USP14 Positively Modulates Head and Neck Squamous Carcinoma Tumorigenesis and Potentiates Heat Shock Pathway through HSF1 Stabilization
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Materials
2.2. Data Acquisition
2.3. HNSCC Tissue Microarray and Samples from Individuals with HNSCC
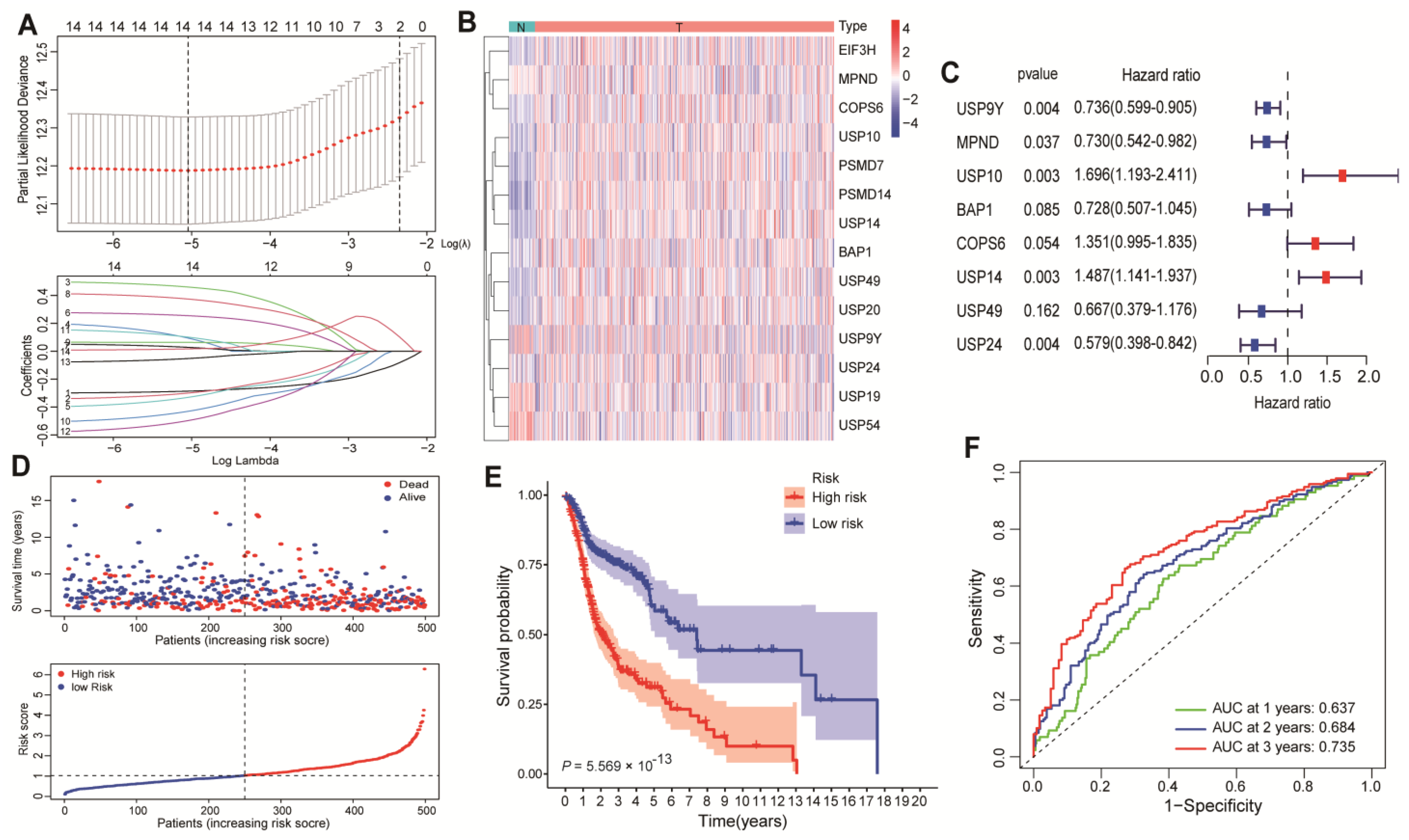
2.4. Establishment of the DAG-Related Risk Signature
2.5. Plasmids and Reagents
2.6. Real-Time RT-PCR
2.7. Fluorescence Microscopy
2.8. CCK8 Assay
2.9. Colony Formation
2.10. Cell Migration
2.11. Wound-Healing Assay
2.12. Dual-Luciferase Reporter Assay
2.13. Flow Cytometry
2.14. Coimmunoprecipitation Assays
2.15. Lung Metastasis and Tumor Xenograft Model
2.16. Deubiquitination Assay
2.17. Luciferase Live Imaging
2.18. Statistical Analysis
3. Results
3.1. Identification of USP14 as a Potent Promoter of HNSCC Patient Survival
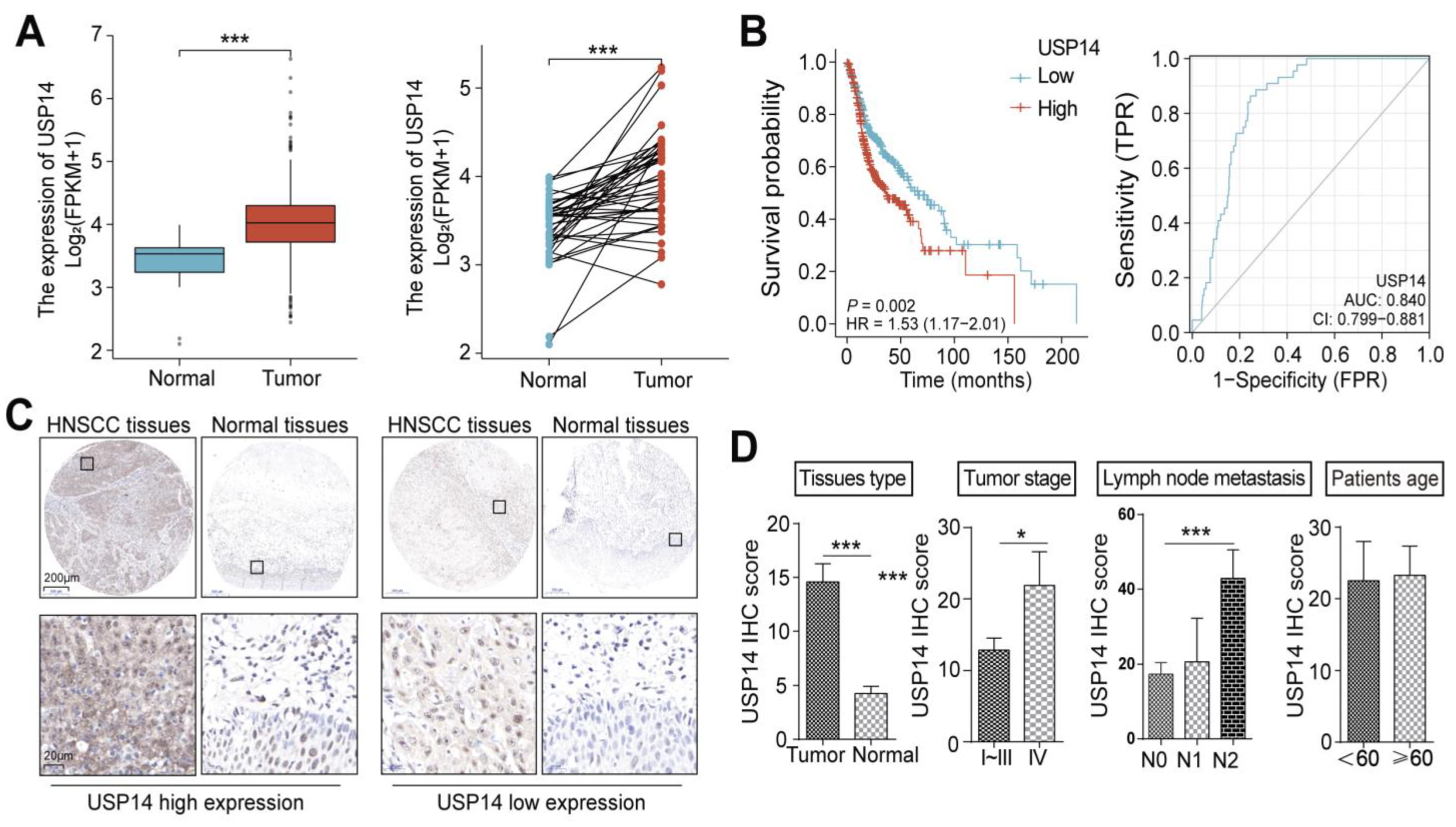
3.2. USP14 Is Highly Expressed in HNSCC Tumors and Predicts Poor Clinical Outcomes in HNSCC Patients
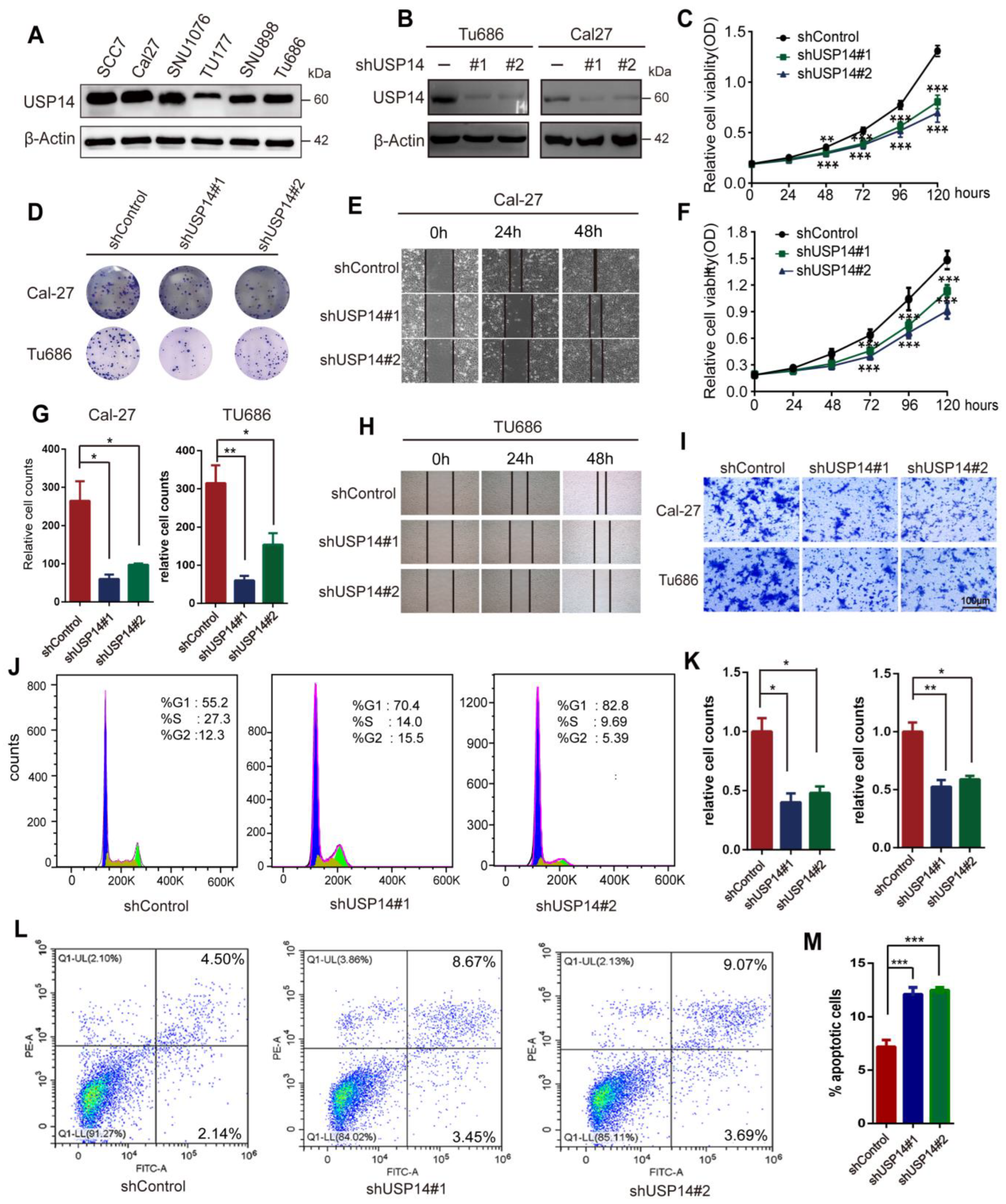
3.3. Knockdown of USP14 Weakened HNSCC Cell Proliferation and Metastasis
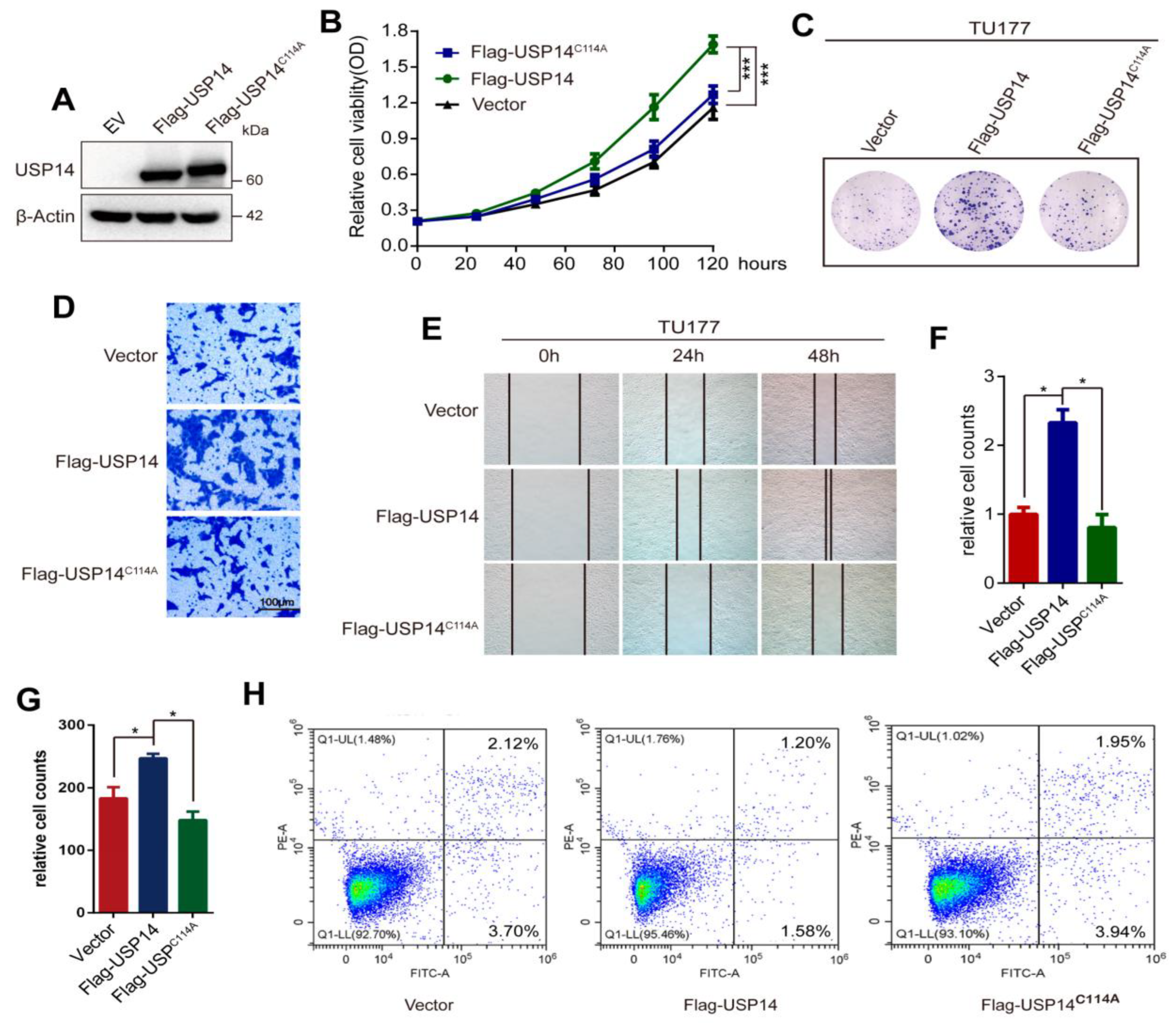
3.4. Overexpression of USP14 Promoted HNSCC Cell Proliferation and Metastasis
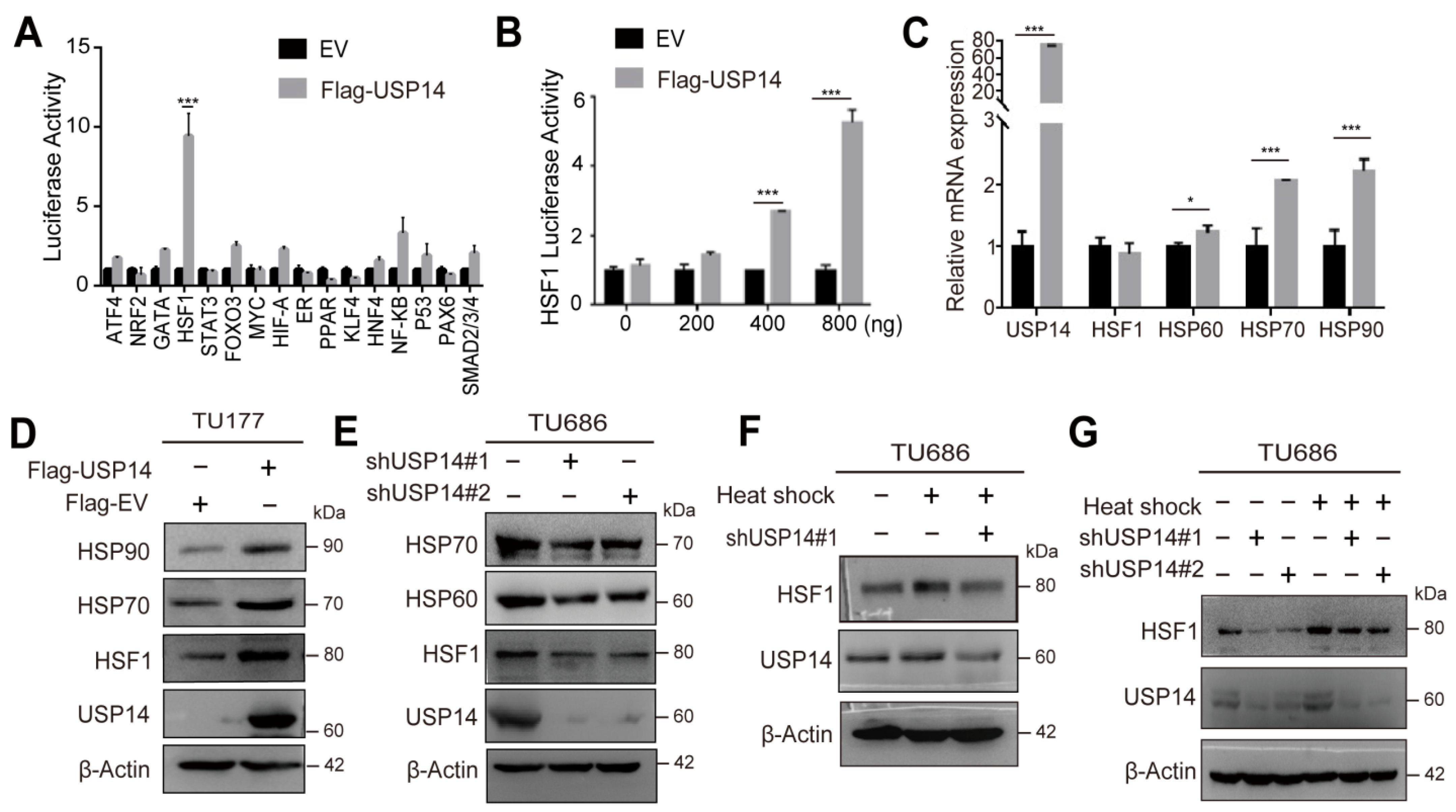
3.5. USP14 Regulates the HSF1 Signaling Cascade
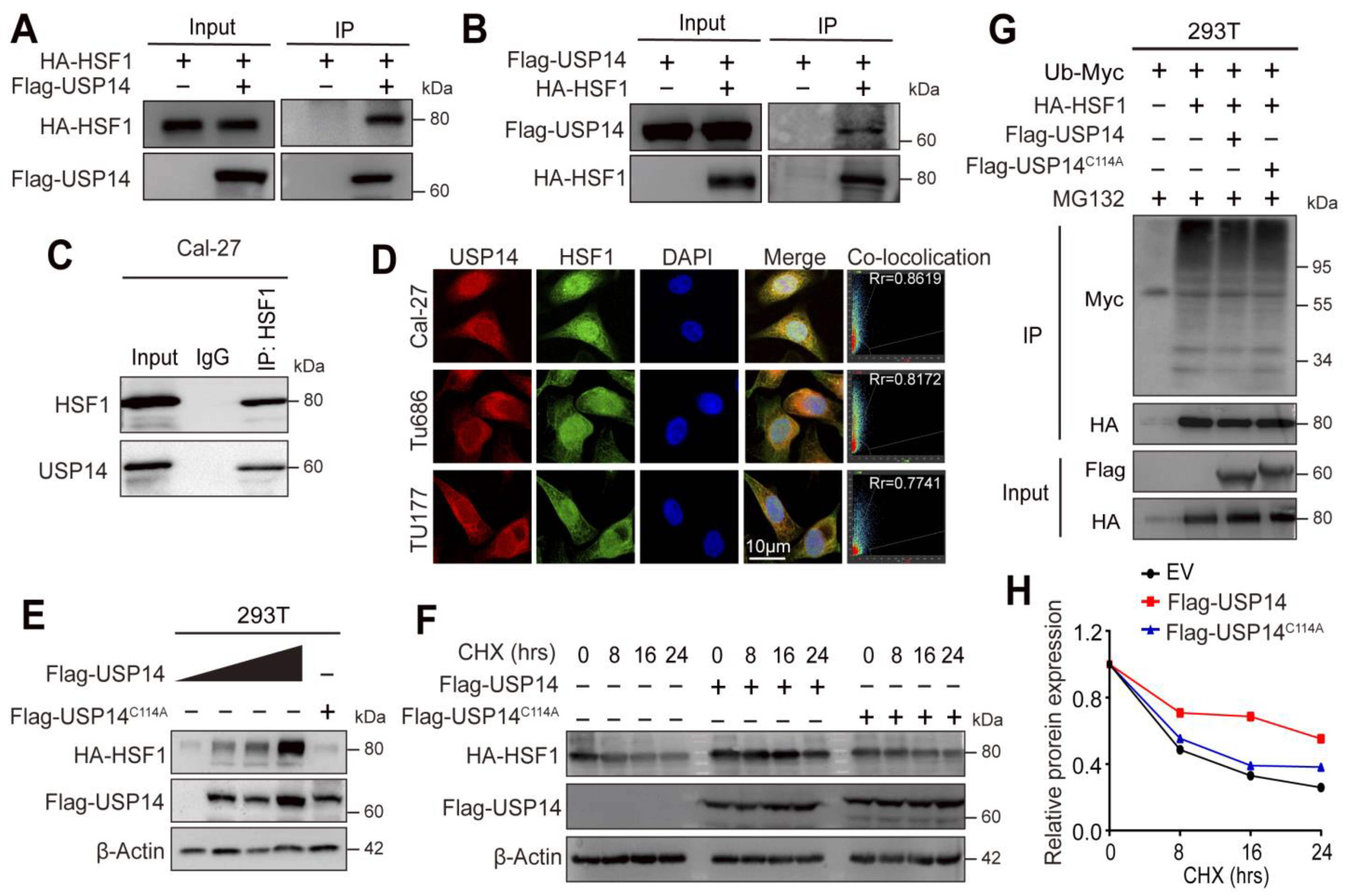
3.6. USP14 Stabilizes the HSF1 Protein by Deubiquitylation of HSF1
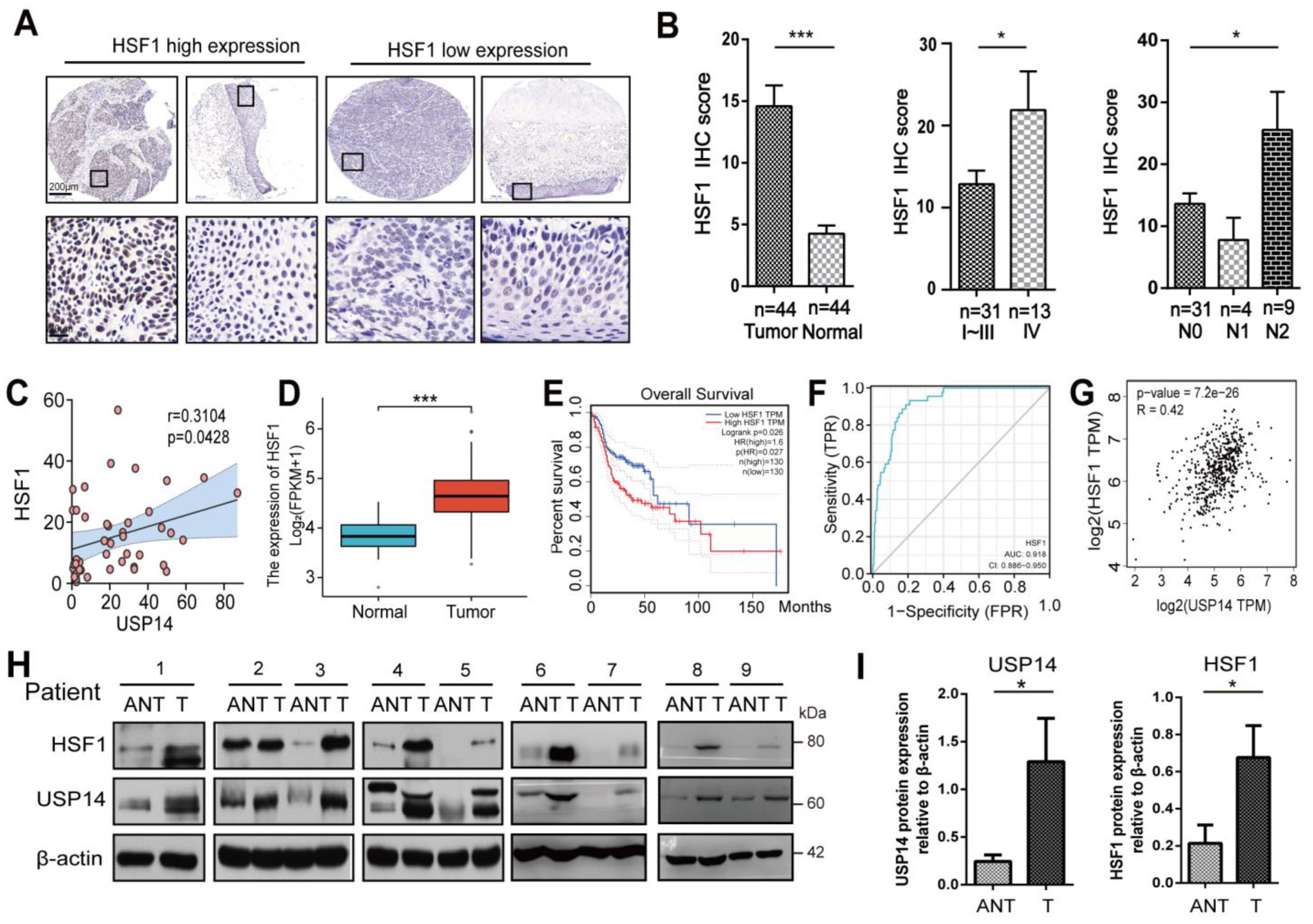
3.7. HSF1 Correlated with USP14 Protein Levels in Human HNSCC Tissues
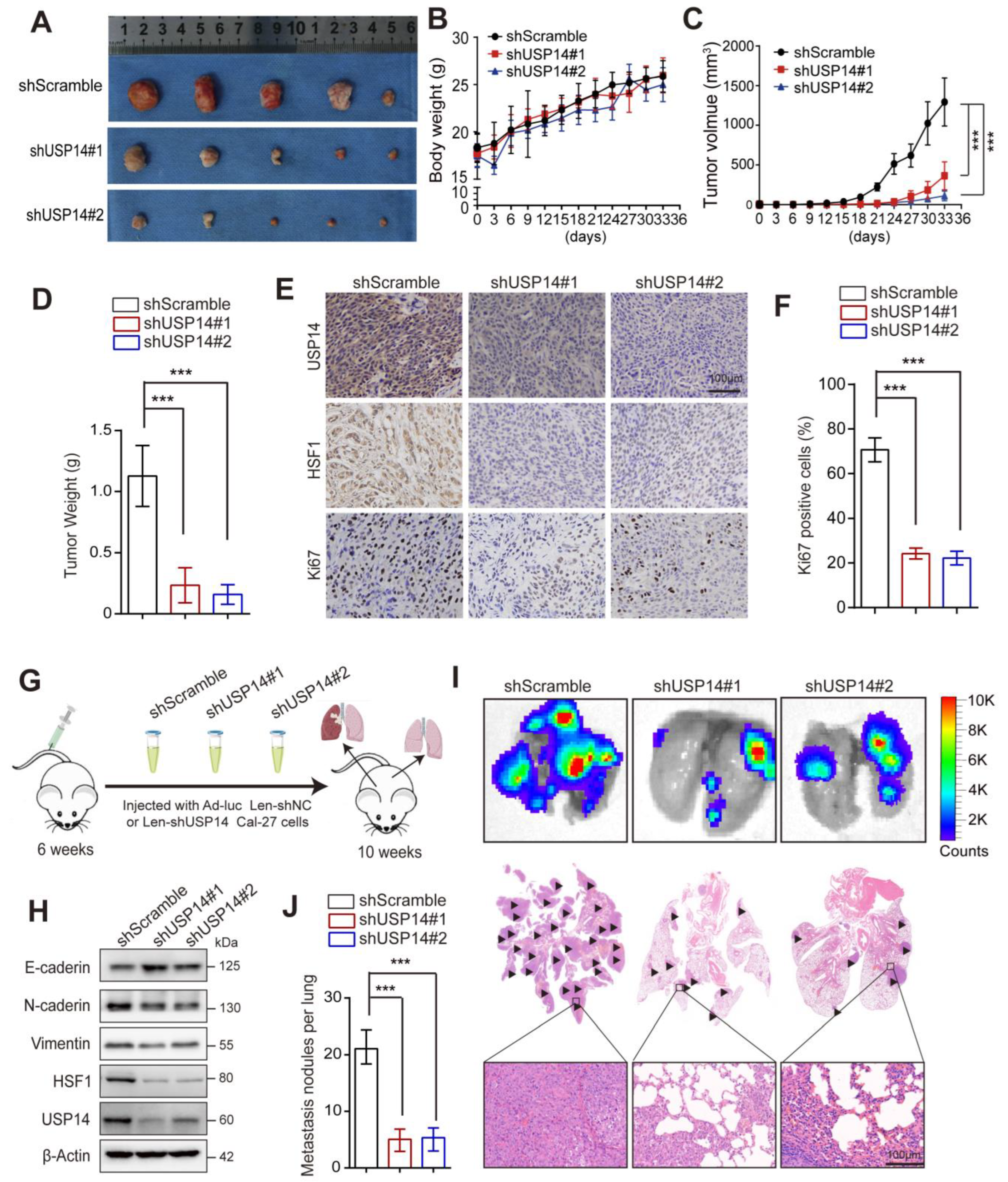
3.8. USP14 Deficiency Represses Tumor Growth and Lung Metastasis In Vivo
3.9. Increased Expression of HSF1 Reverses the Inhibitory Effect of USP14 Depletion on the Proliferation and Metastasis of HNSCC in Mice
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gupta, B.; Johnson, N.W.; Kumar, N. Global Epidemiology of Head and Neck Cancers: A Continuing Challenge. Oncology 2016, 91, 13–23. [Google Scholar] [CrossRef]
- Deshpande, A.M.; Wong, D.T. Molecular mechanisms of head and neck cancer. Expert Rev. Anticancer Ther. 2008, 8, 799–809. [Google Scholar] [CrossRef]
- Economopoulou, P.; Kotsantis, I.; Kyrodimos, E.; Lianidou, E.; Psyrri, A. Liquid biopsy: An emerging prognostic and predictive tool in Head and Neck Squamous Cell Carcinoma (HNSCC). Focus Circ. Tumor Cells (CTCs) 2017, 74, 83–89. [Google Scholar]
- Komander, D.; Rape, M. The ubiquitin code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef]
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct. Target Ther. 2020, 5, 11. [Google Scholar] [CrossRef]
- Mevissen, T.E.T.; Komander, D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 2017, 86, 159–192. [Google Scholar] [CrossRef]
- Du, X.-H.; Ke, S.-B.; Liang, X.-Y.; Gao, J.; Xie, X.-X.; Qi, L.-Z.; Liu, X.-Y.; Xu, G.-Y.; Zhang, X.-D.; Du, R.-L.; et al. USP14 promotes colorectal cancer progression by targeting JNK for stabilization. Cell Death Dis. 2023, 14, 56. [Google Scholar] [CrossRef]
- Hang, C.; Gong, C.; Fang, Y.; Chen, L.; Zhu, J. Ubiquitin-specific protease 14 (USP14) promotes proliferation and metastasis in pancreatic ductal adenocarcinoma. J. Mol. Histol. 2021, 52, 187–196. [Google Scholar] [CrossRef]
- Moghadami, A.-A.; Beilankouhi, E.A.V.; Kalantary-Charvadeh, A.; Hamzavi, M.; Mosayyebi, B.; Sedghi, H.; Haghjo, A.G.; Ahmad, S.N.S. Inhibition of USP14 induces ER stress-mediated autophagy without apoptosis in lung cancer cell line A549. Cell Stress Chaperones 2020, 25, 909–917. [Google Scholar] [CrossRef]
- Min, Y.; Lee, S.; Kim, M.-J.; Chun, E.; Lee, K.-Y. Ubiquitin-Specific Protease 14 Negatively Regulates Toll-like Receptor 4-Mediated Signaling and Autophagy Induction by Inhibiting Ubiquitination of TAK1-Binding Protein 2 and Beclin 1. Front. Immunol. 2017, 8, 1827. [Google Scholar] [CrossRef]
- He, H.; Soncin, F.; Grammatikakis, N.; Li, Y.; Siganou, A.; Gong, J.; Brown, S.A.; Kingston, R.E.; Calderwood, S.K. Elevated expression of heat shock factor (HSF) 2A stimulates HSF1-induced transcription during stress. J. Biol. Chem. 2003, 278, 35465–35475. [Google Scholar] [CrossRef]
- McMillan, D.R.; Xiao, X.; Shao, L.; Graves, K.; Benjamin, I.J. Targeted disruption of heat shock transcription factor 1 abolishes thermotolerance and protection against heat-inducible apoptosis. J. Biol. Chem. 1998, 273, 7523–7528. [Google Scholar] [CrossRef] [PubMed]
- Morange, M. HSFs in development. Handb. Exp. Pharmacol. 2006, 172, 153–169. [Google Scholar]
- Manerba, M.; Di Ianni, L.; Govoni, M.; Roberti, M.; Recanatini, M.; Di Stefano, G. LDH inhibition impacts on heat shock response and induces senescence of hepatocellular carcinoma cells. Eur. J. Pharm. Sci. 2017, 105, 91–98. [Google Scholar] [CrossRef]
- Carpenter, R.L.; Sirkisoon, S.; Zhu, D.; Rimkus, T.; Harrison, A.; Anderson, A.; Paw, I.; Qasem, S.; Xing, F.; Liu, Y.; et al. Combined inhibition of AKT and HSF1 suppresses breast cancer stem cells and tumor growth. Oncotarget 2017, 8, 73947–73963. [Google Scholar] [CrossRef] [PubMed]
- Hoang, A.T.; Huang, J.; Rudra-Ganguly, N.; Zheng, J.; Powell, W.C.; Rabindran, S.K.; Wu, C.; Roy-Burman, P. A novel association between the human heat shock transcription factor 1 (HSF1) and prostate adenocarcinoma. Am. J. Pathol. 2000, 156, 857–864. [Google Scholar] [CrossRef]
- Jin, X.; Moskophidis, D.; Mivechi, N.F. Heat shock transcription factor 1 is a key determinant of HCC development by regulating hepatic steatosis and metabolic syndrome. Cell Metab. 2011, 14, 91–103. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Murshid, A.; Prince, T. The shock of aging: Molecular chaperones and the heat shock response in longevity and aging--a mini-review. Gerontology 2009, 55, 550–558. [Google Scholar] [CrossRef]
- Abdul, N.S.; Alrashed, N.A.; Alsubaie, S.; Albluwi, H.; Alsaleh, H.B.; Alageel, N.; Salma, R.G. Role of Extracellular Heat Shock Protein 90 Alpha in the Metastasis of Oral Squamous Cell Carcinoma: A Systematic Review. Cureus 2023, 15, e38514. [Google Scholar] [CrossRef]
- Zhuang, J.; Qin, Y.-T.; Feng, Y.-S.; Su, Z.-C.; He, X.-W.; Li, W.-Y.; Zhang, Y.-K. A Novel Hexokinases Inhibitor Based on Molecularly Imprinted Polymer for Combined Starvation and Enhanced Photothermal Therapy of Malignant Tumors. ACS Appl. Mater. Interfaces 2023, 15, 25898–25908. [Google Scholar] [CrossRef]
- Dai, W.-Y.; Yao, G.-Q.; Deng, X.-C.; Zang, G.-C.; Liu, J.; Zhang, G.-Y.; Chen, Y.-M.; Lv, M.-Q.; Chen, T.-T. Heat shock protein: A double-edged sword linking innate immunity and hepatitis B virus infection. J. Virus Erad. 2023, 9, 100322. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.D.; Caron, A.W.; Bourget, L.; Denis-Larose, C.; Massie, B. Role of the human heat shock protein hsp70 in protection against stress-induced apoptosis. Mol. Cell. Biol. 1997, 17, 5317–5327. [Google Scholar] [CrossRef] [PubMed]
- Price, B.D.; Calderwood, S.K. Heat-induced transcription from RNA polymerases II and III and HSF binding activity are co-ordinately regulated by the products of the heat shock genes. J. Cell. Physiol. 1992, 153, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Xi, C.; Hu, Y.; Buckhaults, P.; Moskophidis, D.; Mivechi, N.F. Heat shock factor Hsf1 cooperates with ErbB2 (Her2/Neu) protein to promote mammary tumorigenesis and metastasis. J. Biol. Chem. 2012, 287, 35646–35657. [Google Scholar] [CrossRef]
- Chou, S.D.; Murshid, A.; Eguchi, T.; Gong, J.; Calderwood, S.K. HSF1 regulation of beta-catenin in mammary cancer cells through control of HuR/elavL1 expression. Oncogene 2015, 34, 2178–2188. [Google Scholar] [CrossRef]
- Yang, T.; Ren, C.; Lu, C.; Qiao, P.; Han, X.; Wang, L.; Wang, D.; Lv, S.; Sun, Y.; Yu, Z. Phosphorylation of HSF1 by PIM2 Induces PD-L1 Expression and Promotes Tumor Growth in Breast Cancer. Cancer Res. 2019, 79, 5233–5244. [Google Scholar] [CrossRef]
- Purwana, I.; Liu, J.J.; Portha, B.; Buteau, J. HSF1 acetylation decreases its transcriptional activity and enhances glucolipotoxicity-induced apoptosis in rat and human beta cells. Diabetologia 2017, 60, 1432–1441. [Google Scholar] [CrossRef]
- Tan, W.; Zhao, H.; Zhang, F.; Li, Z.; Feng, D.; Li, Y.; Zhou, W.; Liu, L.; Yao, J.; Tian, X. Inhibition of the ubiquitination of HSF1 by FBXW7 protects the intestine against ischemia-reperfusion injury. Apoptosis 2018, 23, 667–678. [Google Scholar] [CrossRef]
- Zhou, H.; Sun, W.; Zou, J. Analysis of expression profiles and prognostic value of COP9 signalosome subunits for patients with head and neck squamous cell carcinoma. Oncol. Lett. 2021, 22, 803. [Google Scholar] [CrossRef]
- Nanayakkara, D.M.; Nguyen, M.N.; Wood, S.A. Deubiquitylating enzyme, USP9X, regulates proliferation of cells of head and neck cancer lines. Cell Prolif. 2016, 49, 494–502. [Google Scholar] [CrossRef]
- Dou, Y.; Lin, J.; Shu, H.; Jiang, N. Role of ubiquitin-specific peptidase 22 in carcinogenesis of human pharyngeal squamous cell carcinoma. Mol. Med. Rep. 2014, 10, 2973–2978. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Kang, H.; Grandis, J.R.; Johnson, D.E. CYLD Alterations in the Tumorigenesis and Progression of Human Papillomavirus-Associated Head and Neck Cancers. Mol. Cancer Res. 2021, 19, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Dyson, O.F.; Pagano, J.S.; Whitehurst, C.B. The Translesion Polymerase Pol eta Is Required for Efficient Epstein-Barr Virus Infectivity and Is Regulated by the Viral Deubiquitinating Enzyme BPLF1. J. Virol. 2017, 91, 10–1128. [Google Scholar] [CrossRef]
- Scherz-Shouval, R.; Santagata, S.; Mendillo, M.L.; Sholl, L.M.; Ben-Aharon, I.; Beck, A.H.; Dias-Santagata, D.; Koeva, M.; Stemmer, S.M.; Whitesell, L.; et al. The reprogramming of tumor stroma by HSF1 is a potent enabler of malignancy. Cell 2014, 158, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Carpenter, R.L.; Gokmen-Polar, Y. HSF1 as a Cancer Biomarker and Therapeutic Target. Curr. Cancer Drug Targets 2019, 19, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Cao, P.; Fan, Y.; Tan, K. Emerging roles of HSF1 in cancer: Cellular and molecular episodes. Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188390. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kim, E.H.; Lee, J.S.; Jeoung, D.; Bae, S.; Kwon, S.H.; Lee, Y.S. HSF1 as a mitotic regulator: Phosphorylation of HSF1 by Plk1 is essential for mitotic progression. Cancer Res. 2008, 68, 7550–7560. [Google Scholar] [CrossRef]
- Westerheide, S.D.; Anckar, J.; Stevens, S.M., Jr.; Sistonen, L.; Morimoto, R.I. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 2009, 323, 1063–1066. [Google Scholar] [CrossRef]
- Hietakangas, V.; Anckar, J.; Blomster, H.A.; Fujimoto, M.; Palvimo, J.J.; Nakai, A.; Sistonen, L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA 2006, 103, 45–50. [Google Scholar] [CrossRef]
- Kourtis, N.; Moubarak, R.S.; Aranda-Orgilles, B.; Lui, K.; Aydin, I.T.; Trimarchi, T.; Darvishian, F.; Salvaggio, C.; Zhong, J.; Bhatt, K.; et al. FBXW7 modulates cellular stress response and metastatic potential through HSF1 post-translational modification. Nat. Cell Biol. 2015, 17, 322–332. [Google Scholar] [CrossRef]
- Ling, X.; Lu, J.; Wang, X.; Liu, L.; Liu, L.; Wang, Y.; Sun, Y.; Ren, C.; Lu, C.; Yu, Z. Ovarian tumorB1-mediated heat shock transcription factor 1 deubiquitination is critical for glycolysis and development of endometriosis. iScience 2022, 25, 105363. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-H.; Lu, Y.; Prado, M.A.; Shi, Y.; Tian, G.; Sun, S.; Elsasser, S.; Gygi, S.P.; King, R.W.; Finley, D. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature 2016, 532, 398–401. [Google Scholar] [CrossRef] [PubMed]
- Shinji, S.; Naito, Z.; Ishiwata, S.; Ishiwata, T.; Tanaka, N.; Furukawa, K.; Suzuki, H.; Seya, T.; Matsuda, A.; Katsuta, M.; et al. Ubiquitin-specific protease 14 expression in colorectal cancer is associated with liver and lymph node metastases. Oncol. Rep. 2006, 15, 539–543. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Zhong, J.; Deng, Y.; Xi, Q.; He, S.; Yang, S.; Jiang, L.; Huang, M.; Tang, C.; et al. Ubiquitin-specific protease 14 (USP14) regulates cellular proliferation and apoptosis in epithelial ovarian cancer. Med. Oncol. 2015, 32, 379. [Google Scholar] [CrossRef]
- Huang, G.; Li, L.M.; Zhou, W.P. USP14 activation promotes tumor progression in hepatocellular carcinoma. Oncol. Rep. 2015, 34, 2917–2924. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Li, M.; Huang, P.; Guan, X.-Y.; Zhu, Y.-H. Overexpression of ubiquitin specific peptidase 14 predicts unfavorable prognosis in esophageal squamous cell carcinoma. Thorac. Cancer 2017, 8, 344–349. [Google Scholar] [CrossRef]
- Geng, L.; Chen, X.; Zhang, M.; Luo, Z. Ubiquitin-specific protease 14 promotes prostate cancer progression through deubiquitinating the transcriptional factor ATF2. Biochem. Biophys. Res. Commun. 2020, 524, 16–21. [Google Scholar] [CrossRef]
- Wu, N.; Liu, C.; Bai, C.; Han, Y.P.; Cho, W.C.; Li, Q. Over-expression of deubiquitinating enzyme USP14 in lung adenocarcinoma promotes proliferation through the accumulation of beta-catenin. Int. J. Mol. Sci. 2013, 14, 10749–10760. [Google Scholar] [CrossRef]
- Yao, L.; Yan, D.; Jiang, B.; Xue, Q.; Huang, Q.; Qi, L.; Tang, D.; Chen, X.; Liu, J. Plumbagin is a novel GPX4 protein degrader that induces apoptosis in hepatocellular carcinoma cells. Free Radic. Biol. Med. 2023, 203, 1–10. [Google Scholar] [CrossRef]
- Lee, M.Y.; Kim, M.-J.; Jin, J.-O.; Lee, P.C.-W. USP14 inhibition regulates tumorigenesis by inducing apoptosis in gastric cancer. BMB Rep. 2023. [Google Scholar] [CrossRef]
- Sharma, A.; Almasan, A. USP14 Regulates DNA Damage Response and Is a Target for Radiosensitization in Non-Small Cell Lung Cancer. Int. J. Mol. Sci. 2020, 21, 6383. [Google Scholar] [CrossRef] [PubMed]
- Jain, N.; Rai, A.; Mishra, R.; Ganesh, S. Loss of malin, but not laforin, results in compromised autophagic flux and proteasomal dysfunction in cells exposed to heat shock. Cell Stress Chaperones 2017, 22, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Uçar, E.Ö.; Şengelen, A.; Kamalı, E.M. Hsp27, Hsp60, Hsp70, or Hsp90 depletion enhances the antitumor effects of resveratrol via oxidative and ER stress response in human glioblastoma cells. Biochem. Pharmacol. 2023, 208, 115409. [Google Scholar] [CrossRef]
- Ahmed, K.; Zaidi, S.F.; Rehman, M.U.; Rehman, R.; Kondo, T. Hyperthermia and protein homeostasis: Cytoprotection and cell death. J. Therm. Biol. 2020, 91, 102615. [Google Scholar] [CrossRef]
- Liang, W.; Liao, Y.; Zhang, J.; Huang, Q.; Luo, W.; Yu, J.; Gong, J.; Zhou, Y.; Li, X.; Tang, B.; et al. Heat shock factor 1 inhibits the mitochondrial apoptosis pathway by regulating second mitochondria-derived activator of caspase to promote pancreatic tumorigenesis. J. Exp. Clin. Cancer Res. 2017, 36, 64. [Google Scholar] [CrossRef]
- Timmons, A.; Fray, E.; Kumar, M.; Wu, F.; Dai, W.; Bullen, C.K.; Kim, P.; Hetzel, C.; Yang, C.; Beg, S.; et al. HSF1 inhibition attenuates HIV-1 latency reversal mediated by several candidate LRAs In Vitro and Ex Vivo. Proc. Natl. Acad. Sci. USA 2020, 117, 15763–15771. [Google Scholar] [CrossRef]
- Morgan, E.L.; Toni, T.; Viswanathan, R.; Robbins, Y.; Yang, X.; Cheng, H.; Gunti, S.; Huynh, A.; Sowers, A.L.; Mitchell, J.B.; et al. Inhibition of USP14 promotes TNFalpha-induced cell death in head and neck squamous cell carcinoma (HNSCC). Cell Death Differ. 2023, 30, 1382–1396. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Xiang, Y.; Xie, Z.; Fan, M.; Fang, S.; Wan, H.; Zhao, R.; Zeng, F.; Hua, Q. USP14 Positively Modulates Head and Neck Squamous Carcinoma Tumorigenesis and Potentiates Heat Shock Pathway through HSF1 Stabilization. Cancers 2023, 15, 4385. https://doi.org/10.3390/cancers15174385
Wang J, Xiang Y, Xie Z, Fan M, Fang S, Wan H, Zhao R, Zeng F, Hua Q. USP14 Positively Modulates Head and Neck Squamous Carcinoma Tumorigenesis and Potentiates Heat Shock Pathway through HSF1 Stabilization. Cancers. 2023; 15(17):4385. https://doi.org/10.3390/cancers15174385
Chicago/Turabian StyleWang, Jie, Yuandi Xiang, Zhanghong Xie, Mengqi Fan, Shizhen Fang, Huanzhi Wan, Rui Zhao, Feng Zeng, and Qingquan Hua. 2023. "USP14 Positively Modulates Head and Neck Squamous Carcinoma Tumorigenesis and Potentiates Heat Shock Pathway through HSF1 Stabilization" Cancers 15, no. 17: 4385. https://doi.org/10.3390/cancers15174385
APA StyleWang, J., Xiang, Y., Xie, Z., Fan, M., Fang, S., Wan, H., Zhao, R., Zeng, F., & Hua, Q. (2023). USP14 Positively Modulates Head and Neck Squamous Carcinoma Tumorigenesis and Potentiates Heat Shock Pathway through HSF1 Stabilization. Cancers, 15(17), 4385. https://doi.org/10.3390/cancers15174385




