A Contrast-Enhanced CT-Based Deep Learning System for Preoperative Prediction of Colorectal Cancer Staging and RAS Mutation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. CT Image Acquisition
2.3. CT Images Collection
2.4. Dataset Construction
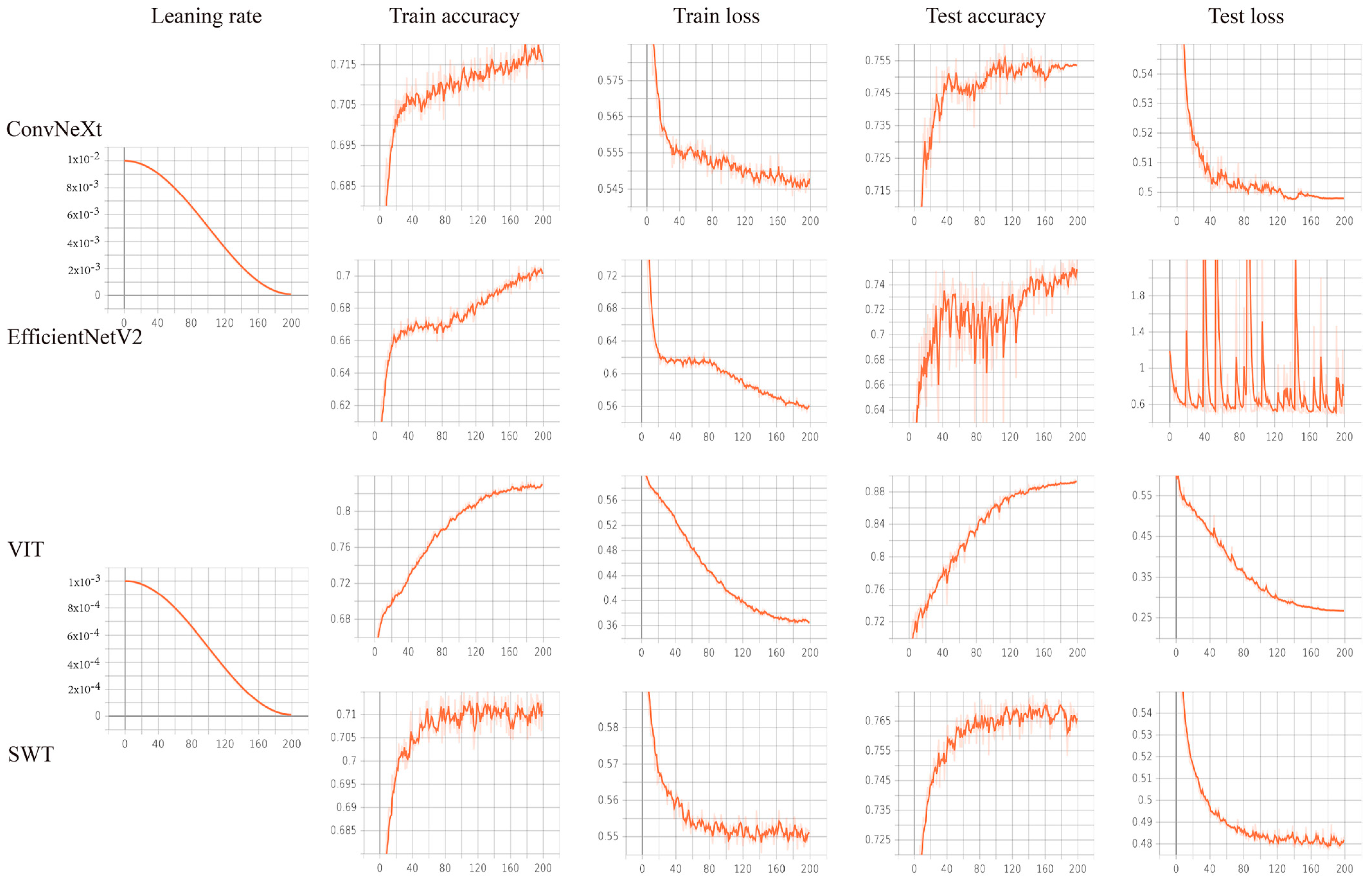
2.5. Model Construction
2.6. Model Evaluation
3. Results
3.1. Patients
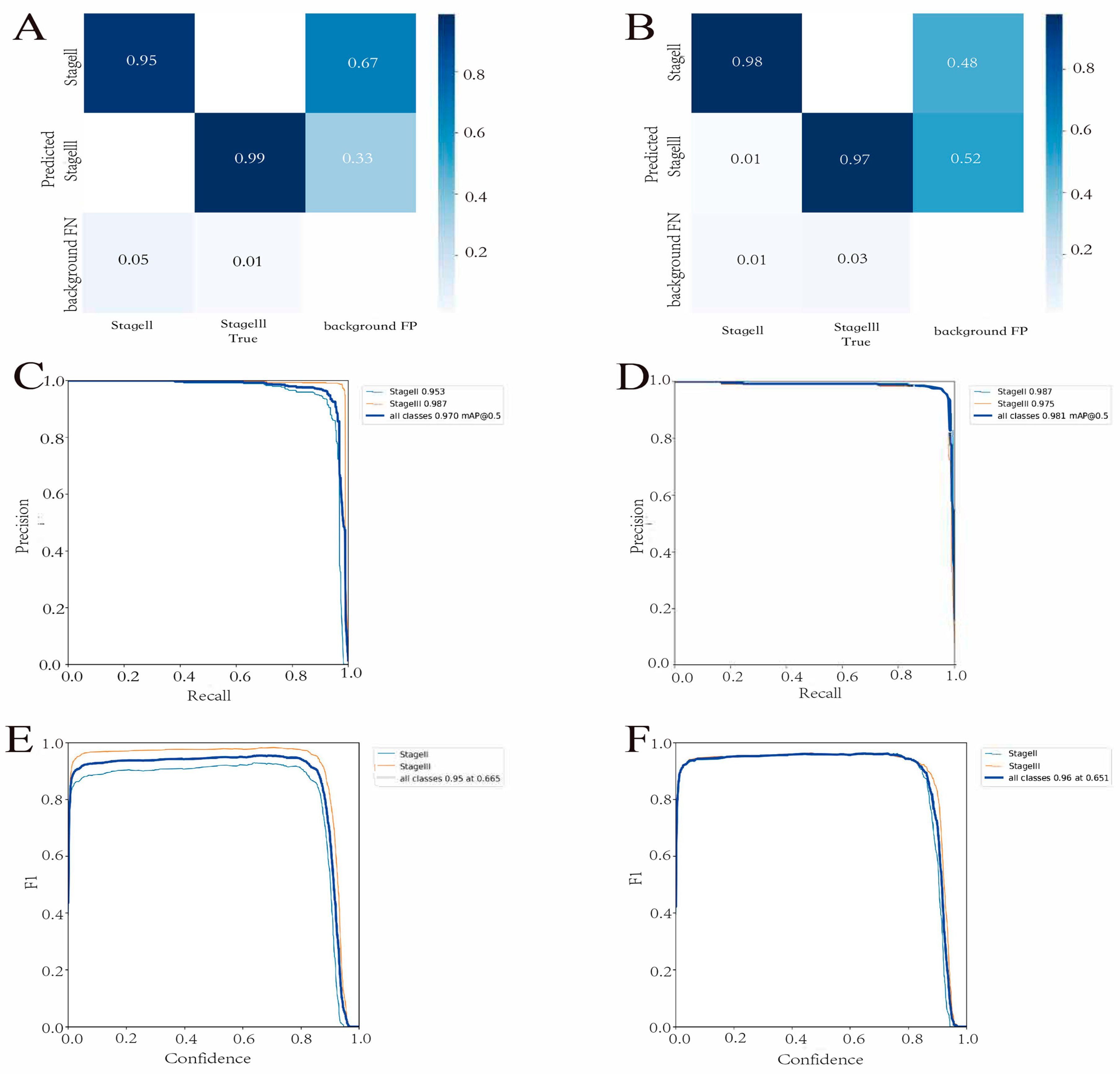
3.2. Detection Model Performance
3.3. Prediction Model Performance
3.4. Deep Learning System
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Cremolini, C.; Loupakis, F.; Antoniotti, C.; Lupi, C.; Sensi, E.; Lonardi, S.; Mezi, S.; Tomasello, G.; Ronzoni, M.; Zaniboni, A.; et al. FOLFOXIRI plus bevacizumab versus FOLFIRI plus bevacizumab as first-line treatment of patients with metastatic colorectal cancer: Updated overall survival and molecular subgroup analyses of the open-label, phase 3 TRIBE study. Lancet Oncol. 2015, 16, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Strickler, J.H.; Wu, C.; Bekaii-Saab, T. Targeting BRAF in metastatic colorectal cancer: Maximizing molecular approaches. Cancer Treat. Rev. 2017, 60, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Sundar, R.; Hong, D.S.; Kopetz, S.; Yap, T.A. Targeting BRAF-Mutant Colorectal Cancer: Progress in Combination Strategies. Cancer Discov. 2017, 7, 558–560. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bibault, J.E.; Giraud, P.; Housset, M.; Durdux, C.; Taieb, J.; Berger, A.; Coriat, R.; Chaussade, S.; Dousset, B.; Nordlinger, B.; et al. Deep Learning and Radiomics predict complete response after neo-adjuvant chemoradiation for locally advanced rectal cancer. Sci. Rep. 2018, 8, 12611. [Google Scholar] [CrossRef]
- Camidge, D.R.; Doebele, R.C.; Kerr, K.M. Comparing and contrasting predictive biomarkers for immunotherapy and targeted therapy of NSCLC. Nature reviews. Clin. Oncol. 2019, 16, 341–355. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Chen, S.; Yi, Z.; Lan, B.; Xing, P.; Fan, Y.; Wang, J.; Luo, Y.; Yuan, P.; et al. Safety, Efficacy, and Biomarker Analysis of Pyrotinib in Combination with Capecitabine in HER2-Positive Metastatic Breast Cancer Patients: A Phase I Clinical Trial. Clin. Cancer Res. 2019, 25, 5212–5220. [Google Scholar] [CrossRef]
- Barras, D.; Missiaglia, E.; Wirapati, P.; Sieber, O.M.; Jorissen, R.N.; Love, C.; Molloy, P.L.; Jones, I.T.; McLaughlin, S.; Gibbs, P.; et al. BRAF V600E Mutant Colorectal Cancer Subtypes Based on Gene Expression. Clin. Cancer Res. 2017, 23, 104–115. [Google Scholar] [CrossRef]
- Peeters, M.; Oliner, K.S.; Price, T.J.; Cervantes, A.; Sobrero, A.F.; Ducreux, M.; Hotko, Y.; André, T.; Chan, E.; Lordick, F.; et al. Analysis of KRAS/NRAS Mutations in a Phase III Study of Panitumumab with FOLFIRI Compared with FOLFIRI Alone as Second-line Treatment for Metastatic Colorectal Cancer. Clin. Cancer Res. 2015, 21, 5469–5479. [Google Scholar] [CrossRef]
- Jia, L.L.; Zhao, J.X.; Zhao, L.P.; Tian, J.H.; Huang, G. Current status and quality of radiomic studies for predicting KRAS mutations in colorectal cancer patients: A systematic review and meta-analysis. Eur. J. Radiol. 2023, 158, 110640. [Google Scholar] [CrossRef]
- European Society of Radiology (ESR). White paper on imaging biomarkers. Insights Imaging 2010, 1, 42–45. [Google Scholar] [CrossRef]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Huang, S.; Yang, J.; Fong, S.; Zhao, Q. Artificial intelligence in cancer diagnosis and prognosis: Opportunities and challenges. Cancer Lett. 2020, 471, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Deo, R.C. Machine Learning in Medicine. Circulation 2015, 132, 1920–1930. [Google Scholar] [CrossRef] [PubMed]
- Wong, D.; Yip, S. Machine learning classifies cancer. Nature 2018, 555, 446–447. [Google Scholar] [CrossRef]
- Cellina, M.; Cè, M.; Irmici, G.; Ascenti, V.; Khenkina, N.; Toto-Brocchi, M.; Martinenghi, C.; Papa, S.; Carrafiello, G. Artificial Intelligence in Lung Cancer Imaging: Unfolding the Future. Diagnostics 2022, 12, 2644. [Google Scholar] [CrossRef]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.S.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying Medical Diagnoses and Treatable Diseases by Image-Based Deep Learning. Cell 2018, 172, 1122–1131.e9. [Google Scholar] [CrossRef] [PubMed]
- Jiang, K.; Jiang, X.; Pan, J.; Wen, Y.; Huang, Y.; Weng, S.; Lan, S.; Nie, K.; Zheng, Z.; Ji, S.; et al. Current Evidence and Future Perspective of Accuracy of Artificial Intelligence Application for Early Gastric Cancer Diagnosis with Endoscopy: A Systematic and Meta-Analysis. Front. Med. 2021, 8, 629080. [Google Scholar] [CrossRef]
- Kim, K.; Kim, S.; Han, K.; Bae, H.; Shin, J.; Lim, J.S. Diagnostic Performance of Deep Learning-Based Lesion Detection Algorithm in CT for Detecting Hepatic Metastasis from Colorectal Cancer. Korean J. Radiol. 2021, 22, 912–921. [Google Scholar] [CrossRef]
- Huang, Y.Q.; Liang, C.H.; He, L.; Tian, J.; Liang, C.S.; Chen, X.; Ma, Z.L.; Liu, Z.Y. Development and Validation of a Radiomics Nomogram for Preoperative Prediction of Lymph Node Metastasis in Colorectal Cancer. J. Clin. Oncol. 2016, 34, 2157–2164. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Z.; Zheng, L.; Kang, G.; Li, S.; Yang, Y. Random Erasing Data Augmentation. In Proceedings of the AAAI Conference on Artificial Intelligence, New York, NY, USA, 7–12 February 2020; Volume 34, pp. 13001–13008. [Google Scholar] [CrossRef]
- Wang, C.Y.; Bochkovskiy, A.; Liao, H.Y.M. YOLOv7: Trainable bag-of-freebies sets new state-of-the-art for real-time object detectors. arXiv 2022, arXiv:2207.02696. [Google Scholar]
- Drozdzal, M.; Chartrand, G.; Vorontsov, E.; Shakeri, M.; Di Jorio, L.; Tang, A.; Romero, A.; Bengio, Y.; Pal, C.; Kadoury, S. Learning normalized inputs for iterative estimation in medical image segmentation. Med. Image Anal. 2018, 44, 1–13. [Google Scholar] [CrossRef]
- Tan, M.; Le, Q.V. Efficientnet: Rethinking Model Scaling for Convolutional Neural Networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A convnet for the 2020s. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, New Orleans, LA, USA, 18–24 June 2022; pp. 11976–11986. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. In Proceedings of the International Conference on Learning Representations, Virtual, 3–7 May 2021. [Google Scholar]
- Liu, Z.; Lin, Y.; Cao, Y.; Hu, H.; Wei, Y.; Zhang, Z.; Lin, S.; Guo, B. Swin transformer: Hierarchical vision transformer using shifted windows. arXiv 2021, arXiv:2103.14030. [Google Scholar]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Proceedings of the 31st International Conference on Neural Information Processing Systems, NIPS’17, Long Beach, CA, USA, 4–9 December 2017; Curran Associates Inc.: Red Hook, NY, USA, 2017; pp. 6000–6010. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Russakovsky, O.; Deng, J.; Su, H.; Krause, J.; Satheesh, S.; Ma, S.; Huang, Z.; Karpathy, A.; Khosla, A.; Bernstein, M.; et al. ImageNet Large Scale Visual Recognition Challenge. Int. J. Comput. Vis. 2015, 115, 211–252. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Das, A.; Vedantam, R.; Cogswell, M.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 2016, 128, 336–359. [Google Scholar] [CrossRef]
- National Comprehensive Cancer Network (NCCN) Guidelines. Available online: http://www.nccn.org/ (accessed on 11 January 2022).
- Nasseri, Y.; Langenfeld, S.J. Imaging for Colorectal Cancer. Surg. Clin. N. Am. 2017, 97, 503–513. [Google Scholar] [CrossRef]
- Levy, I.; Gralnek, I.M. Complications of diagnostic colonoscopy, upper endoscopy, and enteroscopy. Best Pract. Res. Clin. Gastroenterol. 2016, 30, 705–718. [Google Scholar] [CrossRef]
- He, P.; Zou, Y.; Qiu, J.; Yang, T.; Peng, L.; Zhang, X. Pretreatment (18)F-FDG PET/CT Imaging Predicts the KRAS/NRAS/BRAF Gene Mutational Status in Colorectal Cancer. J. Oncol. 2021, 2021, 6687291. [Google Scholar] [CrossRef]
- Obaro, A.E.; Plumb, A.A.; Fanshawe, T.R.; Torres, U.S.; Baldwin-Cleland, R.; Taylor, S.A.; Halligan, S.; Burling, D.N. Post-imaging colorectal cancer or interval cancer rates after CT colonography: A systematic review and meta-analysis. Lancet Gastroenterol. Hepatol. 2018, 3, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Dong, D.; Fang, M.-J.; Tang, L.; Shan, X.-H.; Gao, J.-B.; Giganti, F.; Wang, R.-P.; Chen, X.; Wang, X.-X.; Palumbo, D.; et al. Deep Learning Radiomic Nomogram Can Predict the Number of Lymph Node Metastasis in Locally Advanced Gastric Cancer: An International Multicenter Study. Ann. Oncol. 2020, 31, 912–920. [Google Scholar] [CrossRef] [PubMed]
- Pacal, I.; Karaboga, D.; Basturk, A.; Akay, B.; Nalbantoglu, U. A comprehensive review of deep learning in colon cancer. Comput. Biol. Med. 2020, 126, 104003. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Wang, S.; Zhang, K.; Liu, T.J.; Li, J.N. Development of artificial intelligence technology in diagnosis, treatment, and prognosis of colorectal cancer. World J. Gastrointest. Oncol. 2022, 14, 124–152. [Google Scholar] [CrossRef] [PubMed]
- Bedrikovetski, S.; Dudi-Venkata, N.N.; Kroon, H.M.; Seow, W.; Vather, R.; Carneiro, G.; Moore, J.W.; Sammour, T. Artificial intelligence for pre-operative lymph node staging in colorectal cancer: A systematic review and meta-analysis. BMC Cancer 2021, 21, 1058. [Google Scholar] [CrossRef]
- Minami, S.; Saso, K.; Miyoshi, N.; Fujino, S.; Kato, S.; Sekido, Y.; Hata, T.; Ogino, T.; Takahashi, H.; Uemura, M.; et al. Diagnosis of Depth of Submucosal Invasion in Colorectal Cancer with AI Using Deep Learning. Cancers 2022, 14, 5361. [Google Scholar] [CrossRef]
- Wu, Q.Y.; Liu, S.L.; Sun, P.; Li, Y.; Liu, G.W.; Liu, S.S.; Hu, J.L.; Niu, T.Y.; Lu, Y. Establishment and clinical application value of an automatic diagnosis platform for rectal cancer T-staging based on a deep neural network. Chin. Med. J. 2021, 134, 821–828. [Google Scholar] [CrossRef]
- Hou, M.; Zhou, L.; Sun, J. Deep-learning-based 3D super-resolution MRI radiomics model: Superior predictive performance in preoperative T-staging of rectal cancer. Eur. Radiol. 2023, 33, 1–10. [Google Scholar] [CrossRef]
- AK, A.A.; Garvin, J.H.; Redd, A.; Carter, M.E.; Sweeny, C.; Meystre, S.M. Automated Extraction and Classification of Cancer Stage Mentions from Unstructured Text Fields in a Central Cancer Registry. AMIA Jt. Summits Transl. Sci. Proc. 2018, 2017, 16–25. [Google Scholar]
- Lu, Y.; Yu, Q.; Gao, Y.; Zhou, Y.; Liu, G.; Dong, Q.; Ma, J.; Ding, L.; Yao, H.; Zhang, Z.; et al. Identification of Metastatic Lymph Nodes in MR Imaging with Faster Region-Based Convolutional Neural Networks. Cancer Res. 2018, 78, 5135–5143. [Google Scholar] [CrossRef]
- Kubota, K.; Suzuki, A.; Shiozaki, H.; Wada, T.; Kyosaka, T.; Kishida, A. Accuracy of Multidetector-Row Computed Tomography in the Preoperative Diagnosis of Lymph Node Metastasis in Patients with Gastric Cancer. Gastrointest. Tumors 2017, 3, 163–170. [Google Scholar] [CrossRef]
- Joo, I.; Lee, J.M.; Kim, J.H.; Shin, C.-I.; Han, J.K.; Choi, B.I. Prospective Comparison of 3T MRI with Diffusion-Weighted Imaging and MDCT for the Preoperative TNM Staging of Gastric Cancer. J. Magn. Reson. Imaging 2015, 41, 814–821. [Google Scholar] [CrossRef]
- Zheng, L.; Zhang, X.; Hu, J.; Gao, Y.; Zhang, X.; Zhang, M.; Li, S.; Zhou, X.; Niu, T.; Lu, Y.; et al. Establishment and Applicability of a Diagnostic System for Advanced Gastric Cancer T Staging Based on a Faster Region-Based Convolutional Neural Network. Front. Oncol. 2020, 10, 1238. [Google Scholar] [CrossRef]
- Tang, Y.L.; Li, D.D.; Duan, J.Y.; Sheng, L.M.; Wang, X. Resistance to targeted therapy in metastatic colorectal cancer: Current status and new developments. World J. Gastroenterol. 2023, 29, 926–948. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Jiang, Q.; Guo, T.; Wu, X.; Wang, Q.; Feng, Y.; Zhang, S.; Fang, W.; Zhou, W.; Yang, A. Endoscopic characteristics in predicting prognosis of biopsy-diagnosed gastric low-grade intraepithelial neoplasia. Chin. Med. J. 2022, 135, 26–35. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Wang, X.; Li, W.; Ye, H.; Bai, H.; Wu, J.; Chen, M. Contrast-Enhanced CT Parameters of Gastric Adenocarcinoma: Can Radiomic Features Be Surrogate Biomarkers for HER2 over-Expression Status? Cancer Manag. Res. 2020, 12, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Kalligosfyri, P.M.; Nikou, S.; Karteri, S.; Kalofonos, H.P.; Bravou, V.; Kalogianni, D.P. Rapid Multiplex Strip Test for the Detection of Circulating Tumor DNA Mutations for Liquid Biopsy Applications. Biosensors 2022, 12, 97. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wuethrich, A.; Sina, A.A.; Lane, R.E.; Lin, L.L.; Wang, Y.; Cebon, J.; Behren, A.; Trau, M. Tracking extracellular vesicle phenotypic changes enables treatment monitoring in melanoma. Sci. Adv. 2020, 6, eaax3223. [Google Scholar] [CrossRef]
- Chang, X.; Guo, X.; Li, X.; Han, X.; Li, X.; Liu, X.; Ren, J. Potential Value of Radiomics in the Identification of Stage T3 and T4a Esophagogastric Junction Adenocarcinoma Based on Contrast-Enhanced CT Images. Front. Oncol. 2021, 11, 627947. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Kim, J.; Balagurunathan, Y.; Li, Q.; Garcia, A.L.; Stringfield, O.; Ye, Z.; Gillies, R.J. Radiomic Features Are Associated with EGFR Mutation Status in Lung Adenocarcinomas. Clin. Lung Cancer 2016, 17, 441–448.e6. [Google Scholar] [CrossRef]
- Russo, M.; Crisafulli, G.; Sogari, A.; Reilly, N.M.; Arena, S.; Lamba, S.; Bartolini, A.; Amodio, V.; Magrì, A.; Novara, L.; et al. Adaptive mutability of colorectal cancers in response to targeted therapies. Science 2019, 366, 1473–1480. [Google Scholar] [CrossRef]
- Russo, M.; Siravegna, G.; Blaszkowsky, L.S.; Corti, G.; Crisafulli, G.; Ahronian, L.G.; Mussolin, B.; Kwak, E.L.; Buscarino, M.; Lazzari, L.; et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016, 6, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Liu, S.; Ji, C.; Zheng, H.; Pan, X.; Zhang, Y.; Guan, W.; Chen, L.; Guan, Y.; Li, W.; et al. Application of CT texture analysis in predicting histopathological characteristics of gastric cancers. Eur. Radiol. 2017, 27, 4951–4959. [Google Scholar] [CrossRef]
- Li, M.; Zhang, J.; Dan, Y.; Yao, Y.; Dai, W.; Cai, G.; Yang, G.; Tong, T. A clinical-radiomics nomogram for the preoperative prediction of lymph node metastasis in colorectal cancer. J. Transl. Med. 2020, 18, 46. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Liu, Y.; Kong, X.; Yang, S. T staging with functional and radiomics parameters of computed tomography in colorectal cancer patients. Medicine 2022, 101, e29244. [Google Scholar] [CrossRef] [PubMed]
- Xue, T.; Peng, H.; Chen, Q.; Li, M.; Duan, S.; Feng, F. Preoperative prediction of KRAS mutation status in colorectal cancer using a CT-based radiomics nomogram. Br. J. Radiol. 2022, 95, 20211014. [Google Scholar] [CrossRef]
- Yun, J.; Park, J.E.; Lee, H.; Ham, S.; Kim, N.; Kim, H.S. Radiomic features and multilayer perceptron network classifier: A robust MRI classification strategy for distinguishing glioblastoma from primary central nervous system lymphoma. Sci. Rep. 2019, 9, 5746. [Google Scholar] [CrossRef]
- Chalkidou, A.; O’Doherty, M.J.; Marsden, P.K. False Discovery Rates in PET and CT Studies with Texture Features: A Systematic Review. PLoS ONE 2015, 10, e0124165. [Google Scholar] [CrossRef]
- Traverso, A.; Wee, L.; Dekker, A.; Gillies, R. Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1143–1158. [Google Scholar] [CrossRef]
- Xu, Y.; Hosny, A.; Zeleznik, R.; Parmar, C.; Coroller, T.; Franco, I.; Mak, R.H.; Aerts, H. Deep Learning Predicts Lung Cancer Treatment Response from Serial Medical Imaging. Clin. Cancer Res. 2019, 25, 3266–3275. [Google Scholar] [CrossRef]
- Hosny, A.; Parmar, C.; Coroller, T.P.; Grossmann, P.; Zeleznik, R.; Kumar, A.; Bussink, J.; Gillies, R.J.; Mak, R.H.; Aerts, H. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med. 2018, 15, e1002711. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Chen, Y.; Shen, Z.; Zhang, X.; Sang, J.; Ding, Y.; Yang, X.; Li, J.; Chen, M.; Jin, C.; et al. Convolutional neural network for the diagnosis of early gastric cancer based on magnifying narrow band imaging. Gastric Cancer 2020, 23, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Ueyama, H.; Kato, Y.; Akazawa, Y.; Yatagai, N.; Komori, H.; Takeda, T.; Matsumoto, K.; Ueda, K.; Matsumoto, K.; Hojo, M.; et al. Application of artificial intelligence using a convolutional neural network for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging. J. Gastroenterol. Hepatol. 2021, 36, 482–489. [Google Scholar] [CrossRef]
- Balachandran, V.P.; Gonen, M.; Smith, J.J.; DeMatteo, R.P. Nomograms in oncology: More than meets the eye. Lancet Oncol. 2015, 16, e173–e180. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, D.R.; Patel, R.; Kirsch, D.G.; Lewis, C.A.; Vander Heiden, M.G.; Locasale, J.W. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J. Clin. 2021, 71, 333–358. [Google Scholar] [CrossRef] [PubMed]
- de Boer, L.L.; Spliethoff, J.W.; Sterenborg, H.; Ruers, T.J.M. Review: In vivo optical spectral tissue sensing-how to go from research to routine clinical application? Lasers Med. Sci. 2017, 32, 711–719. [Google Scholar] [CrossRef]
- Cabitza, F.; Rasoini, R.; Gensini, G.F. Unintended Consequences of Machine Learning in Medicine. JAMA 2017, 318, 517–518. [Google Scholar] [CrossRef]




| Clinical Characteristics | ||
|---|---|---|
| Age (mean ± SD) | 63.97 ± 11.086 | |
| Gender, NO (%) | ||
| Male | 170 | |
| Female | 61 | |
| Laboratory tests, median (IQR) | ||
| Albumin | 40.60 (37.30, 42.90) | |
| Neutrophil | 4.49 (3.23, 6.52) | |
| Lymphocyte | 1.29 (0.94, 1.71) | |
| CEA level, NO (%) | ||
| Normal | 179 | |
| Abnormal | 52 | |
| CA125 level, NO (%) | ||
| Normal | 203 | |
| Abnormal | 28 | |
| CA199 level, NO (%) | ||
| Normal | 186 |
| Clinical Characteristics | ||
|---|---|---|
| Age (mean ± SD) | 63.79 ± 11.143 | |
| Gender, NO (%) | ||
| Male | 148 | |
| Female | 49 | |
| Laboratory tests, median (IQR) | ||
| Albumin | 40.50 (37.85, 43.65) | |
| Neutrophil | 4.11 (3.15, 5.86) | |
| Lymphocyte | 1.34 (1.05, 1.74) | |
| CEA level, NO (%) | ||
| Normal | 145 | |
| Abnormal | 52 | |
| CA125 level, NO (%) | ||
| Normal | 165 | |
| Abnormal | 32 | |
| CA199 level, NO (%) | ||
| Normal | 158 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, N.; Guan, X.; Zhu, J.; Li, Y.; Zhang, J. A Contrast-Enhanced CT-Based Deep Learning System for Preoperative Prediction of Colorectal Cancer Staging and RAS Mutation. Cancers 2023, 15, 4497. https://doi.org/10.3390/cancers15184497
Lu N, Guan X, Zhu J, Li Y, Zhang J. A Contrast-Enhanced CT-Based Deep Learning System for Preoperative Prediction of Colorectal Cancer Staging and RAS Mutation. Cancers. 2023; 15(18):4497. https://doi.org/10.3390/cancers15184497
Chicago/Turabian StyleLu, Na, Xiao Guan, Jianguo Zhu, Yuan Li, and Jianping Zhang. 2023. "A Contrast-Enhanced CT-Based Deep Learning System for Preoperative Prediction of Colorectal Cancer Staging and RAS Mutation" Cancers 15, no. 18: 4497. https://doi.org/10.3390/cancers15184497
APA StyleLu, N., Guan, X., Zhu, J., Li, Y., & Zhang, J. (2023). A Contrast-Enhanced CT-Based Deep Learning System for Preoperative Prediction of Colorectal Cancer Staging and RAS Mutation. Cancers, 15(18), 4497. https://doi.org/10.3390/cancers15184497






