Studies in Cancer Epigenetics through a Sex and Gendered Lens: A Comprehensive Scoping Review
Abstract
Simple Summary
Abstract
1. Introduction
1.1. Rationale
1.2. Objectives
- SEX refers to the different biological and physiological characteristics of females, males, and intersex persons, such as chromosomes, hormones, and reproductive organs. Gender and sex are related to but different from gender identity.
- GENDER refers to the characteristics of women, men, girls, and boys that are socially constructed. This includes norms, behaviors, and roles associated with being a woman, man, girl, or boy, as well as relationships with each other. As a social construct, gender varies from society to society and can change over time.
2. Methods
3. Results
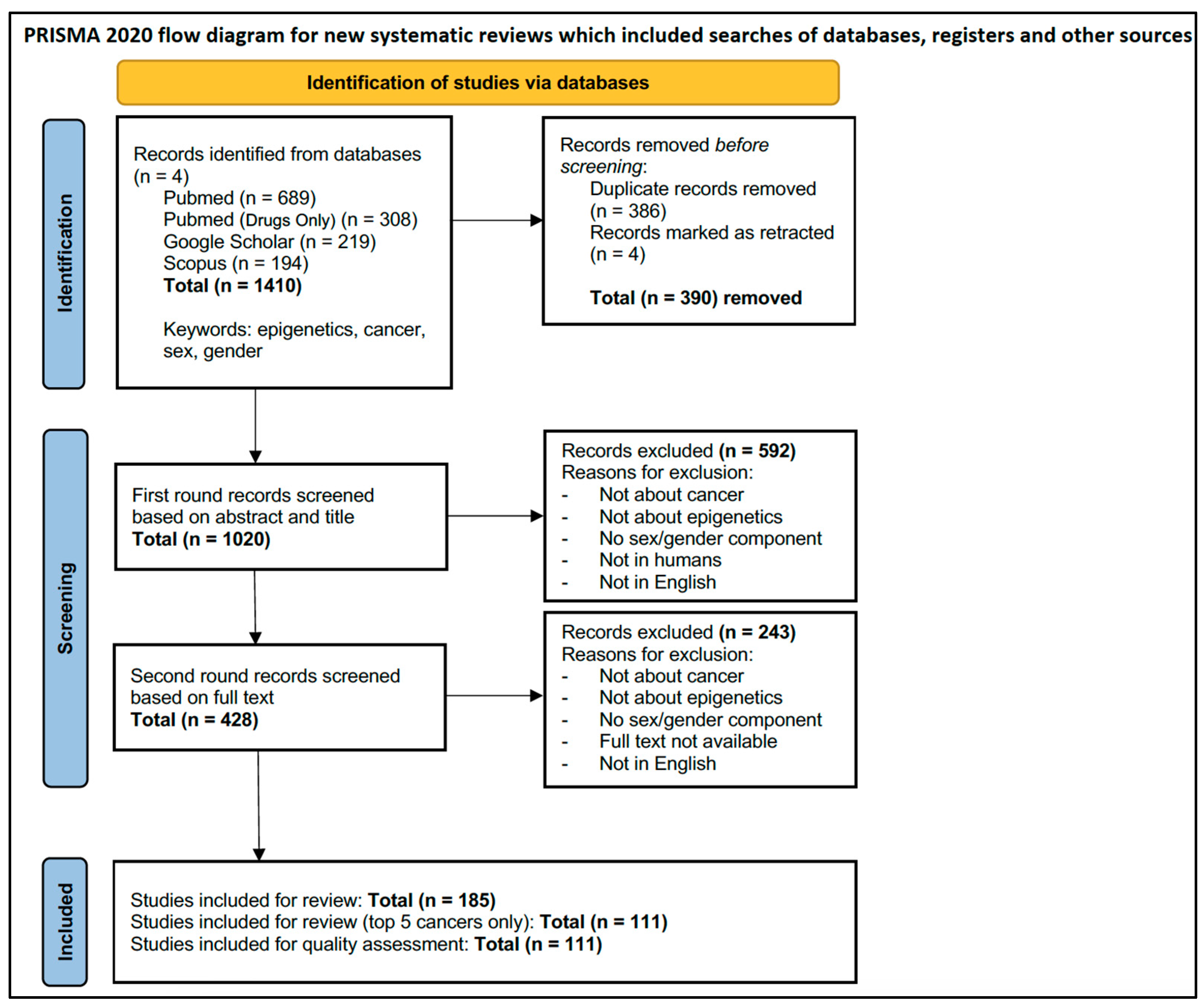
3.1. Selection of Sources of Evidence
3.2. Characteristics of Sources of Evidence
3.2.1. Country of Origin
3.2.2. Publication Year
3.2.3. Cancer Type
3.2.4. Study Type
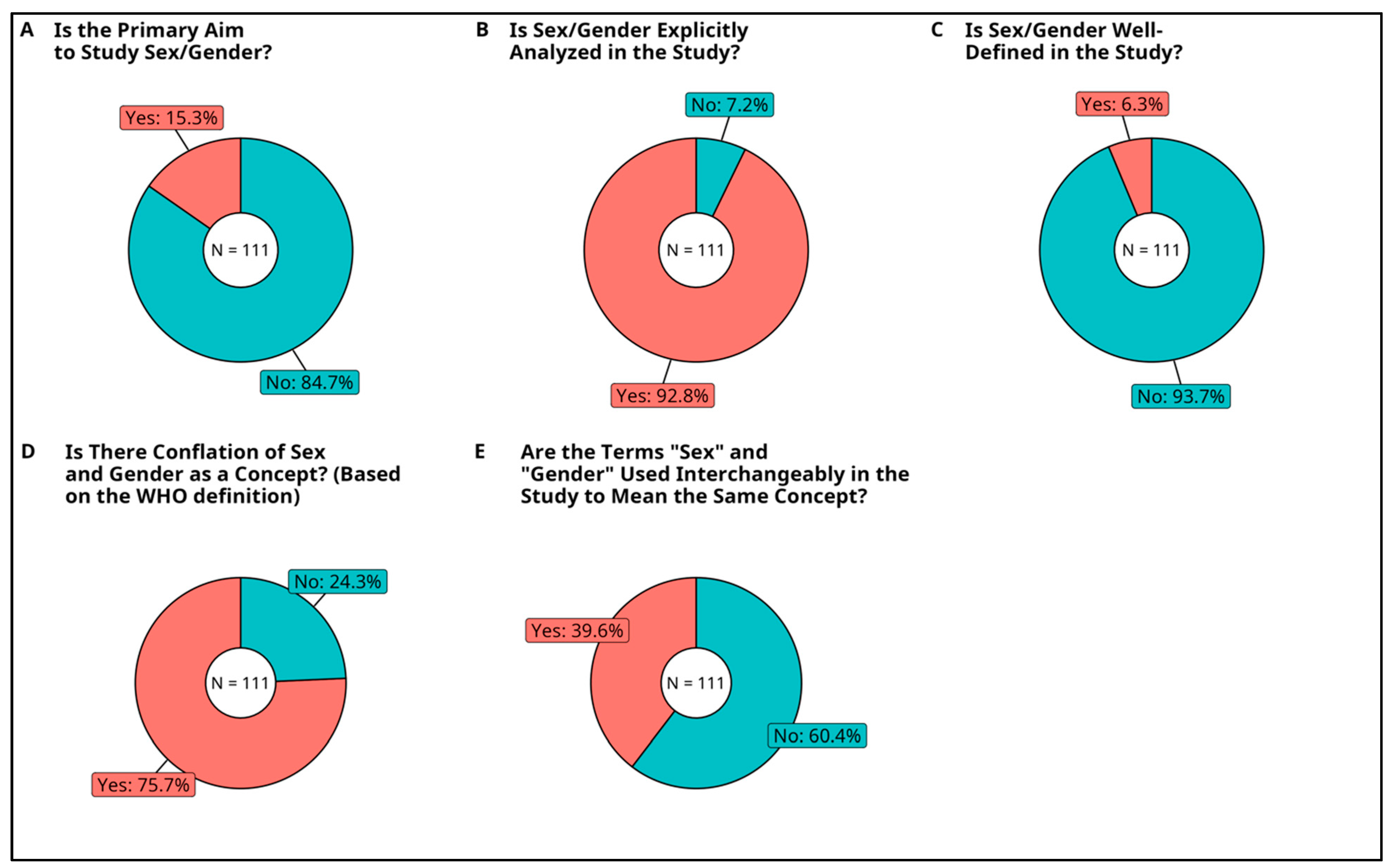
3.2.5. Primary Aim to Study Sex and/or Gender
3.2.6. Analysis of Sex or Gender
3.2.7. Inclusion of Sex and Gender Definitions
3.2.8. Conflation of Sex and Gender as Concepts
3.2.9. Usage of Sex and Gender Terms Interchangeably
3.2.10. Inclusion of Non-Binary Sex/Gender Minorities
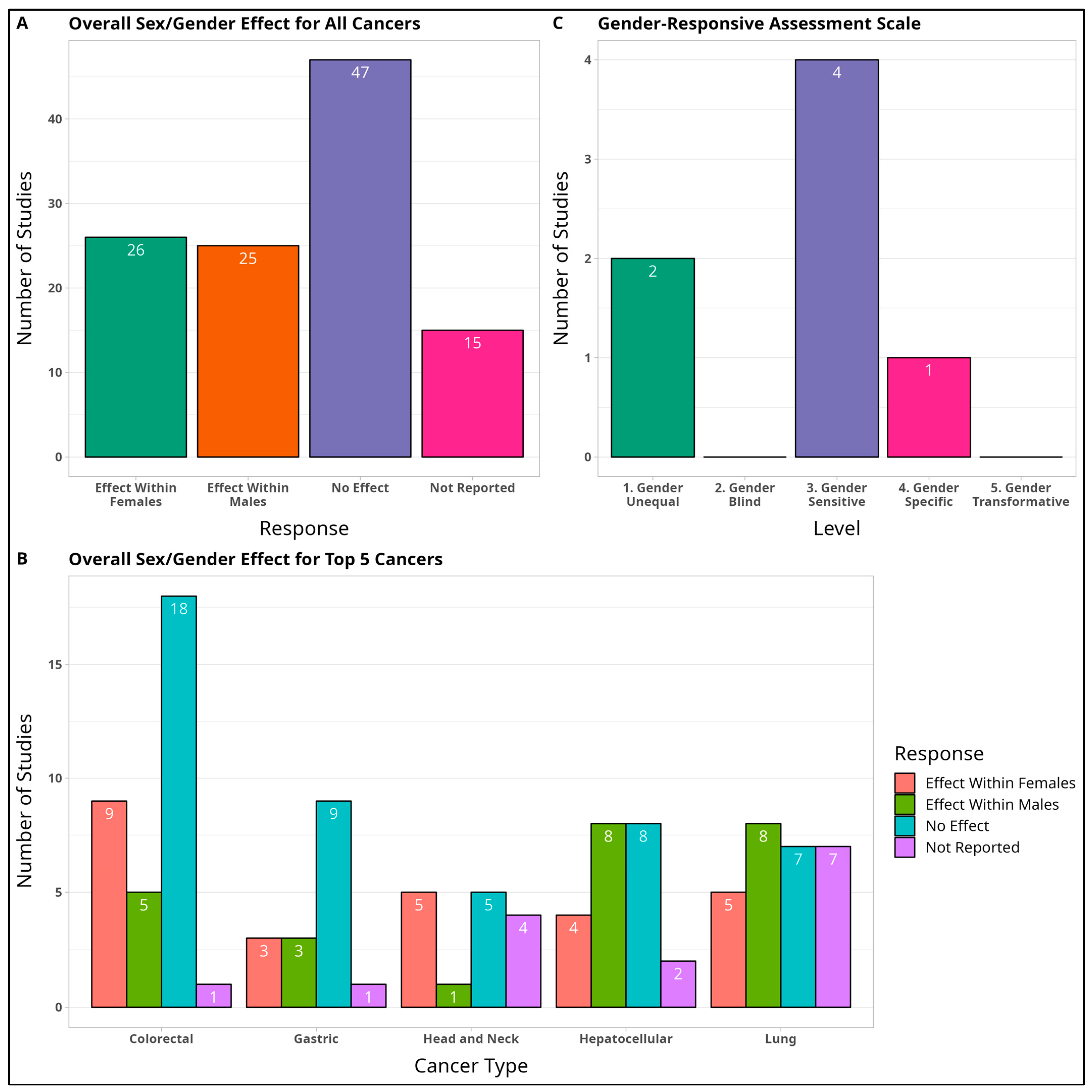
3.2.11. Overall Sex Effect
3.2.12. Gender-Responsive Assessment Scale
3.3. Critical Appraisal within Sources of Evidence
3.4. Results of Individual Sources of Evidence
3.4.1. Colorectal Cancer (CRC)
3.4.2. Gastric Cancer (GC)
3.4.3. Head and Neck Cancers (HNC)
3.4.4. Hepatocellular Carcinoma (HCC)
3.4.5. Lung Cancer
3.5. Synthesis of Results
4. Discussion
4.1. Summary of Evidence
4.2. Limitations
4.3. Future Directions
5. Conclusions
- ❖
- Define a clear conceptual framework for the sex/gender terms used and list the variables for each sex/gender category (e.g., male, female, intersex) in the study.
- ❖
- Include a variety of sex or gender-based covariates in the methodological analysis or discussion sections of the study, with considerations of sex and gender minorities treated distinctively.
- ➢
- For example, gender could be incorporated into statistical analysis by controlling for lifestyle factors (diet, access to screening and diagnosis, smoking habits).
- ❖
- Ensure that all researchers are trained in sex and gender-based analysis methods to create awareness of an-alytical and reporting bias.
- ❖
- Inquire and motivate international scientific organizations to delineate standards for the usage and inter-pretation of sex/gender data in the epigenetics and genetics field.
- ❖
- For authors and journals, ensure higher standards of reporting and publishing sex and gender-based data.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lopes-Ramos, C.M.; Quackenbush, J.; DeMeo, D.L. Genome-Wide Sex and Gender Differences in Cancer. Front. Oncol. 2020, 10, 597788. [Google Scholar] [CrossRef] [PubMed]
- Fazzari, M.J.; Greally, J.M. Introduction to Epigenomics and Epigenome-Wide Analysis. Methods Mol. Biol. 2010, 620, 243–265. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization; Kari, A. Gender and Health. Available online: https://www.who.int/health-topics/gender#tab=tab_1 (accessed on 22 September 2022).
- Heidari, S.; Babor, T.F.; De Castro, P.; Tort, S.; Curno, M. Sex and Gender Equity in Research: Rationale for the SAGER Guidelines and Recommended Use. Res. Integr. Peer Rev. 2016, 1, 2. [Google Scholar] [CrossRef] [PubMed]
- Sex/Gender Analysis of Cancer Epigenetics. Available online: https://osf.io/huz7s (accessed on 17 November 2021).
- WHO Regional Office for Europe. WHO Regional Office for Europe. WHO Gender Responsive Assessment Scale: Criteria for Assessing Programmes and Policies. In Mental Health, Men and Culture: How Do Sociocultural Constructions of Masculinities Relate to Men’s Mental Health Help-Seeking Behaviour in the WHO European Region? Health Evidence Network Synthesis Report; WHO Regional Office for Europe: Copenhagen, Denmark, 2020. [Google Scholar]
- Park, J.-O.; Nam, I.-C.; Kim, C.-S.; Park, S.-J.; Lee, D.-H.; Kim, H.-B.; Han, K.-D.; Joo, Y.-H. Sex Differences in the Prevalence of Head and Neck Cancers: A 10-Year Follow-Up Study of 10 Million Healthy People. Cancers 2022, 14, 2521. [Google Scholar] [CrossRef]
- Thomas, T.H. What Are the Most Common Types of Cancers? Available online: https://www.medicalnewstoday.com/articles/what-are-the-most-common-types-of-cancers (accessed on 27 October 2022).
- Lou, L.; Wang, L.; Zhang, Y.; Chen, G.; Lin, L.; Jin, X.; Huang, Y.; Chen, J. Sex Difference in Incidence of Gastric Cancer: An International Comparative Study Based on the Global Burden of Disease Study 2017. BMJ Open 2020, 10, e033323. [Google Scholar] [CrossRef]
- Wu, E.M.; Wong, L.L.; Hernandez, B.Y.; Ji, J.-F.; Jia, W.; Kwee, S.A.; Kalathil, S. Gender Differences in Hepatocellular Cancer: Disparities in Nonalcoholic Fatty Liver Disease/Steatohepatitis and Liver Transplantation. Hepatoma Res. 2018, 4, 66. [Google Scholar] [CrossRef]
- Zheng, R.; Zeng, H.; Zhang, S.; Gu, X.; Sun, K.; Xia, C.; Yang, Z.; Li, H.; Chen, W. The Epidemiology of Colorectal Cancer in China. Glob. Health J. 2018, 2, 8–20. [Google Scholar] [CrossRef]
- Jia, M.; Gao, X.; Zhang, Y.; Hoffmeister, M.; Brenner, H. Different Definitions of CpG Island Methylator Phenotype and Outcomes of Colorectal Cancer: A Systematic Review. Clin. Epigenet. 2016, 8, 25. [Google Scholar] [CrossRef]
- Kim, S.-E.; Paik, H.Y.; Yoon, H.; Lee, J.E.; Kim, N.; Sung, M.-K. Sex-and Gender-Specific Disparities in Colorectal Cancer Risk. World J. Gastroenterol. 2015, 21, 5167. [Google Scholar] [CrossRef]
- Liang, T.-J.; Wang, H.-X.; Zheng, Y.-Y.; Cao, Y.-Q.; Wu, X.; Zhou, X.; Dong, S.-X. APC Hypermethylation for Early Diagnosis of Colorectal Cancer: A Meta-Analysis and Literature Review. Oncotarget 2017, 8, 46468–46479. [Google Scholar] [CrossRef]
- Fennell, L.; Dumenil, T.; Wockner, L.; Hartel, G.; Nones, K.; Bond, C.; Borowsky, J.; Liu, C.; McKeone, D.; Bowdler, L.; et al. Integrative Genome-Scale DNA Methylation Analysis of a Large and Unselected Cohort Reveals 5 Distinct Subtypes of Colorectal Adenocarcinomas. Cell Mol. Gastroenterol. Hepatol. 2019, 8, 269–290. [Google Scholar] [CrossRef]
- Bi, H.; Liu, Y.; Pu, R.; Xia, T.; Sun, H.; Huang, H.; Zhang, L.; Zhang, Y.; Liu, Y.; Xu, J. CHST7 Gene Methylation and Sex-Specific Effects on Colorectal Cancer Risk. Dig. Dis. Sci. 2019, 64, 2158–2166. [Google Scholar] [CrossRef] [PubMed]
- Kashani, E.; Hadizadeh, M.; Chaleshi, V.; Mirfakhraie, R.; Young, C.; Savabkar, S.; Irani, S.; Aghdaei, H.A.; Bonab, M.A. The Differential DNA Hypermethylation Patterns of MicroRNA-137 and MicroRNA-342 Locus in Early Colorectal Lesions and Tumours. Biomolecules 2019, 9, 519. [Google Scholar] [CrossRef]
- Rawłuszko, A.A.; Horbacka, K.; Krokowicz, P.; Jagodziński, P.P. Decreased Expression of 17β-Hydroxysteroid Dehydrogenase Type 1 Is Associated with DNA Hypermethylation in Colorectal Cancer Located in the Proximal Colon. BMC Cancer 2011, 11, 522. [Google Scholar] [CrossRef]
- Hu, G.; Qin, L.; Zhang, X.; Ye, G.; Huang, T. Epigenetic Silencing of the MLH1 Promoter in Relation to the Development of Gastric Cancer and Its Use as a Biomarker for Patients with Microsatellite Instability: A Systematic Analysis. Cell Physiol. Biochem. 2018, 45, 148–162. [Google Scholar] [CrossRef] [PubMed]
- Ghadami, E.; Nikbakhsh, N.; Fattahi, S.; Kosari-Monfared, M.; Ranaee, M.; Taheri, H.; Amjadi-Moheb, F.; Godazandeh, G.; Shafaei, S.; Nosrati, A.; et al. Epigenetic Alterations of CYLD Promoter Modulate Its Expression in Gastric Adenocarcinoma: A Footprint of Infections. J. Cell Physiol. 2019, 234, 4115–4124. [Google Scholar] [CrossRef]
- Qu, X.; Zhao, L.; Zhang, R.; Wei, Q.; Wang, M. Differential MicroRNA Expression Profiles Associated with Microsatellite Status Reveal Possible Epigenetic Regulation of Microsatellite Instability in Gastric Adenocarcinoma. Ann. Transl. Med. 2020, 8, 484. [Google Scholar] [CrossRef] [PubMed]
- Waraya, M.; Yamashita, K.; Ema, A.; Katada, N.; Kikuchi, S.; Watanabe, M. Exclusive Association of P53 Mutation with Super-High Methylation of Tumor Suppressor Genes in the P53 Pathway in a Unique Gastric Cancer Phenotype. PLoS ONE 2015, 10, e0139902. [Google Scholar] [CrossRef]
- Uesugi, N.; Sugai, T.; Sugimoto, R.; Eizuka, M.; Fujita, Y.; Sato, A.; Osakabe, M.; Ishida, K.; Shiomi, E.; Toya, Y.; et al. Clinicopathological and Molecular Findings of Differentiated-Type Minute Gastric Intramucosal Neoplasia. Digestion 2020, 101, 287–297. [Google Scholar] [CrossRef]
- Shen, S.; Wang, G.; Shi, Q.; Zhang, R.; Zhao, Y.; Wei, Y.; Chen, F.; Christiani, D.C. Seven-CpG-Based Prognostic Signature Coupled with Gene Expression Predicts Survival of Oral Squamous Cell Carcinoma. Clin. Epigenet. 2017, 9, 88. [Google Scholar] [CrossRef]
- Misawa, K.; Mochizuki, D.; Endo, S.; Mima, M.; Misawa, Y.; Imai, A.; Shinmura, K.; Kanazawa, T.; Carey, T.E.; Mineta, H. Site-Specific Methylation Patterns of the GAL and GALR1/2 Genes in Head and Neck Cancer: Potential Utility as Biomarkers for Prognosis. Mol. Carcinog. 2017, 56, 1107–1116. [Google Scholar] [CrossRef] [PubMed]
- Langevin, S.M.; Stone, R.A.; Bunker, C.H.; Grandis, J.R.; Sobol, R.W.; Taioli, E. MicroRNA-137 Promoter Methylation in Oral Rinses from Patients with Squamous Cell Carcinoma of the Head and Neck Is Associated with Gender and Body Mass Index. Carcinogenesis 2010, 31, 864–870. [Google Scholar] [CrossRef] [PubMed]
- Challouf, S.; Ziadi, S.; Zaghdoudi, R.; Ksiaa, F.; Ben Gacem, R.; Trimeche, M. Patterns of Aberrant DNA Hypermethylation in Nasopharyngeal Carcinoma in Tunisian Patients. Clin. Chim. Acta 2012, 413, 795–802. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.X.; Zhu, L.Q.; Pang, F.; Sun, W.; Ou, C.; Li, Y.; Cao, J.; Hu, Y.L. Relationship between RUNX3 Methylation and Hepatocellular Carcinoma in Asian Populations: A Systematic Review. Genet. Mol. Res. 2014, 13, 5182–5189. [Google Scholar] [CrossRef]
- Zhang, H.; Nie, W.; Huang, F. The Correlation Relationship between P14ARF Gene DNA Methylation and Primary Liver Cancer. Med. Sci. Monit. 2015, 21, 3077–3082. [Google Scholar] [CrossRef][Green Version]
- Meunier, L.; Hirsch, T.Z.; Caruso, S.; Imbeaud, S.; Bayard, Q.; Roehrig, A.; Couchy, G.; Nault, J.-C.; Llovet, J.M.; Blanc, J.-F.; et al. DNA Methylation Signatures Reveal the Diversity of Processes Remodeling Hepatocellular Carcinoma Methylomes. Hepatology 2021, 74, 816–834. [Google Scholar] [CrossRef]
- Zhou, Y.; Qiu, X.-P.; Li, Z.-H.; Zhang, S.; Rong, Y.; Yang, G.-H.; Zheng, F. Clinical Significance of Aberrant Cyclin-Dependent Kinase-like 2 Methylation in Hepatocellular Carcinoma. Gene 2019, 683, 35–40. [Google Scholar] [CrossRef]
- Nishida, N.; Kudo, M.; Nagasaka, T.; Ikai, I.; Goel, A. Characteristic Patterns of Altered DNA Methylation Predict Emergence of Human Hepatocellular Carcinoma. Hepatology 2012, 56, 994–1003. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Xie, X.; Chen, Z.; Wu, L.; Yu, Z.; Guo, X.; Chen, G. Association of TNFRSF12A Methylation with Prognosis in Hepatocellular Carcinoma With History of Alcohol Consumption. Front. Genet. 2019, 10, 1299. [Google Scholar] [CrossRef]
- Ye, W.; Siwko, S.; Tsai, R.Y.L. Sex and Race-Related DNA Methylation Changes in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2021, 22, 3820. [Google Scholar] [CrossRef]
- Li, W.; Deng, J.; Wang, S.-S.; Ma, L.; Pei, J.; Zeng, X.-X.; Tang, J.-X. Association of Methylation of the RAR-β Gene with Cigarette Smoking in Non-Small Cell Lung Cancer with Southern-Central Chinese Population. Asian Pac. J. Cancer Prev. 2014, 15, 10937–10941. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-A.; Kim, Y.; Hong, S.-H.; Lee, J.; Cho, Y.G.; Han, J.-Y.; Kim, Y.-H.; Han, J.; Shim, Y.M.; Lee, Y.-S.; et al. Epigenetic Inactivation of Heparan Sulfate (Glucosamine) 3-O-Sulfotransferase 2 in Lung Cancer and Its Role in Tumorigenesis. PLoS ONE 2013, 8, e79634. [Google Scholar] [CrossRef] [PubMed]
- Peddireddy, V. Lung Cancer Incidence in Never Smokers: Genetic and Gender Basis. Gene Rep. 2016, 4, 198–207. [Google Scholar] [CrossRef]
- Nguyen, Q.N.; Vuong, L.D.; Truong, V.-L.; Ta, T.V.; Nguyen, N.T.; Nguyen, H.P.; Chu, H.H. Genetic and Epigenetic Alterations of the EGFR and Mutually Independent Association with BRCA1, MGMT, and RASSF1A Methylations in Vietnamese Lung Adenocarcinomas. Pathol. Res. Pract. 2019, 215, 885–892. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, M.; Yang, L.; Li, Y.; Wang, L.; Huang, Z.; Wang, L.; Chen, Z.; Zhou, M. Prevalence and Patterns of Tobacco Smoking among Chinese Adult Men and Women: Findings of the 2010 National Smoking Survey. J. Epidemiol. Community Health 2017, 71, 154–161. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.P.; Levy, D.T.; Cheng, T.Y.; Hsu, C.-C.; Tsai, S.P. Smoking Behaviour in Taiwan, 2001. Tob. Control 2005, 14, i51. [Google Scholar] [CrossRef]
- Lu, Y.; Liu, Y.; Liao, S.; Tu, W.; Shen, Y.; Yan, Y.; Tao, D.; Lu, Y.; Ma, Y.; Yang, Y.; et al. Epigenetic Modifications Promote the Expression of the Orphan Nuclear Receptor NR0B1 in Human Lung Adenocarcinoma Cells. Oncotarget 2016, 7, 43162–43176. [Google Scholar] [CrossRef]
- Li, N.; Zhan, X. Identification of Pathology-Specific Regulators of M6A RNA Modification to Optimize Lung Cancer Management in the Context of Predictive, Preventive, and Personalized Medicine. EPMA J. 2020, 11, 485–504. [Google Scholar] [CrossRef]
- Arnegard, M.E.; Whitten, L.A.; Hunter, C.; Clayton, J.A. Sex as a Biological Variable: A 5-Year Progress Report and Call to Action. J. Women’s Health 2020, 29, 858–864. [Google Scholar] [CrossRef]

| Cancer Type | Findings |
|---|---|
| Colorectal Cancer | 32 total studies
|
| Gastric Cancer | 16 total studies
|
| Head and Neck Cancers | 15 total studies
|
| Hepatocellular Carcinoma | 22 total studies
|
| Lung Cancer | 26 total studies
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huerne, K.; Jackson, S.S.; Lall, R.; Palmour, N.; Berner, A.M.; Dupras, C.; Joly, Y. Studies in Cancer Epigenetics through a Sex and Gendered Lens: A Comprehensive Scoping Review. Cancers 2023, 15, 4207. https://doi.org/10.3390/cancers15174207
Huerne K, Jackson SS, Lall R, Palmour N, Berner AM, Dupras C, Joly Y. Studies in Cancer Epigenetics through a Sex and Gendered Lens: A Comprehensive Scoping Review. Cancers. 2023; 15(17):4207. https://doi.org/10.3390/cancers15174207
Chicago/Turabian StyleHuerne, Katherine, Sarah S. Jackson, Rina Lall, Nicole Palmour, Alison May Berner, Charles Dupras, and Yann Joly. 2023. "Studies in Cancer Epigenetics through a Sex and Gendered Lens: A Comprehensive Scoping Review" Cancers 15, no. 17: 4207. https://doi.org/10.3390/cancers15174207
APA StyleHuerne, K., Jackson, S. S., Lall, R., Palmour, N., Berner, A. M., Dupras, C., & Joly, Y. (2023). Studies in Cancer Epigenetics through a Sex and Gendered Lens: A Comprehensive Scoping Review. Cancers, 15(17), 4207. https://doi.org/10.3390/cancers15174207






