Tumor-Promoting Role of GNA14 in Colon Cancer Development
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture
2.2. Transient Transfection of siRNA
2.3. Cell Proliferation Assay
2.4. Wound Healing Assay
2.5. RNA Extraction and Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-qPCR)
2.6. Western Blot Analysis
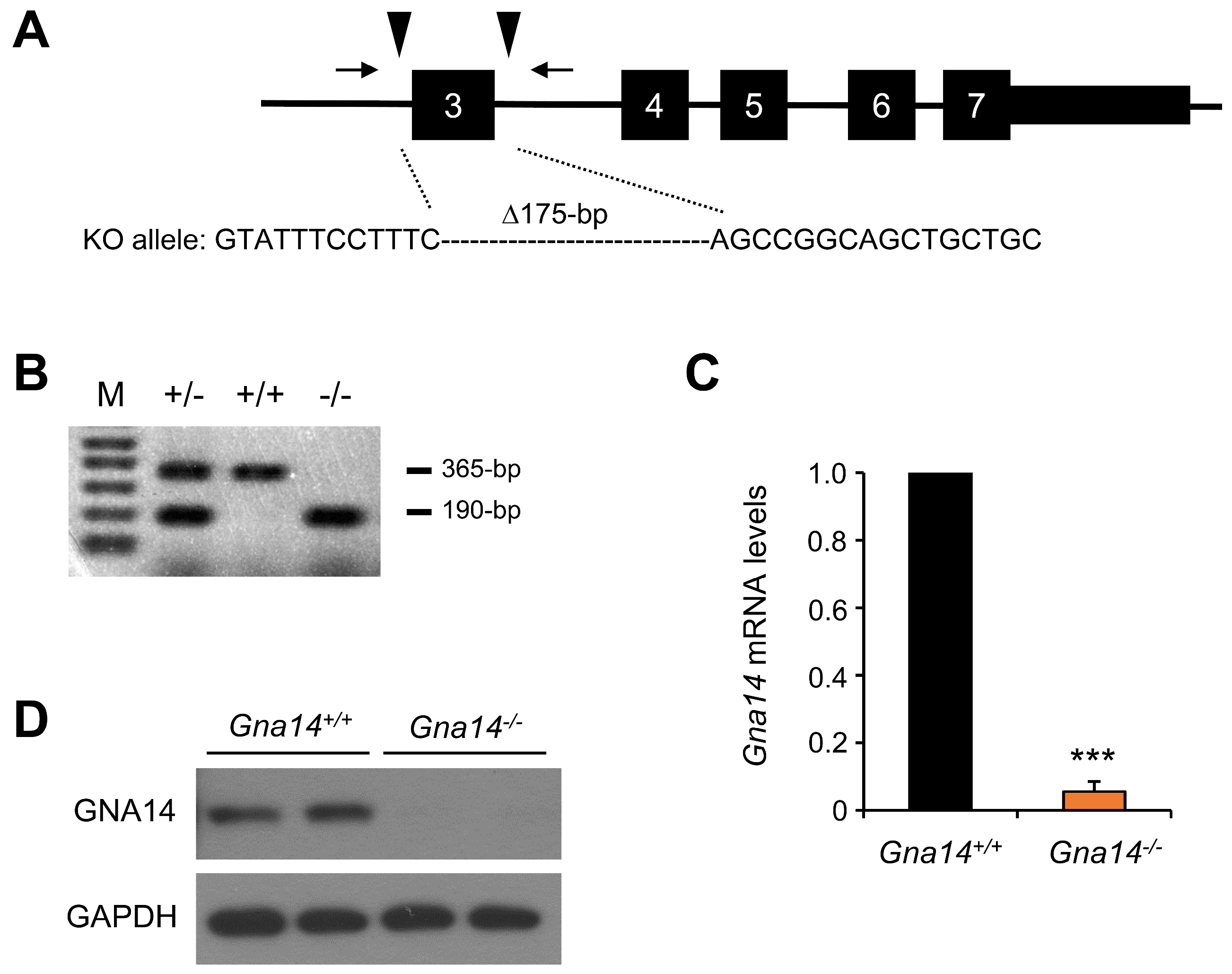
2.7. Construction of Gna14 Knockout Mice Using CRISPR-Cas9 System and Crossing of Mice
2.8. Polyps Counting and Histological Evaluation
2.9. BrdU, TUNEL Staining, and Immunohistochemistry
2.10. Statistics Analysis
3. Results
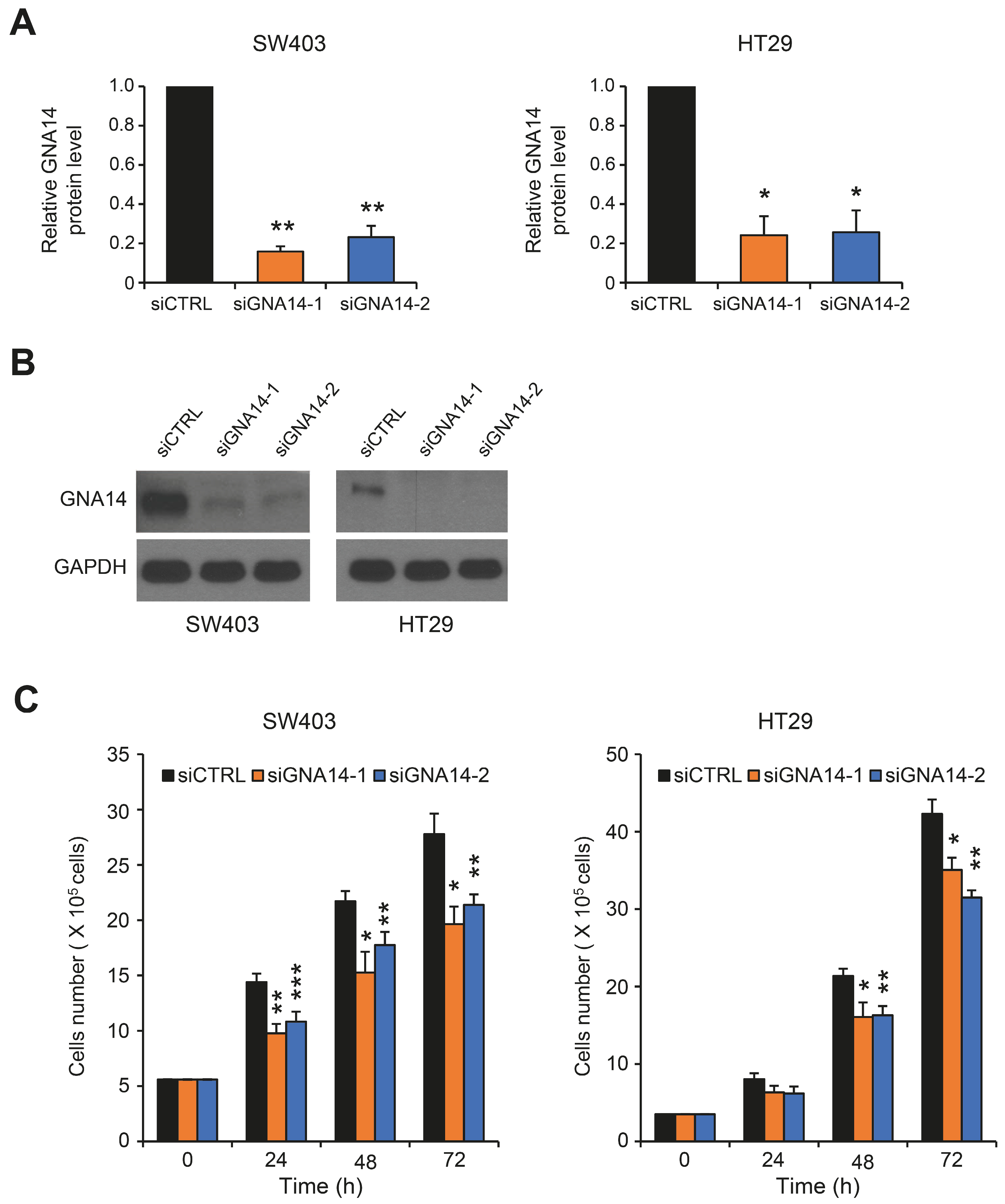
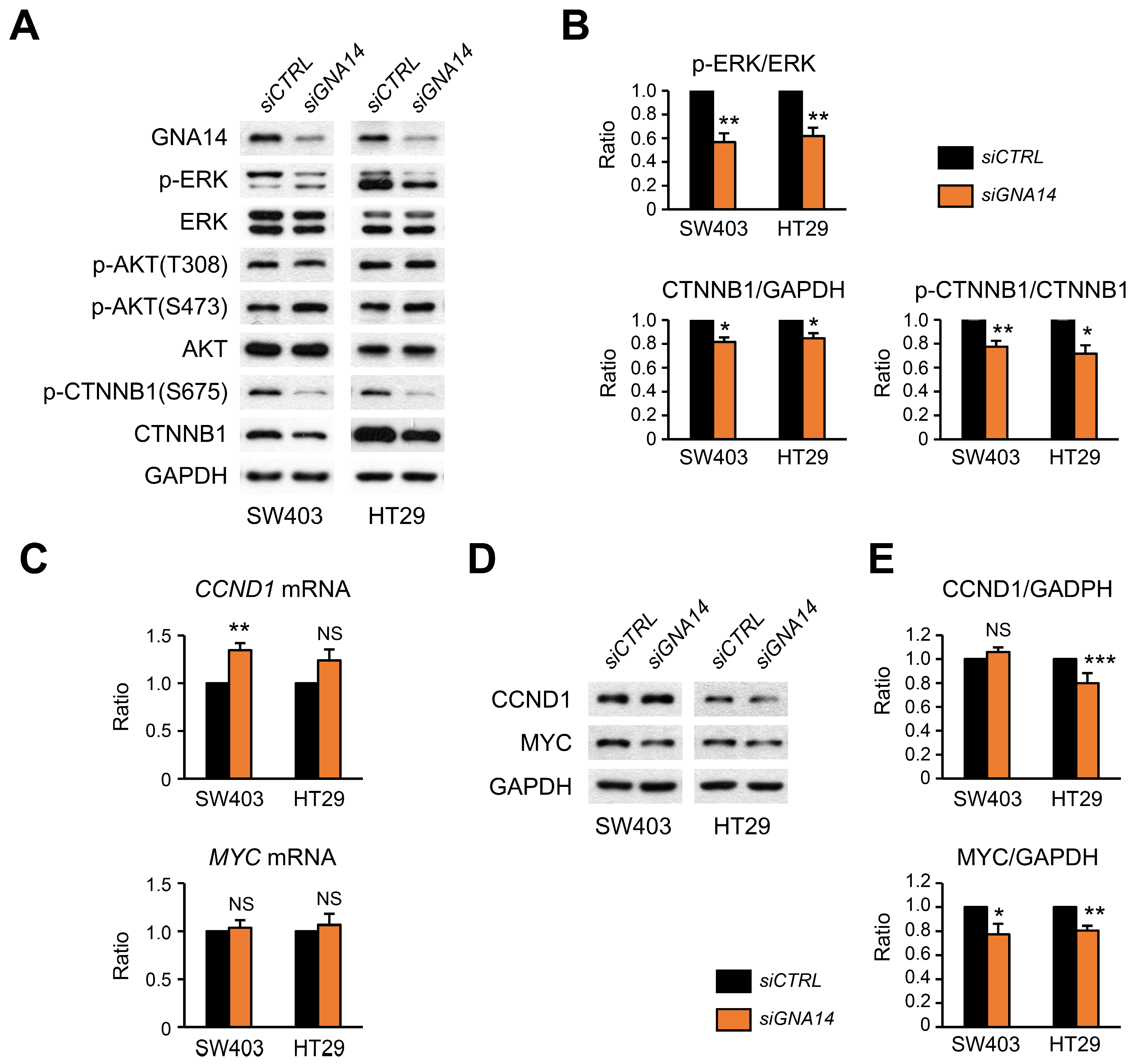
3.1. GNA14 Knockdown Inhibited the Proliferation of Colorectal Cancer (CRC) Cells, but Did Not Affect Cell Migration
3.2. Gna14 Knockout Mice Developed Normally
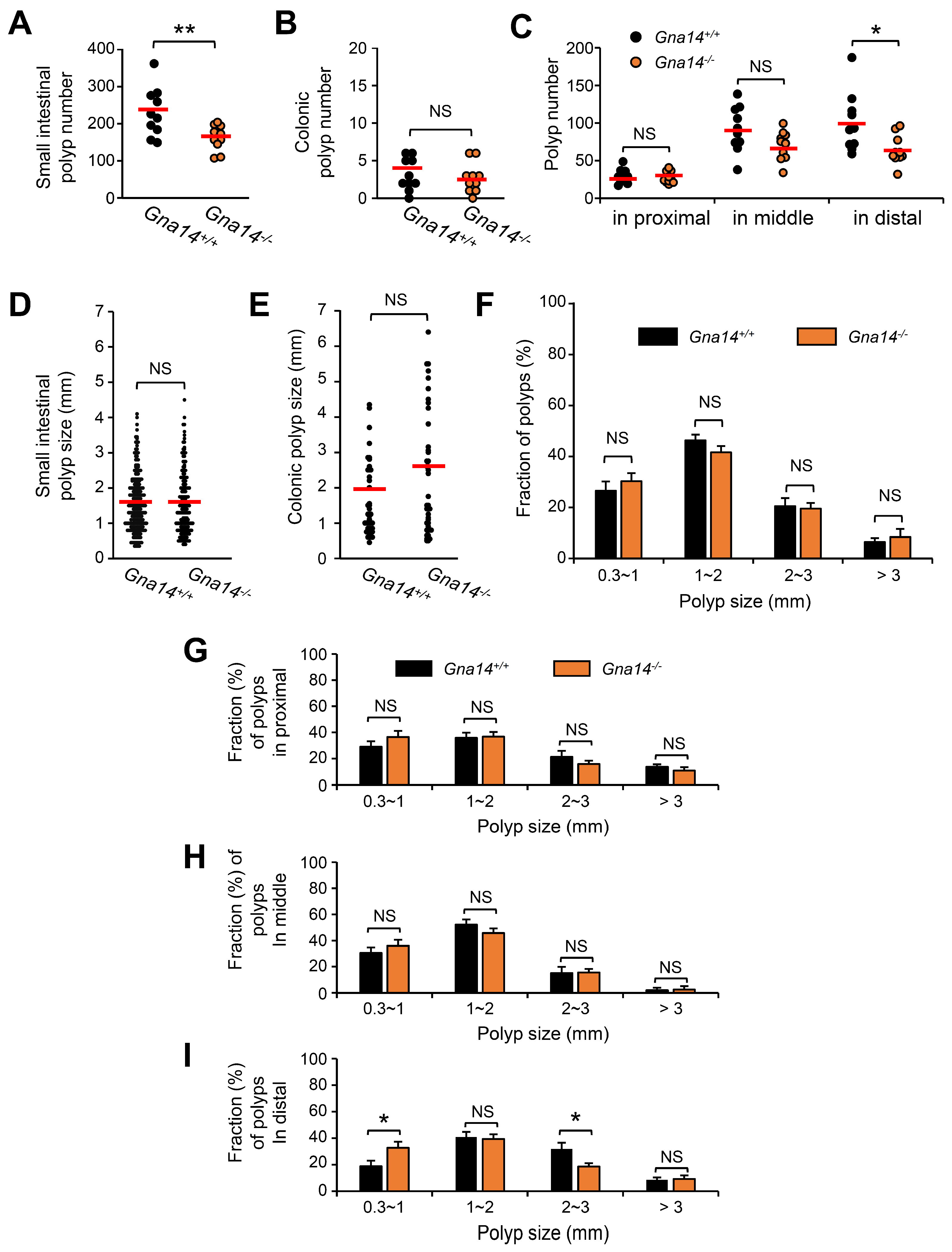
3.3. The Number of Small Intestine Polyps Was Reduced in Gna14 Knockout Mice
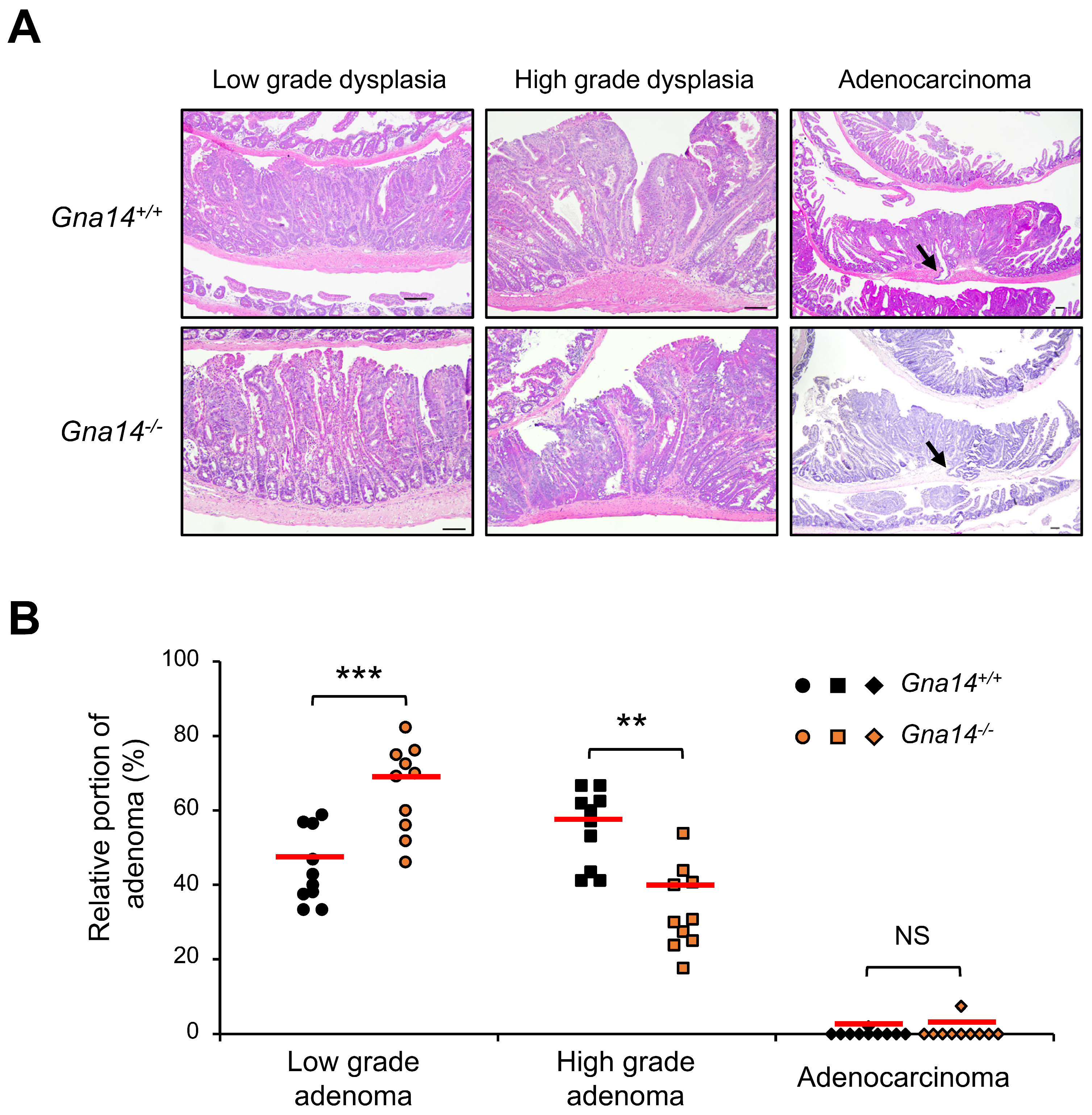
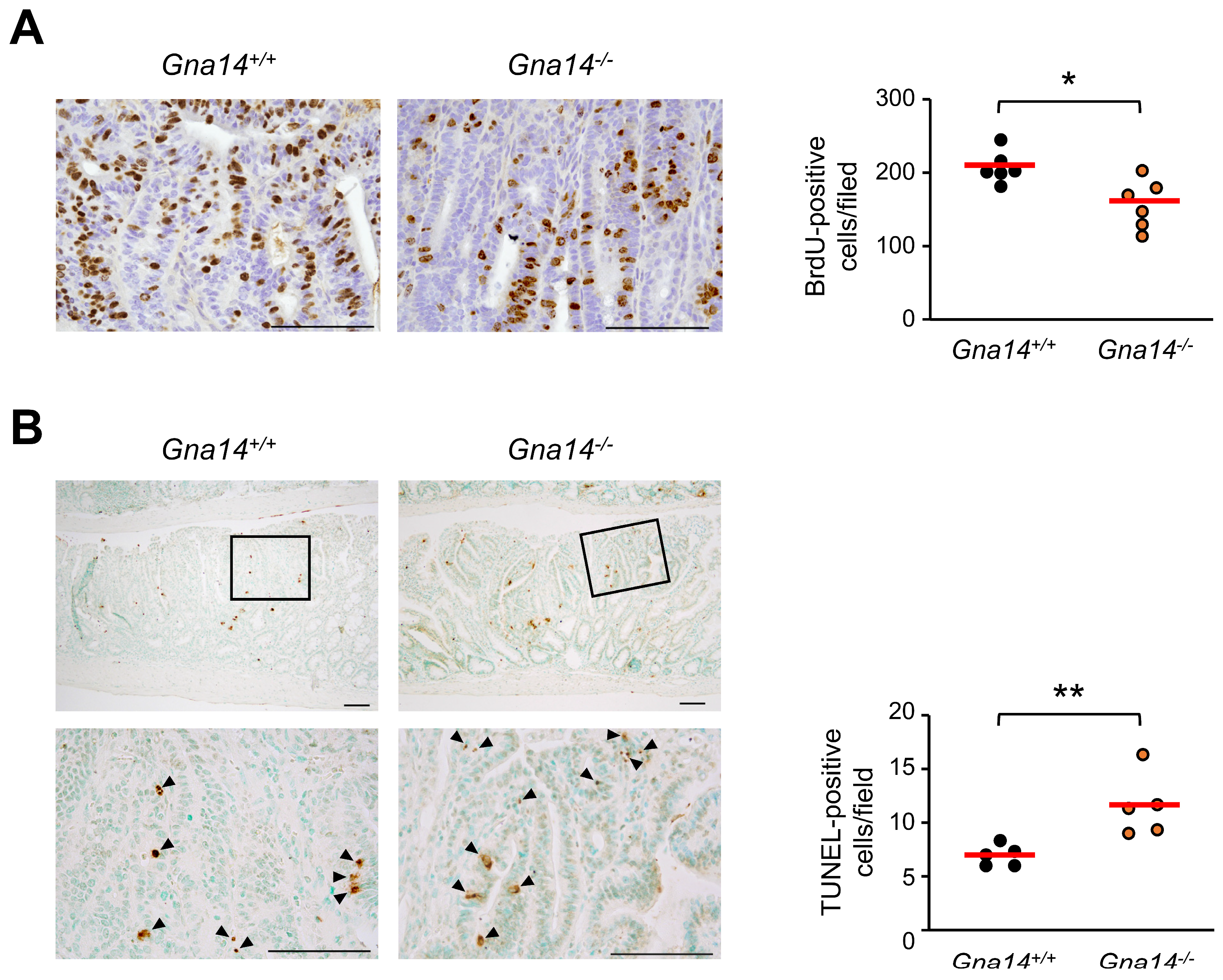
3.4. Tumor Progression was Attenuated in Gna14 Knockout Mice
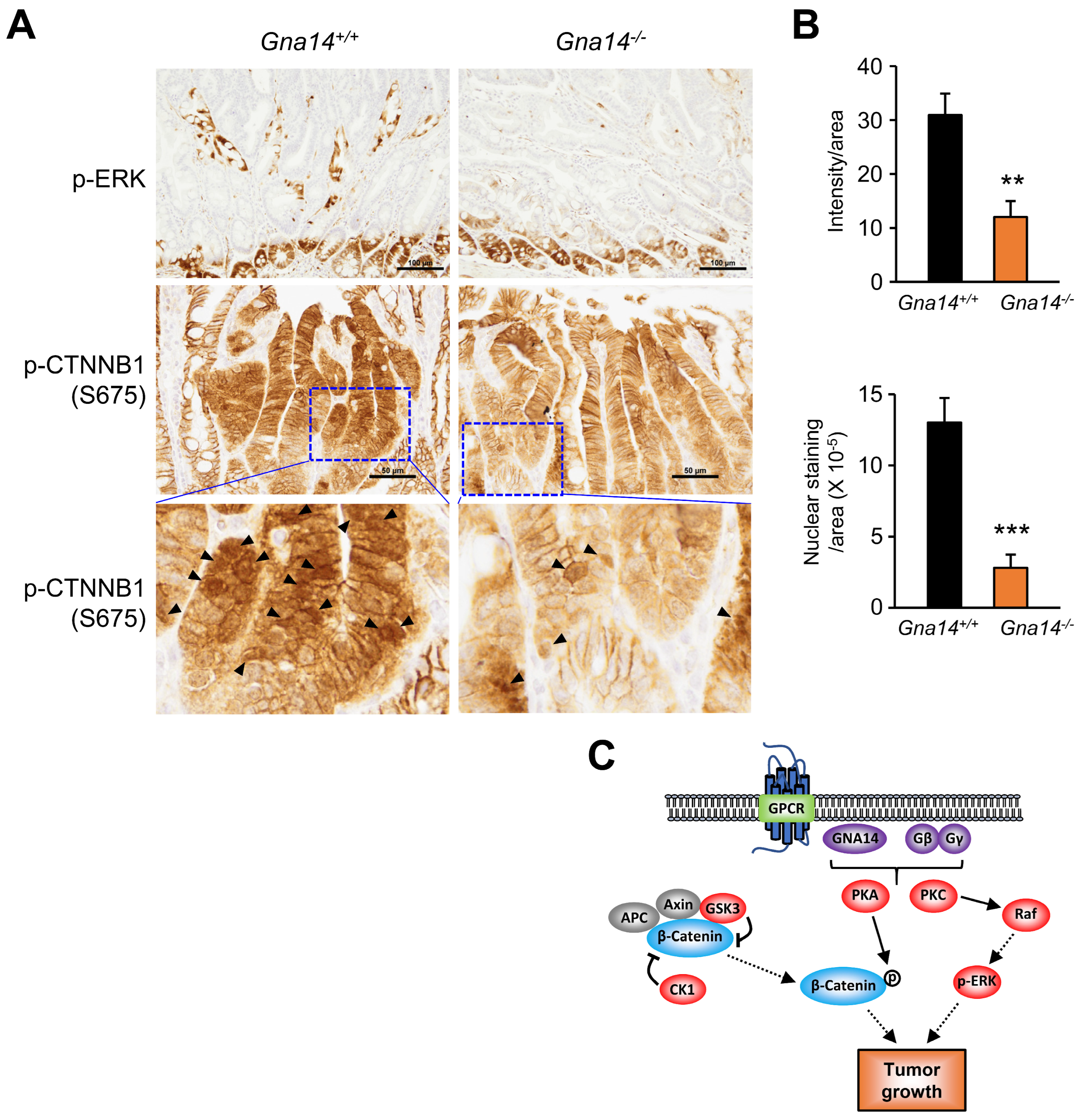
3.5. Decreased Phosphorylation of ERK and CTNNB1 upon GNA14 Reduction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gutkind, J.S. Cell growth control by G protein-coupled receptors: From signal transduction to signal integration. Oncogene 1998, 17, 1331–1342. [Google Scholar] [CrossRef] [PubMed]
- Dorsam, R.T.; Gutkind, J.S. G-protein-coupled receptors and cancer. Nat. Rev. Cancer 2007, 7, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Syrovatkina, V.; Alegre, K.O.; Dey, R.; Huang, X.Y. Regulation, Signaling, and Physiological Functions of G-Proteins. J. Mol. Biol. 2016, 428, 3850–3868. [Google Scholar] [CrossRef]
- Lappano, R.; Maggiolini, M. G protein-coupled receptors: Novel targets for drug discovery in cancer. Nat. Rev. Drug Discov. 2011, 10, 47–60. [Google Scholar] [CrossRef]
- Tennakoon, M.; Senarath, K.; Kankanamge, D.; Ratnayake, K.; Wijayaratna, D.; Olupothage, K.; Ubeysinghe, S.; Martins-Cannavino, K.; Hebert, T.E.; Karunarathne, A. Subtype-dependent regulation of Gbetagamma signalling. Cell Signal. 2021, 82, 109947. [Google Scholar] [CrossRef]
- Arang, N.; Gutkind, J.S. G Protein-Coupled receptors and heterotrimeric G proteins as cancer drivers. FEBS Lett. 2020, 594, 4201–4232. [Google Scholar] [CrossRef]
- Chaudhary, P.K.; Kim, S. An Insight into GPCR and G-Proteins as Cancer Drivers. Cells 2021, 10, 3288. [Google Scholar] [CrossRef]
- Silva-Rodriguez, P.; Fernandez-Diaz, D.; Bande, M.; Pardo, M.; Loidi, L.; Blanco-Teijeiro, M.J. GNAQ and GNA11 Genes: A Comprehensive Review on Oncogenesis, Prognosis and Therapeutic Opportunities in Uveal Melanoma. Cancers 2022, 14, 3066. [Google Scholar] [CrossRef]
- Lim, Y.H.; Bacchiocchi, A.; Qiu, J.; Straub, R.; Bruckner, A.; Bercovitch, L.; Narayan, D.; McNiff, J.; Ko, C.; Robinson-Bostom, L.; et al. GNA14 Somatic Mutation Causes Congenital and Sporadic Vascular Tumors by MAPK Activation. Am. J. Hum. Genet. 2016, 99, 443–450. [Google Scholar] [CrossRef]
- Liau, J.Y.; Lee, J.C.; Tsai, J.H.; Chen, C.C.; Chung, Y.C.; Wang, Y.H. High frequency of GNA14, GNAQ, and GNA11 mutations in cherry hemangioma: A histopathological and molecular study of 85 cases indicating GNA14 as the most commonly mutated gene in vascular neoplasms. Mod. Pathol. 2019, 32, 1657–1665. [Google Scholar] [CrossRef]
- Jansen, P.; Muller, H.; Lodde, G.C.; Zaremba, A.; Moller, I.; Sucker, A.; Paschen, A.; Esser, S.; Schaller, J.; Gunzer, M.; et al. GNA14, GNA11, and GNAQ Mutations Are Frequent in Benign but Not Malignant Cutaneous Vascular Tumors. Front. Genet. 2021, 12, 663272. [Google Scholar] [CrossRef] [PubMed]
- Joseph, N.M.; Brunt, E.M.; Marginean, C.; Nalbantoglu, I.; Snover, D.C.; Thung, S.N.; Yeh, M.M.; Umetsu, S.E.; Ferrell, L.D.; Gill, R.M. Frequent GNAQ and GNA14 Mutations in Hepatic Small Vessel Neoplasm. Am. J. Surg. Pathol. 2018, 42, 1201–1207. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Lv, X.; Xu, F.; Wei, M.; Liu, C.; Yang, Y. GNA14 silencing suppresses the proliferation of endometrial carcinoma cells through inducing apoptosis and G(2)/M cell cycle arrest. Biosci. Rep. 2018, 38, BSR20180574. [Google Scholar] [CrossRef] [PubMed]
- Kwan, D.H.; Yung, L.Y.; Ye, R.D.; Wong, Y.H. Activation of Ras-dependent signaling pathways by G(14) -coupled receptors requires the adaptor protein TPR1. J. Cell. Biochem. 2012, 113, 3486–3497. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Lu, S.; Xie, W. Downregulation of GNA14 in hepatocellular carcinoma indicates an unfavorable prognosis. Oncol. Lett. 2020, 20, 165–172. [Google Scholar] [CrossRef]
- Song, G.; Zhu, X.; Xuan, Z.; Zhao, L.; Dong, H.; Chen, J.; Li, Z.; Song, W.; Jin, C.; Zhou, M.; et al. Hypermethylation of GNA14 and its tumor-suppressive role in hepatitis B virus-related hepatocellular carcinoma. Theranostics 2021, 11, 2318–2333. [Google Scholar] [CrossRef]
- Zanini, S.; Giovinazzo, F.; Alaimo, D.; Lawrence, B.; Pfragner, R.; Bassi, C.; Modlin, I.; Kidd, M. GNA15 expression in small intestinal neuroendocrine neoplasia: Functional and signalling pathway analyses. Cell Signal. 2015, 27, 899–907. [Google Scholar] [CrossRef]
- Innamorati, G.; Wilkie, T.M.; Malpeli, G.; Paiella, S.; Grasso, S.; Rusev, B.; Leone, B.E.; Valenti, M.T.; Carbonare, L.D.; Cheri, S.; et al. Galpha15 in early onset of pancreatic ductal adenocarcinoma. Sci. Rep. 2021, 11, 14922. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- O’Hayre, M.; Vázquez-Prado, J.; Kufareva, I.; Stawiski, E.W.; Handel, T.M.; Seshagiri, S.; Gutkind, J.S. The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat. Rev. Cancer 2013, 13, 412–424. [Google Scholar] [CrossRef]
- Paoni, N.F.; Feldman, M.W.; Gutierrez, L.S.; Ploplis, V.A.; Castellino, F.J. Transcriptional profiling of the transition from normal intestinal epithelia to adenomas and carcinomas in the APCMin/+ mouse. Physiol. Genom. 2003, 15, 228–235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lee, H.; Lee, H.; Chin, H.; Kim, K.; Lee, D. ERBB3 knockdown induces cell cycle arrest and activation of Bak and Bax-dependent apoptosis in colon cancer cells. Oncotarget 2014, 5, 5138–5152. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, A.T.; Lee, S.Y.; Chin, H.J.; Le, Q.V.; Lee, D. Kinase activity of ERBB3 contributes to intestinal organoids growth and intestinal tumorigenesis. Cancer Sci. 2020, 111, 137–147. [Google Scholar] [CrossRef]
- Hashimoto, M.; Yamashita, Y.; Takemoto, T. Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev. Biol. 2016, 418, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Oh, T.I.; Lee, M.; Lee, Y.M.; Kim, G.H.; Lee, D.; You, J.S.; Kim, S.H.; Choi, M.; Jang, H.; Park, Y.M.; et al. PGC1 alpha Loss Promotes Lung Cancer Metastasis through Epithelial-Mesenchymal Transition. Cancers 2021, 13, 1772. [Google Scholar] [CrossRef]
- Berg, K.C.G.; Eide, P.W.; Eilertsen, I.A.; Johannessen, B.; Bruun, J.; Danielsen, S.A.; Bjornslett, M.; Meza-Zepeda, L.A.; Eknaes, M.; Lind, G.E.; et al. Multi-omics of 34 colorectal cancer cell lines—A resource for biomedical studies. Mol. Cancer 2017, 16, 116. [Google Scholar] [CrossRef]
- Zeilstra, J.; Joosten, S.P.J.; Dokter, M.; Verwiel, E.; Spaargaren, M.; Pals, S.T. Deletion of the WNT target and cancer stem cell marker CD44 in Apc(Min/plus) mice attenuates intestinal tumorigenesis. Cancer Res. 2008, 68, 3655–3661. [Google Scholar] [CrossRef]
- Rajamanickam, S.; Velmurugan, B.; Kaur, M.; Singh, R.P.; Agarwal, R. Chemoprevention of intestinal tumorigenesis in APCmin/+ mice by silibinin. Cancer Res. 2010, 70, 2368–2378. [Google Scholar] [CrossRef]
- Smith, J.S.; Lefkowitz, R.J.; Rajagopal, S. Biased signalling: From simple switches to allosteric microprocessors. Nat. Rev. Drug. Discov. 2018, 17, 243–260. [Google Scholar] [CrossRef]
- Wootten, D.; Christopoulos, A.; Marti-Solano, M.; Babu, M.M.; Sexton, P.M. Mechanisms of signalling and biased agonism in G protein-coupled receptors. Nat. Rev. Mol. Cell Biol. 2018, 19, 638–653. [Google Scholar] [CrossRef]
- Nieto Gutierrez, A.; McDonald, P.H. GPCRs: Emerging anti-cancer drug targets. Cell Signal. 2018, 41, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Deka, S.J.; Trivedi, V. Potentials of PKC in Cancer Progression and Anticancer Drug Development. Curr. Drug Discov. Technol. 2019, 16, 135–147. [Google Scholar] [CrossRef] [PubMed]
- Graness, A.; Adomeit, A.; Heinze, R.; Wetzker, R.; Liebmann, C. A novel mitogenic signaling pathway of bradykinin in the human colon carcinoma cell line SW-480 involves sequential activation of a Gq/11 protein, phosphatidylinositol 3-kinase beta, and protein kinase Cepsilon. J. Biol. Chem. 1998, 273, 32016–32022. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.M.; Pei, L.J.; Xia, H.W.; Tang, Q.L.; Bi, F. Role of oncogenic KRAS in the prognosis, diagnosis and treatment of colorectal cancer. Mol. Cancer 2021, 20, 143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Liu, H.T. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002, 12, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shay, J.W. Multiple Roles of APC and its Therapeutic Implications in Colorectal Cancer. J. Natl. Cancer Inst. 2017, 109, djw332. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Q.; Xiao, J.; Niu, C.; Li, Y.; Zhang, X.; Zhou, Z.; Shu, G.; Yin, G. Wnt/beta-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal. Transduct. Target. Ther. 2022, 7, 3. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, X. Advances of Wnt Signalling Pathway in Colorectal Cancer. Cells 2023, 12, 447. [Google Scholar] [CrossRef]
- Luongo, C.; Moser, A.R.; Gledhill, S.; Dove, W.F. Loss of Apc+ in intestinal adenomas from Min mice. Cancer Res. 1994, 54, 5947–5952. [Google Scholar]
- Ren, J.Z.; Sui, H.; Fang, F.F.; Li, Q.; Li, B. The application of Apc(Min/+) mouse model in colorectal tumor researches. J. Cancer Res. Clin. 2019, 145, 1111–1122. [Google Scholar] [CrossRef]
- Yang, M.; Zhong, W.W.; Srivastava, N.; Slavin, A.; Yang, J.; Hoey, T.; An, S. G protein-coupled lysophosphatidic acid receptors stimulate proliferation of colon cancer cells through the beta-catenin pathway. Proc. Natl. Acad. Sci. USA 2005, 102, 6027–6032. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Yu, S.X.; Lu, Y.; Bast, R.C., Jr.; Woodgett, J.R.; Mills, G.B. Phosphorylation and inactivation of glycogen synthase kinase 3 by protein kinase A. Proc. Natl. Acad. Sci. USA 2000, 97, 11960–11965. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Yu, S.; Tanyi, J.L.; Lu, Y.; Woodgett, J.R.; Mills, G.B. Convergence of multiple signaling cascades at glycogen synthase kinase 3: Edg receptor-mediated phosphorylation and inactivation by lysophosphatidic acid through a protein kinase C-dependent intracellular pathway. Mol. Cell. Biol. 2002, 22, 2099–2110. [Google Scholar] [CrossRef] [PubMed]
- Hino, S.; Tanji, C.; Nakayama, K.I.; Kikuchi, A. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase stabilizes beta-catenin through inhibition of its ubiquitination. Mol. Cell. Biol. 2005, 25, 9063–9072. [Google Scholar] [CrossRef] [PubMed]
- Taurin, S.; Sandbo, N.; Qin, Y.; Browning, D.; Dulin, N.O. Phosphorylation of beta-catenin by cyclic AMP-dependent protein kinase. J. Biol. Chem. 2006, 281, 9971–9976. [Google Scholar] [CrossRef]
- Herbst, A.; Jurinovic, V.; Krebs, S.; Thieme, S.E.; Blum, H.; Goke, B.; Kolligs, F.T. Comprehensive analysis of beta-catenin target genes in colorectal carcinoma cell lines with deregulated Wnt/beta-catenin signaling. BMC Genom. 2014, 15, 74. [Google Scholar] [CrossRef]
- Ehyai, S.; Miyake, T.; Williams, D.; Vinayak, J.; Bayfield, M.A.; McDermott, J.C. FMRP recruitment of beta-catenin to the translation pre-initiation complex represses translation. EMBO Rep. 2018, 19, e45536. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Park, R.; Lee, S.; Chin, H.; Nguyen, A.T.-Q.; Lee, D. Tumor-Promoting Role of GNA14 in Colon Cancer Development. Cancers 2023, 15, 4572. https://doi.org/10.3390/cancers15184572
Park R, Lee S, Chin H, Nguyen AT-Q, Lee D. Tumor-Promoting Role of GNA14 in Colon Cancer Development. Cancers. 2023; 15(18):4572. https://doi.org/10.3390/cancers15184572
Chicago/Turabian StylePark, Rahui, Seungmin Lee, Hyunjung Chin, Anh Thai-Quynh Nguyen, and Daekee Lee. 2023. "Tumor-Promoting Role of GNA14 in Colon Cancer Development" Cancers 15, no. 18: 4572. https://doi.org/10.3390/cancers15184572
APA StylePark, R., Lee, S., Chin, H., Nguyen, A. T.-Q., & Lee, D. (2023). Tumor-Promoting Role of GNA14 in Colon Cancer Development. Cancers, 15(18), 4572. https://doi.org/10.3390/cancers15184572




