Active Autophagy Is Associated with Favorable Outcome in Patients with Surgically Resected Cholangiocarcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Immunohistochemistry (IHC)
2.3. Ribonucleic Acid (RNA) Isolation and Immune-Exhaustion Expression Analysis
2.4. Statistical Analysis
3. Results
3.1. Patients and Clinical Characteristics
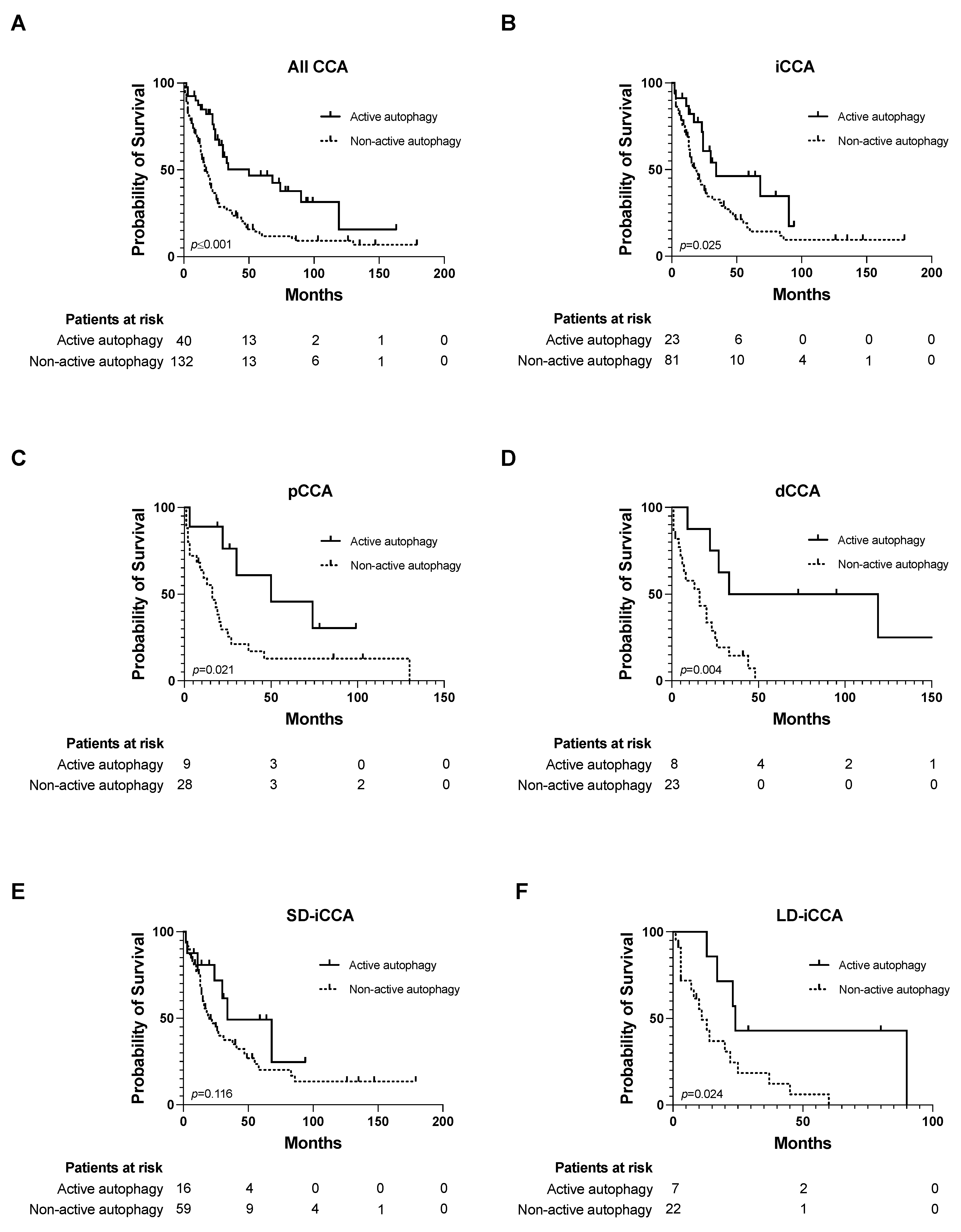
3.2. Impact of Autophagic Activity on Overall Survival
3.3. Active Autophagy as an Independent Risk Factor for OS
3.4. iCCA with Active Autophagy Displays a Differential Expression Profile
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kendall, T.; Verheij, J.; Gaudio, E.; Evert, M.; Guido, M.; Goeppert, B.; Carpino, G. Anatomical, histomorphological and molecular classification of cholangiocarcinoma. Liver Int. 2019, 39 (Suppl. S1), 7–18. [Google Scholar] [CrossRef]
- WHO Classification of Tumours 2019, 5th ed. Vol. 1. Digestive System Tumours. 2019. Available online: https://publications.iarc.fr/Book-And-Report-Series/Who-Classification-Of-Tumours/Digestive-System-Tumours-2019 (accessed on 1 June 2023).
- Bertuccio, P.; Bosetti, C.; Levi, F.; Decarli, A.; Negri, E.; La Vecchia, C. A comparison of trends in mortality from primary liver cancer and intrahepatic cholangiocarcinoma in Europe. Ann. Oncol. 2013, 24, 1667–1674. [Google Scholar] [CrossRef] [PubMed]
- Shaib, Y.H.; Davila, J.A.; McGlynn, K.; El-Serag, H.B. Rising incidence of intrahepatic cholangiocarcinoma in the United States: A true increase? J. Hepatol. 2004, 40, 472–477. [Google Scholar] [CrossRef]
- Groot Koerkamp, B.; Fong, Y. Outcomes in biliary malignancy. J. Surg. Oncol. 2014, 110, 585–591. [Google Scholar] [CrossRef]
- Valle, J.; Wasan, H.; Palmer, D.H.; Cunningham, D.; Anthoney, A.; Maraveyas, A.; Madhusudan, S.; Iveson, T.; Hughes, S.; Pereira, S.P.; et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010, 362, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Oh, D.-Y.; He, A.R.; Qin, S.; Chen, L.-T.; Okusaka, T.; Vogel, A.; Kim, J.W.; Suksombooncharoen, T.; Lee, M.A.; Kitano, M.; et al. A phase 3 randomized, double-blind, placebo-controlled study of durvalumab in combination with gemcitabine plus cisplatin (GemCis) in patients (pts) with advanced biliary tract cancer (BTC): TOPAZ-1. J. Clin. Oncol. 2022, 40, 378. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Doherty, J.; Baehrecke, E.H. Life, death and autophagy. Nat. Cell Biol. 2018, 20, 1110–1117. [Google Scholar] [CrossRef]
- Bialik, S.; Dasari, S.K.; Kimchi, A. Autophagy-dependent cell death—Where, how and why a cell eats itself to death. J. Cell Sci. 2018, 131, jcs215152. [Google Scholar] [CrossRef]
- Gewirtz, D.A. Cytoprotective and nonprotective autophagy in cancer therapy. Autophagy 2013, 9, 1263–1265. [Google Scholar] [CrossRef]
- Galluzzi, L.; Pietrocola, F.; Bravo-San Pedro, J.M.; Amaravadi, R.K.; Baehrecke, E.H.; Cecconi, F.; Codogno, P.; Debnath, J.; Gewirtz, D.A.; Karantza, V.; et al. Autophagy in malignant transformation and cancer progression. EMBO J. 2015, 34, 856–880. [Google Scholar] [CrossRef]
- Ma, Y.; Galluzzi, L.; Zitvogel, L.; Kroemer, G. Autophagy and cellular immune responses. Immunity 2013, 39, 211–227. [Google Scholar] [CrossRef]
- Bantug, G.R.; Galluzzi, L.; Kroemer, G.; Hess, C. The spectrum of T cell metabolism in health and disease. Nat. Rev. Immunol. 2017, 18, 19. [Google Scholar] [CrossRef]
- Sasaki, M.; Nitta, T.; Sato, Y.; Nakanuma, Y. Autophagy may occur at an early stage of cholangiocarcinogenesis via biliary intraepithelial neoplasia. Hum. Pathol. 2015, 46, 202–209. [Google Scholar] [CrossRef]
- Perng, D.-S.; Hung, C.-M.; Lin, H.-Y.; Morgan, P.; Hsu, Y.-C.; Wu, T.-C.; Hsieh, P.-M.; Yeh, J.-H.; Hsiao, P.; Lee, C.-Y.; et al. Role of autophagy-related protein in the prognosis of combined hepatocellular carcinoma and cholangiocarcinoma after surgical resection. BMC Cancer 2021, 21, 828. [Google Scholar] [CrossRef]
- Dong, L.W.; Hou, Y.J.; Tan, Y.X.; Tang, L.; Pan, Y.F.; Wang, M.; Wang, H.Y. Prognostic significance of Beclin 1 in intrahepatic cholangiocellular carcinoma. Autophagy 2011, 7, 1222–1229. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Cao, Q.H.; Chen, M.Y.; Xia, Q.; Fan, X.J.; Ma, X.K.; Lin, Q.; Jia, C.C.; Dong, M.; Ruan, D.Y.; et al. Beclin 1 deficiency correlated with lymph node metastasis, predicts a distinct outcome in intrahepatic and extrahepatic cholangiocarcinoma. PLoS ONE 2013, 8, e80317. [Google Scholar] [CrossRef] [PubMed]
- Lendvai, G.; Szekerczés, T.; Illyés, I.; Csengeri, M.; Schlachter, K.; Szabó, E.; Lotz, G.; Kiss, A.; Borka, K.; Schaff, Z. Autophagy activity in cholangiocarcinoma is associated with anatomical localization of the tumor. PLoS ONE 2021, 16, e0253065. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Fu, H.; Lu, T.; Cai, J.; Liu, W.; Yao, J.; Liang, J.; Zhao, H.; Zhang, J.; Zheng, J.; et al. An Integrated Nomogram Combining Clinical Factors and Microtubule-Associated Protein 1 Light Chain 3B Expression to Predict Postoperative Prognosis in Patients with Intrahepatic Cholangiocarcinoma. Cancer Res. Treat. 2020, 52, 469–480. [Google Scholar] [CrossRef]
- Thongchot, S.; Vidoni, C.; Ferraresi, A.; Loilome, W.; Khuntikeo, N.; Sangkhamanon, S.; Titapun, A.; Isidoro, C.; Namwat, N. Cancer-Associated Fibroblast-Derived IL-6 Determines Unfavorable Prognosis in Cholangiocarcinoma by Affecting Autophagy-Associated Chemoresponse. Cancers 2021, 13, 2134. [Google Scholar] [CrossRef]
- Chung, S.J.; Nagaraju, G.P.; Nagalingam, A.; Muniraj, N.; Kuppusamy, P.; Walker, A.; Woo, J.; Győrffy, B.; Gabrielson, E.; Saxena, N.K.; et al. ADIPOQ/adiponectin induces cytotoxic autophagy in breast cancer cells through STK11/LKB1-mediated activation of the AMPK-ULK1 axis. Autophagy 2017, 13, 1386–1403. [Google Scholar] [CrossRef]
- Donadelli, M.; Dando, I.; Zaniboni, T.; Costanzo, C.; Dalla Pozza, E.; Scupoli, M.T.; Scarpa, A.; Zappavigna, S.; Marra, M.; Abbruzzese, A.; et al. Gemcitabine/cannabinoid combination triggers autophagy in pancreatic cancer cells through a ROS-mediated mechanism. Cell Death Dis. 2011, 2, e152. [Google Scholar] [CrossRef]
- Vara, D.; Salazar, M.; Olea-Herrero, N.; Guzman, M.; Velasco, G.; Diaz-Laviada, I. Anti-tumoral action of cannabinoids on hepatocellular carcinoma: Role of AMPK-dependent activation of autophagy. Cell Death Differ. 2011, 18, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Perez-Montoyo, H. Therapeutic Potential of Autophagy Modulation in Cholangiocarcinoma. Cells 2020, 9, 614. [Google Scholar] [CrossRef]
- Kinzler, M.N.; Schulze, F.; Gretser, S.; Abedin, N.; Trojan, J.; Zeuzem, S.; Schnitzbauer, A.A.; Walter, D.; Wild, P.J.; Bankov, K. Expression of MUC16/CA125 Is Associated with Impaired Survival in Patients with Surgically Resected Cholangiocarcinoma. Cancers 2022, 14, 4703. [Google Scholar] [CrossRef]
- Kinzler, M.N.; Schulze, F.; Bankov, K.; Gretser, S.; Becker, N.; Leichner, R.; Stehle, A.; Abedin, N.; Trojan, J.; Zeuzem, S.; et al. Impact of small duct- and large duct type on survival in patients with intrahepatic cholangiocarcinoma: Results from a German tertiary center. Pathol. Res. Pract. 2022, 238, 154126. [Google Scholar] [CrossRef]
- Liu, J.L.; Chen, F.F.; Lung, J.; Lo, C.H.; Lee, F.H.; Lu, Y.C.; Hung, C.H. Prognostic significance of p62/SQSTM1 subcellular localization and LC3B in oral squamous cell carcinoma. Br. J. Cancer 2014, 111, 944–954. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Bindea, G.; Galon, J.; Mlecnik, B. CluePedia Cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef] [PubMed]
- Goedhart, J.; Luijsterburg, M.S. VolcaNoseR is a web app for creating, exploring, labeling and sharing volcano plots. Sci. Rep. 2020, 10, 20560. [Google Scholar] [CrossRef] [PubMed]
- Hamasaki, M.; Furuta, N.; Matsuda, A.; Nezu, A.; Yamamoto, A.; Fujita, N.; Oomori, H.; Noda, T.; Haraguchi, T.; Hiraoka, Y.; et al. Autophagosomes form at ER-mitochondria contact sites. Nature 2013, 495, 389–393. [Google Scholar] [CrossRef]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Overvatn, A.; Bjorkoy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef]
- Scarlatti, F.; Maffei, R.; Beau, I.; Codogno, P.; Ghidoni, R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ. 2008, 15, 1318–1329. [Google Scholar] [CrossRef] [PubMed]
- Grishchuk, Y.; Ginet, V.; Truttmann, A.C.; Clarke, P.G.; Puyal, J. Beclin 1-independent autophagy contributes to apoptosis in cortical neurons. Autophagy 2011, 7, 1115–1131. [Google Scholar] [CrossRef]
- White, E. Deconvoluting the context-dependent role for autophagy in cancer. Nat. Rev. Cancer 2012, 12, 401–410. [Google Scholar] [CrossRef]
- Di Lernia, G.; Leone, P.; Solimando, A.G.; Buonavoglia, A.; Saltarella, I.; Ria, R.; Ditonno, P.; Silvestris, N.; Crudele, L.; Vacca, A.; et al. Bortezomib Treatment Modulates Autophagy in Multiple Myeloma. J. Clin. Med. 2020, 9, 552. [Google Scholar] [CrossRef]
- Liang, C.; Dong, Z.; Cai, X.; Shen, J.; Xu, Y.; Zhang, M.; Li, H.; Yu, W.; Chen, W. Hypoxia induces sorafenib resistance mediated by autophagy via activating FOXO3a in hepatocellular carcinoma. Cell Death Dis. 2020, 11, 1017. [Google Scholar] [CrossRef]
- Gao, W.; Wang, X.; Zhou, Y.; Wang, X.; Yu, Y. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy. Signal Transduct. Target. Ther. 2022, 7, 196. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.-J.; Dong, L.-W.; Tan, Y.-X.; Yang, G.-Z.; Pan, Y.-F.; Li, Z.; Tang, L.; Wang, M.; Wang, Q.; Wang, H.-Y. Inhibition of active autophagy induces apoptosis and increases chemosensitivity in cholangiocarcinoma. Lab. Investig. 2011, 91, 1146–1157. [Google Scholar] [CrossRef]
- Malik, S.A.; Orhon, I.; Morselli, E.; Criollo, A.; Shen, S.; Marino, G.; BenYounes, A.; Benit, P.; Rustin, P.; Maiuri, M.C.; et al. BH3 mimetics activate multiple pro-autophagic pathways. Oncogene 2011, 30, 3918–3929. [Google Scholar] [CrossRef]
- Mott, J.L.; Bronk, S.F.; Mesa, R.A.; Kaufmann, S.H.; Gores, G.J. BH3-only protein mimetic obatoclax sensitizes cholangiocarcinoma cells to Apo2L/TRAIL-induced apoptosis. Mol. Cancer Ther. 2008, 7, 2339–2347. [Google Scholar] [CrossRef]
- Yi, C.; Ma, M.; Ran, L.; Zheng, J.; Tong, J.; Zhu, J.; Ma, C.; Sun, Y.; Zhang, S.; Feng, W.; et al. Function and molecular mechanism of acetylation in autophagy regulation. Science 2012, 336, 474–477. [Google Scholar] [CrossRef] [PubMed]
- Pant, K.; Peixoto, E.; Richard, S.; Gradilone, S.A. Role of Histone Deacetylases in Carcinogenesis: Potential Role in Cholangiocarcinoma. Cells 2020, 9, 780. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-L.; Yang, P.-M.; Shun, C.-T.; Wu, M.-S.; Weng, J.-R.; Chen, C.-C. Autophagy potentiates the anti-cancer effects of the histone deacetylase inhibitors in hepatocellular carcinoma. Autophagy 2010, 6, 1057–1065. [Google Scholar] [CrossRef] [PubMed]
- Jung, D.E.; Park, S.B.; Kim, K.; Kim, C.; Song, S.Y. CG200745, an HDAC inhibitor, induces anti-tumour effects in cholangiocarcinoma cell lines via miRNAs targeting the Hippo pathway. Sci. Rep. 2017, 7, 10921. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Yang, W.; Che, J.; Li, D.; Wang, H.; Li, Y.; Zhou, W. Suppression of histone deacetylase 1 by JSL-1 attenuates the progression and metastasis of cholangiocarcinoma via the TPX2/Snail axis. Cell Death Dis. 2022, 13, 324. [Google Scholar] [CrossRef] [PubMed]
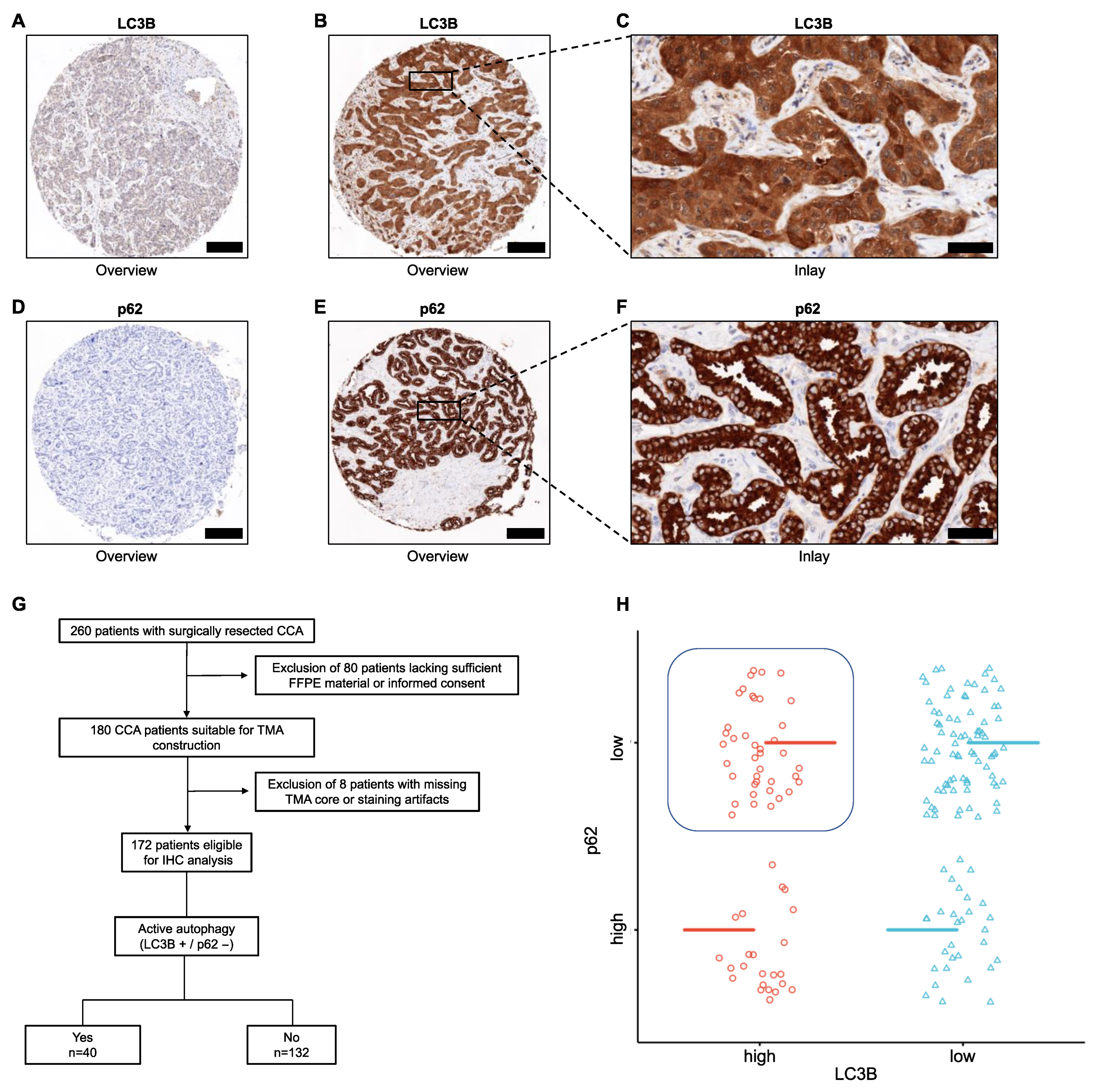



| Active Autophagy | |||
|---|---|---|---|
| Characteristics | Yes (n = 40) No. (%) | No (n = 132) No. (%) | p-Value |
| Sex | 0.573 | ||
| Female | 12 (30) | 46 (34.8) | |
| Male | 28 (70) | 86 (65.2) | |
| Age at initial diagnosis | 0.318 | ||
| Mean, years, (range) | 64.1 (42–86) | 65.9 (38–86) | |
| CCA subtype | 0.649 | ||
| iCCA | 23 (57.5) | 81 (61.4) | |
| pCCA | 9 (22.5) | 28 (21.2) | |
| dCCA | 8 (20) | 23 (17.4) | |
| iCCA subtype | 0.76 | ||
| Small duct | 16 (69.6) | 59 (72.8) | |
| Large duct | 7 (30.4) | 22 (27.2) | |
| ECOG | 0.026 | ||
| 0 | 32 (80) | 82 (62.1) | |
| 1 | 8 (20) | 45 (34.1) | |
| 2 | 0 (0) | 5 (3.8) | |
| CA-19/9 (ng/mL) | 0.23 | ||
| <37 | 16 (40) | 46 (34.8) | |
| ≥37 | 13 (32.5) | 62 (47) | |
| n.a. | 11 (27.5) | 24 (18.2) | |
| Tumor size (cm) | 0.004 | ||
| ≤5 | 31 (77.5) | 69 (52.3) | |
| >5 | 9 (22.5) | 63 (47.7) | |
| Single Tumor | 0.025 | ||
| Yes | 33 (82.5) | 84 (63.6) | |
| No | 7 (17.5) | 48 (36.4) | |
| Pathological grade | 0.009 | ||
| Grade 1 | 2 (5) | 1 (0.8) | |
| Grade 2 | 33 (82.5) | 91 (68.9) | |
| Grade 3 | 5 (12.5) | 40 (30.3) | |
| M status | 0.883 | ||
| M0 | 37 (92.5) | 123 (93.2) | |
| M1 | 3 (7.5) | 9 (6.8) | |
| R status | 0.063 | ||
| R0 | 35 (87.5) | 93 (70.5) | |
| R1 | 5 (12.5) | 34 (25.8) | |
| Rx | 0 (0) | 5 (3.8) | |
| L status | 0.297 | ||
| L0 | 22 (55) | 58 (43.9) | |
| L1 | 13 (32.5) | 52 (39.4) | |
| Lx | 5 (12.5) | 22 (16.7) | |
| Pn status | 0.119 | ||
| Pn0 | 15 (37.5) | 32 (24.2) | |
| Pn1 | 19 (47.5) | 76 (57.6) | |
| Pnx | 6 (15) | 24 (18.2) | |
| Recurrence | 0.723 | ||
| Yes | 16 (40) | 57 (43.2) | |
| No | 24 (60) | 75 (56.8) | |
| Cholelithiasis | 0.746 | ||
| Yes | 3 (7.5) | 8 (6.1) | |
| No | 37 (92.5) | 124 (93.9) | |
| PSC | 0.172 | ||
| Yes | 0 (0) | 6 (4.5) | |
| No | 40 (100) | 126 (95.5) | |
| Viral hepatitis | 0.071 | ||
| Yes | 6 (15) | 8 (6.1) | |
| No | 34 (85) | 124 (93.9) | |
| Diabetes | 0.286 | ||
| Yes | 7 (17.5) | 34 (25.8) | |
| No | 33 (82.5) | 98 (74.2) | |
| Liver cirrhosis | 0.905 | ||
| Yes | 2 (5) | 6 (4.5) | |
| No | 38 (95) | 126 (95.5) | |
| LDH | 0.732 | ||
| <248 | 20 (50) | 68 (51.5) | |
| ≥248 | 8 (20) | 32 (24.2) | |
| n.a. | 12 (30) | 32 (24.2) | |
| Bilirubin | 0.551 | ||
| <1.4 | 28 (70) | 86 (65.2) | |
| ≥1.4 | 11 (27.5) | 43 (32.6) | |
| n.a. | 1 (2.5) | 3 (2.3) | |
| Univariate Analysis | Multivariate Analysis | |||||
|---|---|---|---|---|---|---|
| Characteristics | HR | 95% CI | p-Value | HR | 95% CI | p-Value |
| Sex | ||||||
| Female | ref | |||||
| Male | 1.232 | 0.846–1.794 | 0.276 | |||
| CCA subtype | ||||||
| iCCA | ref | |||||
| pCCA | 1.228 | 0.8–1.885 | 0.348 | |||
| dCCA | 1.234 | 0.786–1.937 | 0.36 | |||
| ECOG | ||||||
| 0 | ref | ref | ||||
| 1 | 2.547 | 1.724–3.763 | <0.001 | 2.209 | 1.403–3.476 | 0.001 |
| 2 | 2.386 | 0.869–6.554 | 0.092 | 1.684 | 0.588–4.829 | 0.332 |
| CA-19/9 (ng/mL) | ||||||
| <37 | ref | ref | ||||
| ≥37 | 2.197 | 1.455–3.317 | <0.001 | 1.814 | 1.176–2.799 | 0.007 |
| Tumor size (cm) | ||||||
| ≤5 | ref | |||||
| >5 | 1.058 | 0.742–1.508 | 0.755 | |||
| Single Tumor | ||||||
| Yes | ref | ref | ||||
| No | 2.09 | 1.438–3.039 | <0.001 | 1.522 | 0.982–2.36 | 0.06 |
| Active autophagy | ||||||
| Yes | ref | ref | ||||
| No | 2.46 | 1.546–3.913 | <0.001 | 1.988 | 1.143–3.459 | 0.015 |
| Pathological grade | ||||||
| Grade 1 | ref | ref | ||||
| Grade 2 | 1.616 | 0.397–6.584 | 0.503 | 0.842 | 0.111–6.381 | 0.868 |
| Grade 3 | 4.596 | 1.101–19.182 | 0.036 | 1.902 | 0.246–14.7 | 0.538 |
| M status | ||||||
| M0 | ref | ref | ||||
| M1 | 2.555 | 1.393–4.686 | 0.002 | 2.05 | 1.004–4.184 | 0.049 |
| R status | ||||||
| R0 | ref | |||||
| R1 | 1.435 | 0.963–2.137 | 0.076 | |||
| Recurrence | ||||||
| No | ref | |||||
| Yes | 1.051 | 0.74–1.494 | 0.781 | |||
| Cholelithiasis | ||||||
| No | ref | |||||
| Yes | 1.643 | 0.881–3.067 | 0.119 | |||
| PSC | ||||||
| No | ref | |||||
| Yes | 1.651 | 0.725–3.759 | 0.232 | |||
| Diabetes | ||||||
| No | ref | |||||
| Yes | 1.197 | 0.794–1.804 | 0.39 | |||
| Viral hepatitis | ||||||
| No | ref | |||||
| Yes | 0.512 | 0.239–1.099 | 0.086 | |||
| Liver cirrhosis | ||||||
| No | ref | |||||
| Yes | 0.713 | 0.263–1.935 | 0.507 | |||
| LDH | ||||||
| <248 | ref | |||||
| ≥248 | 1.211 | 0.779–1.884 | 0.396 | |||
| Bilirubin | ||||||
| <1.4 | ref | ref | ||||
| ≥1.4 | 1.55 | 1.075–2.235 | 0.019 | 1.555 | 1.02–2.369 | 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bankov, K.; Schulze, F.; Gretser, S.; Reis, H.; Abedin, N.; Finkelmeier, F.; Trojan, J.; Zeuzem, S.; Schnitzbauer, A.A.; Walter, D.; et al. Active Autophagy Is Associated with Favorable Outcome in Patients with Surgically Resected Cholangiocarcinoma. Cancers 2023, 15, 4322. https://doi.org/10.3390/cancers15174322
Bankov K, Schulze F, Gretser S, Reis H, Abedin N, Finkelmeier F, Trojan J, Zeuzem S, Schnitzbauer AA, Walter D, et al. Active Autophagy Is Associated with Favorable Outcome in Patients with Surgically Resected Cholangiocarcinoma. Cancers. 2023; 15(17):4322. https://doi.org/10.3390/cancers15174322
Chicago/Turabian StyleBankov, Katrin, Falko Schulze, Steffen Gretser, Henning Reis, Nada Abedin, Fabian Finkelmeier, Jörg Trojan, Stefan Zeuzem, Andreas A. Schnitzbauer, Dirk Walter, and et al. 2023. "Active Autophagy Is Associated with Favorable Outcome in Patients with Surgically Resected Cholangiocarcinoma" Cancers 15, no. 17: 4322. https://doi.org/10.3390/cancers15174322
APA StyleBankov, K., Schulze, F., Gretser, S., Reis, H., Abedin, N., Finkelmeier, F., Trojan, J., Zeuzem, S., Schnitzbauer, A. A., Walter, D., Wild, P. J., & Kinzler, M. N. (2023). Active Autophagy Is Associated with Favorable Outcome in Patients with Surgically Resected Cholangiocarcinoma. Cancers, 15(17), 4322. https://doi.org/10.3390/cancers15174322







