Applications of CRISPR Technology to Breast Cancer and Triple Negative Breast Cancer Research
Abstract
:Simple Summary
Abstract
1. Introduction
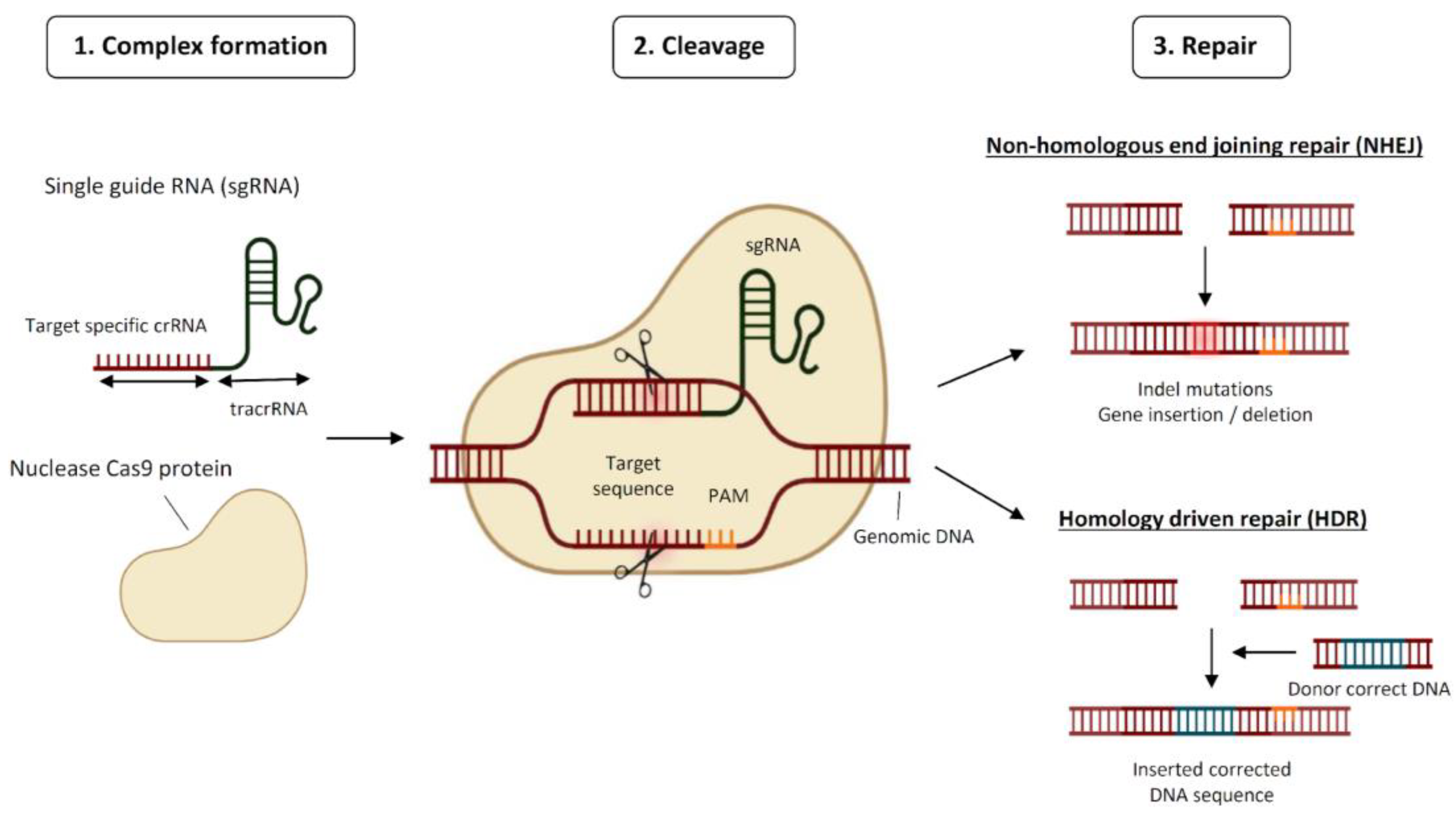
1.1. The CRISPR System: How It Works
1.2. Breast Cancer/Triple Negative Breast Cancer: Incidence, Genetic, and Epigenetic Alterations
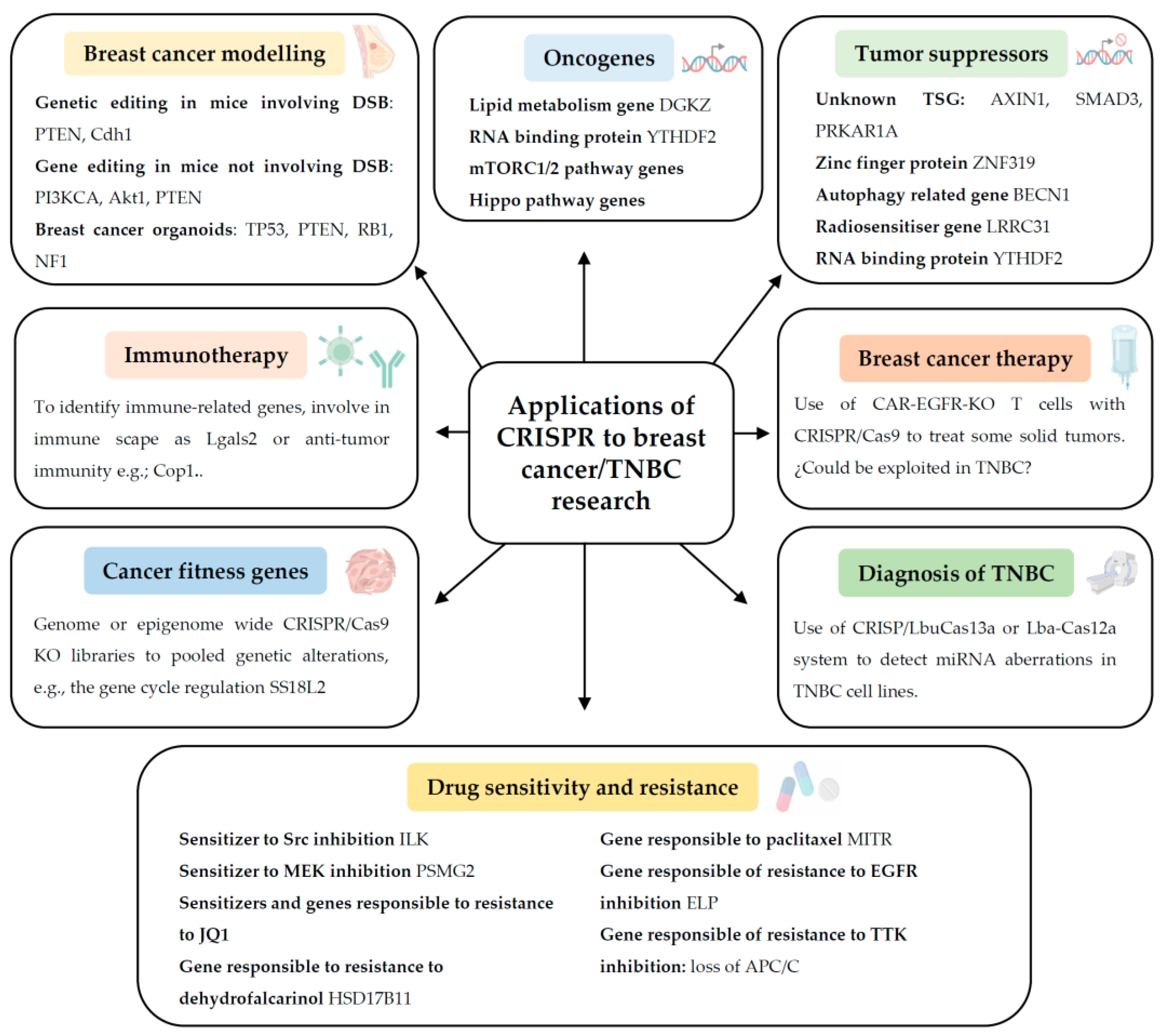
2. Applications of CRISPR to Breast Cancer/TNBC Research
2.1. Modelling TNBC Genetically in Cells, Organoids, and Animals
2.2. Identification of Novel Oncogenes in TNBC
2.3. Identification of New Tumour Suppressor Genes in Breast Cancer in General
2.4. Identification of Genes Responsible for Immunotherapy Response in TNBC
2.5. Identification of Genes Responsible for Drug Sensitivity and Resistance in Breast Cancer and in TNBC
2.6. Determination of Cancer Fitness Genes in TNBC
2.7. Diagnosis of Breast Cancer and TNBC
2.8. Breast Cancer Therapy
3. Delivery Methods for CRISPR Technology for TNBC
4. Limitations and Future Perspective of CRISPR Technology towards TNBC Research
4.1. Extensive Use of In Vivo CRISPR Screens
4.2. Direct Gene Editing in Breast Cancer Tissue
4.3. Novel Delivery Methods for CRISPR Technology
4.4. Modern Methods Coupled with CRISPR Technology
4.5. Unequal Access to CRISPR Medical Advances and Other Ethical Aspects
Author Contributions
Funding
Conflicts of Interest
References
- Horvath, P.; Barrangou, R. CRISPR/Cas, the immune system of bacteria and archaea. Science 2010, 327, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Sorolla, A.; Parisi, E.; Sorolla, M.A.; Marques, M.; Porcel, J.M. Applications of CRISPR technology to lung cancer research. Eur. Respir. J. 2022, 59, 2102610. [Google Scholar] [CrossRef] [PubMed]
- Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014, 507, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Lu, B.; Natarajan, E.; Balaji Raghavendran, H.R.; Markandan, U.D. Molecular Classification, Treatment, and Genetic Biomarkers in Triple-Negative Breast Cancer: A Review. Technol. Cancer Res. Treat. 2023, 22, 15330338221145246. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Yin, L.; Duan, J.J.; Bian, X.W.; Yu, S.C. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020, 22, 61. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, R68. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas, N. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Marques, M.; Sorolla, M.A.; Urdanibia, I.; Parisi, E.; Hidalgo, I.; Morales, S.; Salud, A.; Sorolla, A. Are Transcription Factors Plausible Oncotargets for Triple Negative Breast Cancers? Cancers 2022, 14, 1101. [Google Scholar] [CrossRef]
- Sporikova, Z.; Koudelakova, V.; Trojanec, R.; Hajduch, M. Genetic Markers in Triple-Negative Breast Cancer. Clin. Breast Cancer 2018, 18, e841–e850. [Google Scholar] [CrossRef]
- Peluffo, G.; Subedee, A.; Harper, N.W.; Kingston, N.; Jovanovic, B.; Flores, F.; Stevens, L.E.; Beca, F.; Trinh, A.; Chilamakuri, C.S.R.; et al. EN1 Is a Transcriptional Dependency in Triple-Negative Breast Cancer Associated with Brain Metastasis. Cancer Res. 2019, 79, 4173–4183. [Google Scholar] [CrossRef]
- Sorolla, A.; Wang, E.; Golden, E.; Duffy, C.; Henriques, S.T.; Redfern, A.D.; Blancafort, P. Precision medicine by designer interference peptides: Applications in oncology and molecular therapeutics. Oncogene 2020, 39, 1167–1184. [Google Scholar] [CrossRef]
- Sorolla, A.; Wang, E.; Clemons, T.D.; Evans, C.W.; Plani-Lam, J.H.; Golden, E.; Dessauvagie, B.; Redfern, A.D.; Swaminathan-Iyer, K.; Blancafort, P. Triple-hit therapeutic approach for triple negative breast cancers using docetaxel nanoparticles, EN1-iPeps and RGD peptides. Nanomedicine 2019, 20, 102003. [Google Scholar] [CrossRef]
- Annunziato, S.; Kas, S.M.; Nethe, M.; Yucel, H.; Del Bravo, J.; Pritchard, C.; Bin Ali, R.; van Gerwen, B.; Siteur, B.; Drenth, A.P.; et al. Modeling invasive lobular breast carcinoma by CRISPR/Cas9-mediated somatic genome editing of the mammary gland. Genes Dev. 2016, 30, 1470–1480. [Google Scholar] [CrossRef]
- Annunziato, S.; Lutz, C.; Henneman, L.; Bhin, J.; Wong, K.; Siteur, B.; van Gerwen, B.; de Korte-Grimmerink, R.; Zafra, M.P.; Schatoff, E.M.; et al. In situ CRISPR-Cas9 base editing for the development of genetically engineered mouse models of breast cancer. EMBO J. 2020, 39, e102169. [Google Scholar] [CrossRef]
- Dekkers, J.F.; Whittle, J.R.; Vaillant, F.; Chen, H.R.; Dawson, C.; Liu, K.; Geurts, M.H.; Herold, M.J.; Clevers, H.; Lindeman, G.J.; et al. Modeling Breast Cancer Using CRISPR-Cas9-Mediated Engineering of Human Breast Organoids. J. Natl. Cancer Inst. 2020, 112, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sun, H.; Li, X.; Liu, Q.; Liu, Y.; Hou, Y.; Jin, W. DGKZ promotes TGFbeta signaling pathway and metastasis in triple-negative breast cancer by suppressing lipid raft-dependent endocytosis of TGFbetaR2. Cell Death Dis. 2022, 13, 105. [Google Scholar] [CrossRef] [PubMed]
- Einstein, J.M.; Perelis, M.; Chaim, I.A.; Meena, J.K.; Nussbacher, J.K.; Tankka, A.T.; Yee, B.A.; Li, H.; Madrigal, A.A.; Neill, N.J.; et al. Inhibition of YTHDF2 triggers proteotoxic cell death in MYC-driven breast cancer. Mol. Cell 2021, 81, 3048–3064.e3049. [Google Scholar] [CrossRef] [PubMed]
- Dai, M.; Yan, G.; Wang, N.; Daliah, G.; Edick, A.M.; Poulet, S.; Boudreault, J.; Ali, S.; Burgos, S.A.; Lebrun, J.J. In vivo genome-wide CRISPR screen reveals breast cancer vulnerabilities and synergistic mTOR/Hippo targeted combination therapy. Nat. Commun. 2021, 12, 3055. [Google Scholar] [CrossRef]
- Heitink, L.; Whittle, J.R.; Vaillant, F.; Capaldo, B.D.; Dekkers, J.F.; Dawson, C.A.; Milevskiy, M.J.G.; Surgenor, E.; Tsai, M.; Chen, H.R.; et al. In vivo genome-editing screen identifies tumor suppressor genes that cooperate with Trp53 loss during mammary tumorigenesis. Mol. Oncol. 2022, 16, 1119–1131. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, L.; Li, M.; Zhao, J.; Liu, Y.; Chen, Y.; Qin, X.; Wang, S.; Chen, H.; Piao, Y.; et al. Genome-wide CRISPR/Cas9 knockout screening uncovers ZNF319 as a novel tumor suppressor critical for breast cancer metastasis. Biochem. Biophys. Res. Commun. 2022, 589, 107–115. [Google Scholar] [CrossRef]
- Wijshake, T.; Zou, Z.; Chen, B.; Zhong, L.; Xiao, G.; Xie, Y.; Doench, J.G.; Bennett, L.; Levine, B. Tumor-suppressor function of Beclin 1 in breast cancer cells requires E-cadherin. Proc. Natl. Acad. Sci. USA 2021, 118, e2020478118. [Google Scholar] [CrossRef]
- Oser, M.G.; Fonseca, R.; Chakraborty, A.A.; Brough, R.; Spektor, A.; Jennings, R.B.; Flaifel, A.; Novak, J.S.; Gulati, A.; Buss, E.; et al. Cells Lacking the RB1 Tumor Suppressor Gene Are Hyperdependent on Aurora B Kinase for Survival. Cancer Discov. 2019, 9, 230–247. [Google Scholar] [CrossRef]
- Zhao, X.; Li, J.; Liu, Z.; Powers, S. Combinatorial CRISPR/Cas9 Screening Reveals Epistatic Networks of Interacting Tumor Suppressor Genes and Therapeutic Targets in Human Breast Cancer. Cancer Res. 2021, 81, 6090–6105. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, T.; Zhang, H.; Gou, X.; Han, C.; Wang, J.; Chen, A.T.; Ma, J.; Liu, J.; Chen, Z.; et al. LRRC31 inhibits DNA repair and sensitizes breast cancer brain metastasis to radiation therapy. Nat. Cell Biol. 2020, 22, 1276–1285. [Google Scholar] [CrossRef]
- Ji, P.; Gong, Y.; Jin, M.L.; Wu, H.L.; Guo, L.W.; Pei, Y.C.; Chai, W.J.; Jiang, Y.Z.; Liu, Y.; Ma, X.Y.; et al. In vivo multidimensional CRISPR screens identify Lgals2 as an immunotherapy target in triple-negative breast cancer. Sci. Adv. 2022, 8, eabl8247. [Google Scholar] [CrossRef]
- Wang, X.; Tokheim, C.; Gu, S.S.; Wang, B.; Tang, Q.; Li, Y.; Traugh, N.; Zeng, Z.; Zhang, Y.; Li, Z.; et al. In vivo CRISPR screens identify the E3 ligase Cop1 as a modulator of macrophage infiltration and cancer immunotherapy target. Cell 2021, 184, 5357–5374.e5322. [Google Scholar] [CrossRef]
- Beetham, H.; Griffith, B.G.C.; Murina, O.; Loftus, A.E.P.; Parry, D.A.; Temps, C.; Culley, J.; Muir, M.; Unciti-Broceta, A.; Sims, A.H.; et al. Loss of Integrin-Linked Kinase Sensitizes Breast Cancer to SRC Inhibitors. Cancer Res. 2022, 82, 632–647. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, J.; Liu, X.; Luo, D.; Li, Y.; Song, L.; Jiang, X.; Yin, X.; Wang, Y.; Chai, L.; et al. PSMG2-controlled proteasome-autophagy balance mediates the tolerance for MEK-targeted therapy in triple-negative breast cancer. Cell Rep. Med. 2022, 3, 100741. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.; Wu, H.J.; Ge, J.Y.; Zeid, R.; Harris, I.S.; Jovanovic, B.; Murphy, K.; Wang, B.; Qiu, X.; Endress, J.E.; et al. Synthetic Lethal and Resistance Interactions with BET Bromodomain Inhibitors in Triple-Negative Breast Cancer. Mol. Cell. 2020, 78, 1096–1113.e1098. [Google Scholar] [CrossRef]
- Barkovskaya, A.; Goodwin, C.M.; Seip, K.; Hilmarsdottir, B.; Pettersen, S.; Stalnecker, C.; Engebraaten, O.; Briem, E.; Der, C.J.; Moestue, S.A.; et al. Detection of phenotype-specific therapeutic vulnerabilities in breast cells using a CRISPR loss-of-function screen. Mol. Oncol. 2021, 15, 2026–2045. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.V.; Cai, S.; Risinger, A.L.; Liang, H.; O’Keefe, B.R.; Doench, J.G.; Cichewicz, R.H.; Mooberry, S.L. CRISPR-Cas9 Genome-Wide Knockout Screen Identifies Mechanism of Selective Activity of Dehydrofalcarinol in Mesenchymal Stem-like Triple-Negative Breast Cancer Cells. J. Nat. Prod. 2020, 83, 3080–3092. [Google Scholar] [CrossRef]
- Lian, B.; Pei, Y.C.; Jiang, Y.Z.; Xue, M.Z.; Li, D.Q.; Li, X.G.; Zheng, Y.Z.; Liu, X.Y.; Qiao, F.; Sun, W.L.; et al. Truncated HDAC9 identified by integrated genome-wide screen as the key modulator for paclitaxel resistance in triple-negative breast cancer. Theranostics 2020, 10, 11092–11109. [Google Scholar] [CrossRef]
- Cruz-Gordillo, P.; Honeywell, M.E.; Harper, N.W.; Leete, T.; Lee, M.J. ELP-dependent expression of MCL1 promotes resistance to EGFR inhibition in triple-negative breast cancer cells. Sci. Signal 2020, 13, eabb9820. [Google Scholar] [CrossRef]
- Thu, K.L.; Silvester, J.; Elliott, M.J.; Ba-Alawi, W.; Duncan, M.H.; Elia, A.C.; Mer, A.S.; Smirnov, P.; Safikhani, Z.; Haibe-Kains, B.; et al. Disruption of the anaphase-promoting complex confers resistance to TTK inhibitors in triple-negative breast cancer. Proc. Natl. Acad. Sci. USA 2018, 115, E1570–E1577. [Google Scholar] [CrossRef]
- Eirew, P.; O’Flanagan, C.; Ting, J.; Salehi, S.; Brimhall, J.; Wang, B.; Biele, J.; Algara, T.; Lee, S.R.; Hoang, C.; et al. Accurate determination of CRISPR-mediated gene fitness in transplantable tumours. Nat. Commun. 2022, 13, 4534. [Google Scholar] [CrossRef] [PubMed]
- Yedier-Bayram, O.; Gokbayrak, B.; Kayabolen, A.; Aksu, A.C.; Cavga, A.D.; Cingoz, A.; Kala, E.Y.; Karabiyik, G.; Gunsay, R.; Esin, B.; et al. EPIKOL, a chromatin-focused CRISPR/Cas9-based screening platform, to identify cancer-specific epigenetic vulnerabilities. Cell Death Dis. 2022, 13, 710. [Google Scholar] [CrossRef]
- Shan, Y.; Zhou, X.; Huang, R.; Xing, D. High-Fidelity and Rapid Quantification of miRNA Combining crRNA Programmability and CRISPR/Cas13a trans-Cleavage Activity. Anal. Chem. 2019, 91, 5278–5285. [Google Scholar] [CrossRef]
- Wang, G.; Tian, W.; Liu, X.; Ren, W.; Liu, C. New CRISPR-Derived microRNA Sensing Mechanism Based on Cas12a Self-Powered and Rolling Circle Transcription-Unleashed Real-Time crRNA Recruiting. Anal. Chem. 2020, 92, 6702–6708. [Google Scholar] [CrossRef]
- Hu, Y.; Zhou, Y.; Zhang, M.; Ge, W.; Li, Y.; Yang, L.; Wei, G.; Han, L.; Wang, H.; Yu, S.; et al. CRISPR/Cas9-Engineered Universal CD19/CD22 Dual-Targeted CAR-T Cell Therapy for Relapsed/Refractory B-cell Acute Lymphoblastic Leukemia. Clin. Cancer Res. 2021, 27, 2764–2772. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, N.; Feng, K.; Chen, M.; Zhang, Y.; Liu, Y.; Yang, Q.; Nie, J.; Tang, N.; Zhang, X.; et al. Phase I study of CAR-T cells with PD-1 and TCR disruption in mesothelin-positive solid tumors. Cell. Mol. Immunol. 2021, 18, 2188–2198. [Google Scholar] [CrossRef]
- Yang, W.; Yan, J.; Zhuang, P.; Ding, T.; Chen, Y.; Zhang, Y.; Zhang, H.; Cui, W. Progress of delivery methods for CRISPR-Cas9. Expert Opin. Drug Deliv. 2022, 19, 913–926. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, P.; Zhu, M.; Liang, M.; Zhang, L.; Zong, Y.; Wan, M. High-Performance Delivery of a CRISPR Interference System via Lipid-Polymer Hybrid Nanoparticles Combined with Ultrasound-Mediated Microbubble Destruction for Tumor-Specific Gene Repression. Adv. Healthc. Mater. 2023, 12, e2203082. [Google Scholar] [CrossRef]
- Sun, M.; Shi, W.; Wu, Y.; He, Z.; Sun, J.; Cai, S.; Luo, Q. Immunogenic Nanovesicle-Tandem-Augmented Chemoimmunotherapy via Efficient Cancer-Homing Delivery and Optimized Ordinal-Interval Regime. Adv. Sci. Weinh 2022, 10, e2205247. [Google Scholar] [CrossRef]
- Gao, R.; Luo, Q.; Li, Y.; Song, L.; Cai, J.S.; Xiong, Y.; Yan, F.; Liu, J. Biosynthetic Nanobubble-Mediated CRISPR/Cas9 Gene Editing of Cdh2 Inhibits Breast Cancer Metastasis. Pharmaceutics 2022, 14, 1382. [Google Scholar] [CrossRef]
- Rahimi, H.; Zaboli, K.A.; Thekkiniath, J.; Mousavi, S.H.; Johari, B.; Hashemi, M.R.; Nosrati, H.; Goldschneider, D.; Bernet, A.; Danafar, H.; et al. BSA-PEI Nanoparticle Mediated Efficient Delivery of CRISPR/Cas9 into MDA-MB-231 Cells. Mol. Biotechnol. 2022, 64, 1376–1387. [Google Scholar] [CrossRef]
- Li, F.; Song, N.; Dong, Y.; Li, S.; Li, L.; Liu, Y.; Li, Z.; Yang, D. A Proton-Activatable DNA-Based Nanosystem Enables Co-Delivery of CRISPR/Cas9 and DNAzyme for Combined Gene Therapy. Angew. Chem. Int. Ed. Engl. 2022, 61, e202116569. [Google Scholar] [CrossRef]
- Duan, J.; Bao, C.; Xie, Y.; Guo, H.; Liu, Y.; Li, J.; Liu, R.; Li, P.; Bai, J.; Yan, Y.; et al. Targeted core-shell nanoparticles for precise CTCF gene insert in treatment of metastatic breast cancer. Bioact. Mater. 2022, 11, 1–14. [Google Scholar] [CrossRef]
- Yue, H.; Huang, R.; Shan, Y.; Xing, D. Delivery of Cas13a/crRNA by self-degradable black phosphorus nanosheets to specifically inhibit Mcl-1 for breast cancer therapy. J. Mater. Chem. B 2020, 8, 11096–11106. [Google Scholar] [CrossRef]
- Deng, H.; Tan, S.; Gao, X.; Zou, C.; Xu, C.; Tu, K.; Song, Q.; Fan, F.; Huang, W.; Zhang, Z. Cdk5 knocking out mediated by CRISPR-Cas9 genome editing for PD-L1 attenuation and enhanced antitumor immunity. Acta Pharm. Sin. B 2020, 10, 358–373. [Google Scholar] [CrossRef]
- Kretzmann, J.A.; Evans, C.W.; Moses, C.; Sorolla, A.; Kretzmann, A.L.; Wang, E.; Ho, D.; Hackett, M.J.; Dessauvagie, B.F.; Smith, N.M.; et al. Tumour suppression by targeted intravenous non-viral CRISPRa using dendritic polymers. Chem. Sci. 2019, 10, 7718–7727. [Google Scholar] [CrossRef]
- Guo, P.; Yang, J.; Huang, J.; Auguste, D.T.; Moses, M.A. Therapeutic genome editing of triple-negative breast tumors using a noncationic and deformable nanolipogel. Proc. Natl. Acad. Sci. USA 2019, 116, 18295–18303. [Google Scholar] [CrossRef]
- Lino, C.A.; Harper, J.C.; Carney, J.P.; Timlin, J.A. Delivering CRISPR: A review of the challenges and approaches. Drug Deliv. 2018, 25, 1234–1257. [Google Scholar] [CrossRef]
- Sioson, V.A.; Kim, M.; Joo, J. Challenges in delivery systems for CRISPR-based genome editing and opportunities of nanomedicine. Biomed. Eng. Lett. 2021, 11, 217–233. [Google Scholar] [CrossRef]
- Nakamura, Y.; Kubota, J.; Nishimura, Y.; Nagata, K.; Nishimura, M.; Daino, K.; Ishikawa, A.; Kaneko, T.; Mashimo, T.; Kokubo, T.; et al. Brca1(L63X) (/+) rat is a novel model of human BRCA1 deficiency displaying susceptibility to radiation-induced mammary cancer. Cancer Sci. 2022, 113, 3362–3375. [Google Scholar] [CrossRef]
- Sharif, G.M.; Campbell, M.J.; Nasir, A.; Sengupta, S.; Graham, G.T.; Kushner, M.H.; Kietzman, W.B.; Schmidt, M.O.; Pearson, G.W.; Loudig, O.; et al. An AIB1 Isoform Alters Enhancer Access and Enables Progression of Early-Stage Triple-Negative Breast Cancer. Cancer Res. 2021, 81, 4230–4241. [Google Scholar] [CrossRef]
- Lu, Y.; Xue, J.; Deng, T.; Zhou, X.; Yu, K.; Deng, L.; Huang, M.; Yi, X.; Liang, M.; Wang, Y.; et al. Safety and feasibility of CRISPR-edited T cells in patients with refractory non-small-cell lung cancer. Nat. Med. 2020, 26, 732–740. [Google Scholar] [CrossRef]
- Bhat, A.A.; Nisar, S.; Mukherjee, S.; Saha, N.; Yarravarapu, N.; Lone, S.N.; Masoodi, T.; Chauhan, R.; Maacha, S.; Bagga, P.; et al. Integration of CRISPR/Cas9 with artificial intelligence for improved cancer therapeutics. J. Transl. Med. 2022, 20, 534. [Google Scholar] [CrossRef]
- Lorenzo, D.; Esquerda, M.; Palau, F.; Cambra, F.J.; Bioética, G.I.e. Ethics and Genomic Editing Using the Crispr-Cas9 Technique: Challenges and Conflicts. Nanoethics 2022, 16, 313–321. [Google Scholar] [CrossRef]
| Delivery Method | Subtype | Material | Applicability/ Delivery of | Target Gene/s | In Vivo Model of Breast Cancer/TNBC | Route and Frequency of Administration | Ref. |
|---|---|---|---|---|---|---|---|
| Non-viral delivery | Lipid-polymer hybrid nanoparticles | Phenylboronic acid—functionalised low molecular weight polyethyleneimine PEI 1.8k (PEI-PBA) | dCas9-based CRISPR interference system (CRISPRi) | miR-10b | 4T1 TNBC allograft | Intravenously Every 5 days for 28 days | [48] |
| Non-viral delivery | Nanovesicles | Tumour-derived extracellular vesicles—fusogenic anthracycline doxorubicin liposomes (T-DOX) | CRISPR/Cas9 | PD-L1 | Orthotopic 4T1 TNBC allograft | Subcutaneously 1-day interval | [49] |
| Non-viral delivery | Nanobubbles | Polyethyleneimine (PEI) | CRISPR/Cas9 | Cdh2 | Orthotopic 4T1 TNBC allograft | N/A | [50] |
| Non-viral delivery | Polyethyleneimine–Bovine serum albumin-based nanoparticles | Polyethyleneimine– Bovine serum albumin (PEI-BSA) | CRISPR/Cas9 system in plasmid and ribonucleoprotein format | CD81 | BALB/c mice | Intravenously One injection | [51] |
| Non-viral delivery | DNA-based nanoparticles | Polyglycerol Dimethacrylate | Co-delivery of Cas9/sgRNA ribonucleoprotein and DNAzyme | PLK1 EGR-1 | Breast cancer MCF7 xenografts | Intravenously Days 0 and 6 | [52] |
| Non-viral delivery | Targeted core-shell nanoparticles | Polyacrylaminoester (PAA) | Dual plasmids pHR-pCas9 | CTCF | Female BALB/c nude mice | Intravenously | [53] |
| Non-viral delivery | Nanoparticles | Polylysine functionalised black phosphorus (PLL-PBP) | PBP/Cas13a/ crMcl-1 complex | Mcl-1 | TNBC MDA-MB-231 xenograft | Intratumourally Every two days for a total of 10 injections | [54] |
| Non-viral delivery | Autocatalytic brain tumour-targeted nanoparticles | HDL-DES-MDEA polymer | LRRC31 cDNA loaded NPs | LRRC31 | Female BALB/c nude mice | Intravenously Days 6 and 11 | [30] |
| Non-viral delivery | Polymeric nanoparticles | Poly-β-amino ester (PBAE) | aPBAE/cas9-Cdk5 complex | Cdk5 | TNBC orthotopic 4T1 allograft | Intratumourally Days 7, 10, 13, and 16 post-inoculation of 4T1 cells | [55] |
| Non-viral delivery | Organic polymer | Poly-glycidyl methacrylate (PGMA) | CRISPR/dCas9 conjugated to the effector domains VPR or SAM | MASPIN CCN6 | Breast cancer MCF7 xenograft | Intravenously Every 5 days | [56] |
| Non-viral delivery | Targeted nanolipogel (tNLGs) | Noncationic lipid bilayer and a biodegradable hydrogel core | Three CRISPR plasmids targeted to different DNA sequences of Lnc2 | Lnc2 | Orthotopic TNBC MDA-MB-231 xenograft | Intravenously Weekly, administered for 4 weeks | [57] |
| Viral delivery | Lentiviruses | Lentiviral pSECC vector encoding Cre and CRISPR components | sgRNA encoding vector | PTEN | Cas9-knock-in and mammary tissue-specific Cdh1F/F female mice | Intraductal injection | [19] |
| Viral delivery | Lentiviruses | Lentiviral | sgRNA encoding vector | PI3KCA Akt1 | Mammary tissue specific of the base editor BE3 and Cre, Cas9 knock-in Brca1F/F;Trp53F/F female mice | Intraductal injection | [20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pont, M.; Marqués, M.; Sorolla, M.A.; Parisi, E.; Urdanibia, I.; Morales, S.; Salud, A.; Sorolla, A. Applications of CRISPR Technology to Breast Cancer and Triple Negative Breast Cancer Research. Cancers 2023, 15, 4364. https://doi.org/10.3390/cancers15174364
Pont M, Marqués M, Sorolla MA, Parisi E, Urdanibia I, Morales S, Salud A, Sorolla A. Applications of CRISPR Technology to Breast Cancer and Triple Negative Breast Cancer Research. Cancers. 2023; 15(17):4364. https://doi.org/10.3390/cancers15174364
Chicago/Turabian StylePont, Mariona, Marta Marqués, Maria Alba Sorolla, Eva Parisi, Izaskun Urdanibia, Serafín Morales, Antonieta Salud, and Anabel Sorolla. 2023. "Applications of CRISPR Technology to Breast Cancer and Triple Negative Breast Cancer Research" Cancers 15, no. 17: 4364. https://doi.org/10.3390/cancers15174364
APA StylePont, M., Marqués, M., Sorolla, M. A., Parisi, E., Urdanibia, I., Morales, S., Salud, A., & Sorolla, A. (2023). Applications of CRISPR Technology to Breast Cancer and Triple Negative Breast Cancer Research. Cancers, 15(17), 4364. https://doi.org/10.3390/cancers15174364






