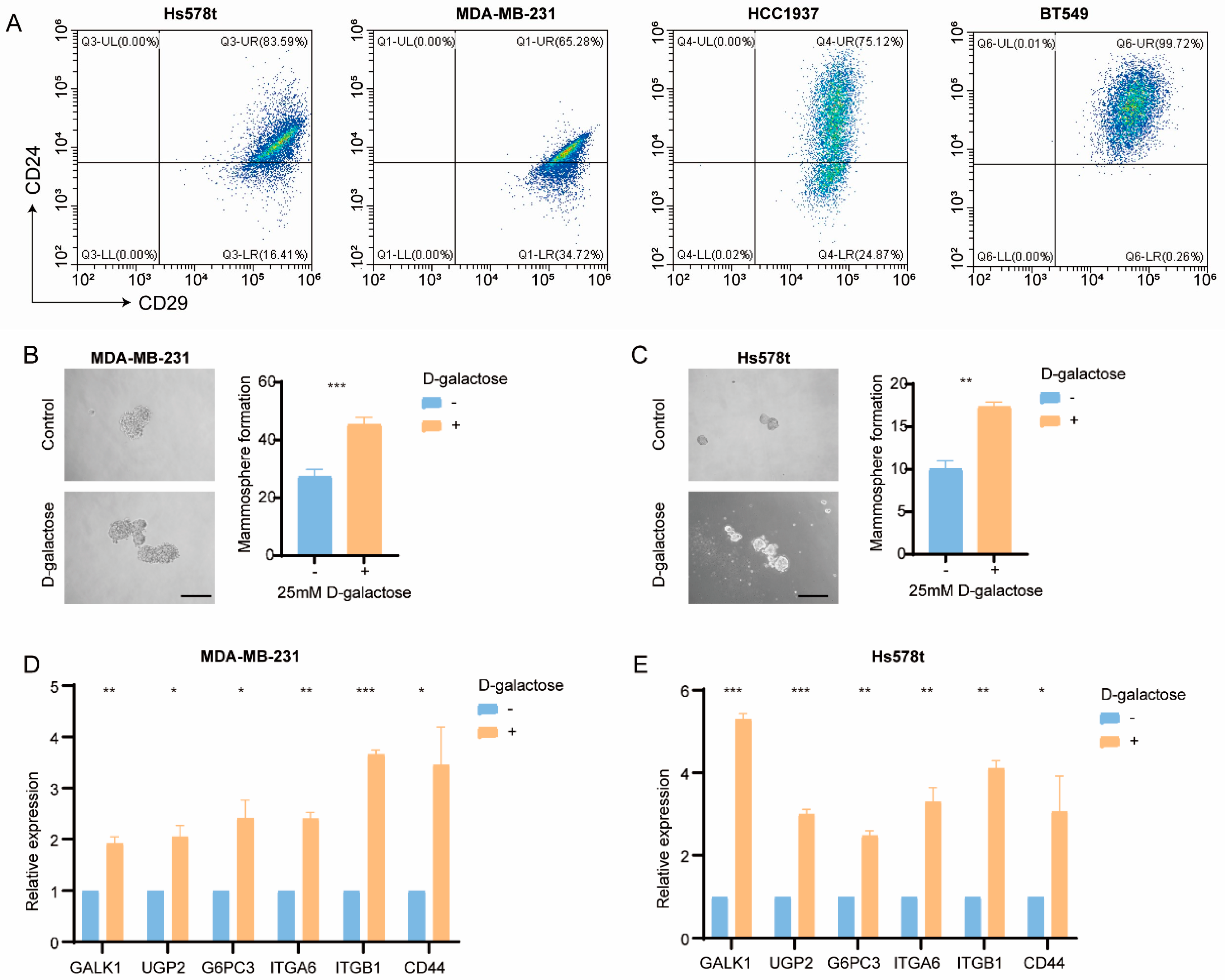
3.5. Galactose Metabolism Regulates Breast Cancer Stemness
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef]
- Tichy, J.R.; Lim, E.; Anders, C.K. Breast Cancer in Adolescents and Young Adults: A Review with a Focus on Biology. J. Natl. Compr. Cancer Netw. 2013, 11, 1060–1069. [Google Scholar] [CrossRef]
- Paluch-Shimon, S.; Cardoso, F.; Partridge, A.; Abulkhair, O.; Azim, H.; Bianchi-Micheli, G.; Cardoso, M.; Curigliano, G.; Gelmon, K.; Gentilini, O.; et al. ESO–ESMO fifth international consensus guidelines for breast cancer in young women (BCY5). Ann. Oncol. 2022, 33, 1097–1118. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Chan, P.S.; Lok, V.; Chen, X.; Ding, H.; Jin, Y.; Yuan, J.; Lao, X.-Q.; Zheng, Z.-J.; Wong, M.C. Global incidence and mortality of breast cancer: A trend analysis. Aging 2021, 13, 5748–5803. [Google Scholar] [CrossRef]
- Eiriz, I.; Batista, M.V.; Tomás, T.C.; Neves, M.; Guerra-Pereira, N.; Braga, S. Breast cancer in very young women—A multicenter 10-year experience. ESMO Open 2021, 6, 100029. [Google Scholar] [CrossRef]
- Hu, X.; Myers, K.S.; Oluyemi, E.T.; Philip, M.; Azizi, A.; Ambinder, E.B. Presentation and characteristics of breast cancer in young women under age 40. Breast Cancer Res. Treat. 2021, 186, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Z.; Al-Eryani, G.; Roden, D.L.; Junankar, S.; Harvey, K.; Andersson, A.; Thennavan, A.; Wang, C.; Torpy, J.R.; Bartonicek, N.; et al. A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 2021, 53, 1334–1347. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-Q.; Gao, Z.-J.; Wu, J.; Zheng, H.-M.; Li, B.; Sun, S.; Meng, X.-Y.; Wu, Q. Single-cell and spatially resolved analysis uncovers cell heterogeneity of breast cancer. J. Hematol. Oncol. 2022, 15, 19. [Google Scholar] [CrossRef]
- Gong, Y.; Ji, P.; Yang, Y.-S.; Xie, S.; Yu, T.-J.; Xiao, Y.; Jin, M.-L.; Ma, D.; Guo, L.-W.; Pei, Y.-C.; et al. Metabolic-Pathway-Based Subtyping of Triple-Negative Breast Cancer Reveals Potential Therapeutic Targets. Cell Metab. 2021, 33, 51.e9–64.e9. [Google Scholar] [CrossRef]
- Yu, T.-J.; Ma, D.; Liu, Y.-Y.; Xiao, Y.; Gong, Y.; Jiang, Y.-Z.; Shao, Z.-M.; Hu, X.; Di, G.-H. Bulk and single-cell transcriptome profiling reveal the metabolic heterogeneity in human breast cancers. Mol. Ther. 2021, 29, 2350–2365. [Google Scholar] [CrossRef]
- Jiang, Y.-Z.; Ma, D.; Suo, C.; Shi, J.; Xue, M.; Hu, X.; Xiao, Y.; Yu, K.-D.; Liu, Y.-R.; Yu, Y.; et al. Genomic and Transcriptomic Landscape of Triple-Negative Breast Cancers: Subtypes and Treatment Strategies. Cancer Cell 2019, 35, 428–440.e5. [Google Scholar] [CrossRef]
- Xiao, Y.; Ma, D.; Yang, Y.-S.; Yang, F.; Ding, J.-H.; Gong, Y.; Jiang, L.; Ge, L.-P.; Wu, S.-Y.; Yu, Q.; et al. Comprehensive metabolomics expands precision medicine for triple-negative breast cancer. Cell Res. 2022, 32, 477–490. [Google Scholar] [CrossRef]
- Yang, F.; Xiao, Y.; Ding, J.-H.; Jin, X.; Ma, D.; Li, D.-Q.; Shi, J.-X.; Huang, W.; Wang, Y.-P.; Jiang, Y.-Z.; et al. Ferroptosis heterogeneity in triple-negative breast cancer reveals an innovative immunotherapy combination strategy. Cell Metab. 2023, 35, 84–100.e8. [Google Scholar] [CrossRef]
- Wang, D.; Hu, X.; Liu, C.; Jia, Y.; Bai, Y.; Cai, C.; Wang, J.; Bai, L.; Yang, R.; Lin, C.; et al. Protein C receptor is a therapeutic stem cell target in a distinct group of breast cancers. Cell Res. 2019, 29, 832–845. [Google Scholar] [CrossRef]
- Hong, S.P.; Chan, T.E.; Lombardo, Y.; Corleone, G.; Rotmensz, N.; Bravaccini, S.; Rocca, A.; Pruneri, G.; McEwen, K.R.; Coombes, R.C.; et al. Single-cell transcriptomics reveals multi-step adaptations to endocrine therapy. Nat. Commun. 2019, 10, 3840. [Google Scholar] [CrossRef]
- Fendler, A.; Bauer, D.; Busch, J.; Jung, K.; Wulf-Goldenberg, A.; Kunz, S.; Song, K.; Myszczyszyn, A.; Elezkurtaj, S.; Erguen, B.; et al. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat. Commun. 2020, 11, 929. [Google Scholar] [CrossRef]
- Liu, S.; Sun, Y.; Hou, Y.; Yang, L.; Wan, X.; Qin, Y.; Liu, Y.; Wang, R.; Zhu, P.; Teng, Y.; et al. A novel lncRNA ROPM-mediated lipid metabolism governs breast cancer stem cell properties. J. Hematol. Oncol. 2021, 14, 178. [Google Scholar] [CrossRef]
- Wang, H.; Xiang, D.; Liu, B.; He, A.; Randle, H.J.; Zhang, K.X.; Dongre, A.; Sachs, N.; Clark, A.P.; Tao, L.; et al. Inadequate DNA Damage Repair Promotes Mammary Transdifferentiation, Leading to BRCA1 Breast Cancer. Cell 2019, 178, 135.e19–151.e19. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.-F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [CrossRef] [PubMed]
- Garofano, L.; Migliozzi, S.; Oh, Y.T.; D’Angelo, F.; Najac, R.D.; Ko, A.; Frangaj, B.; Caruso, F.P.; Yu, K.; Yuan, J.; et al. Pathway-based classification of glioblastoma uncovers a mitochondrial subtype with therapeutic vulnerabilities. Nat. Rev. Cancer 2021, 2, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Gulati, G.S.; Sikandar, S.S.; Wesche, D.J.; Manjunath, A.; Bharadwaj, A.; Berger, M.J.; Ilagan, F.; Kuo, A.H.; Hsieh, R.W.; Cai, S.; et al. Single-cell transcriptional diversity is a hallmark of developmental potential. Science 2020, 367, 405–411. [Google Scholar] [CrossRef]
- Gonzalez, P.S.; O’prey, J.; Cardaci, S.; Barthet, V.J.A.; Sakamaki, J.-I.; Beaumatin, F.; Roseweir, A.; Gay, D.M.; Mackay, G.; Malviya, G.; et al. Mannose impairs tumour growth and enhances chemotherapy. Nature 2018, 563, 719–723. [Google Scholar] [CrossRef]
- Blondeaux, E.; Arecco, L.; Punie, K.; Graffeo, R.; Toss, A.; De Angelis, C.; Trevisan, L.; Buzzatti, G.; Linn, S.C.; Dubsky, P.; et al. Germline TP53 pathogenic variants and breast cancer: A narrative review. Cancer Treat. Rev. 2023, 114, 102522. [Google Scholar] [CrossRef]
- Shahbandi, A.; Nguyen, H.D.; Jackson, J.G. TP53 Mutations and Outcomes in Breast Cancer: Reading beyond the Headlines. Trends Cancer 2020, 6, 98–110. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Wan, Q.; Ouyang, T.; Li, J.; Wang, T.; Fan, Z.; Xie, Y. Risk of ipsilateral breast tumor recurrence and contralateral breast cancer in patients with and without TP53 variant in a large series of breast cancer patients. Breast 2022, 65, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Tung, N.M.; Boughey, J.C.; Pierce, L.J.; Robson, M.E.; Bedrosian, I.; Dietz, J.R.; Dragun, A.; Gelpi, J.B.; Hofstatter, E.W.; Isaacs, C.J.; et al. Management of Hereditary Breast Cancer: American Society of Clinical Oncology, American Society for Radiation Oncology, and Society of Surgical Oncology Guideline. J. Clin. Oncol. 2020, 38, 2080–2106. [Google Scholar] [CrossRef]
- Zardavas, D.; Marvelde, L.T.; Milne, R.L.; Fumagalli, D.; Fountzilas, G.; Kotoula, V.; Razis, E.; Papaxoinis, G.; Joensuu, H.; Moynahan, M.E.; et al. Tumor PIK3CA Genotype and Prognosis in Early-Stage Breast Cancer: A Pooled Analysis of Individual Patient Data. J. Clin. Oncol. 2018, 36, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Mosele, F.; Stefanovska, B.; Lusque, A.; Dien, A.T.; Garberis, I.; Droin, N.; Le Tourneau, C.; Sablin, M.-P.; Lacroix, L.; Enrico, D.; et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann. Oncol. 2020, 31, 377–386. [Google Scholar] [CrossRef]
- Sharpe, M.A.; Ijare, O.B.; Baskin, D.S.; Baskin, A.M.; Baskin, B.N.; Pichumani, K. The Leloir Cycle in Glioblastoma: Galactose Scavenging and Metabolic Remodeling. Cancers 2021, 13, 1815. [Google Scholar] [CrossRef]
- Liu, G.; Wu, X.; Chen, J. Identification and validation of a glycolysis-related gene signature for depicting clinical characteristics and its relationship with tumor immunity in patients with colon cancer. Aging 2022, 14, 8700–8718. [Google Scholar] [CrossRef]
- Wolfe, A.L.; Zhou, Q.; Toska, E.; Galeas, J.; Ku, A.A.; Koche, R.P.; Bandyopadhyay, S.; Scaltriti, M.; Lebrilla, C.B.; McCormick, F.; et al. UDP-glucose pyrophosphorylase 2, a regulator of glycogen synthesis and glycosylation, is critical for pancreatic cancer growth. Proc. Natl. Acad. Sci. USA 2021, 118, e2103592118. [Google Scholar] [CrossRef] [PubMed]
- De Jonge, H.J.M.; Woolthuis, C.M.; Vos, A.Z.; Mulder, A.; Berg, E.v.D.; Kluin, P.M.; van der Weide, K.; de Bont, E.S.J.M.; Huls, G.; Vellenga, E.; et al. Gene expression profiling in the leukemic stem cell-enriched CD34+ fraction identifies target genes that predict prognosis in normal karyotype AML. Leukemia 2011, 25, 1825–1833. [Google Scholar] [CrossRef] [PubMed]
- De Vitis, C.; Corleone, G.; Salvati, V.; Ascenzi, F.; Pallocca, M.; De Nicola, F.; Fanciulli, M.; di Martino, S.; Bruschini, S.; Napoli, C.; et al. B4GALT1 Is a New Candidate to Maintain the Stemness of Lung Cancer Stem Cells. J. Clin. Med. 2019, 8, 1928. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Fang, Z.; Guo, X.; Dong, H.; Zhou, K.; Huang, Z.; Xiao, Z. lncRNA B4GALT1-AS1 promotes colon cancer cell stemness and migration by recruiting YAP to the nucleus and enhancing YAP transcriptional activity. J. Cell. Physiol. 2019, 234, 18524–18534. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, X.; Han, B.; Luo, H.; Ling, H.; Hu, X. Integrated Multi-Omics Profiling of Young Breast Cancer Patients Reveals a Correlation between Galactose Metabolism Pathway and Poor Disease-Free Survival. Cancers 2023, 15, 4637. https://doi.org/10.3390/cancers15184637
Han X, Han B, Luo H, Ling H, Hu X. Integrated Multi-Omics Profiling of Young Breast Cancer Patients Reveals a Correlation between Galactose Metabolism Pathway and Poor Disease-Free Survival. Cancers. 2023; 15(18):4637. https://doi.org/10.3390/cancers15184637
Chicago/Turabian StyleHan, Xiangchen, Boyue Han, Hong Luo, Hong Ling, and Xin Hu. 2023. "Integrated Multi-Omics Profiling of Young Breast Cancer Patients Reveals a Correlation between Galactose Metabolism Pathway and Poor Disease-Free Survival" Cancers 15, no. 18: 4637. https://doi.org/10.3390/cancers15184637
APA StyleHan, X., Han, B., Luo, H., Ling, H., & Hu, X. (2023). Integrated Multi-Omics Profiling of Young Breast Cancer Patients Reveals a Correlation between Galactose Metabolism Pathway and Poor Disease-Free Survival. Cancers, 15(18), 4637. https://doi.org/10.3390/cancers15184637





