Unlocking the Potential of Kinase Targets in Cancer: Insights from CancerOmicsNet, an AI-Driven Approach to Drug Response Prediction in Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Deep Learning Model
2.2. Saliency Graph
3. Results
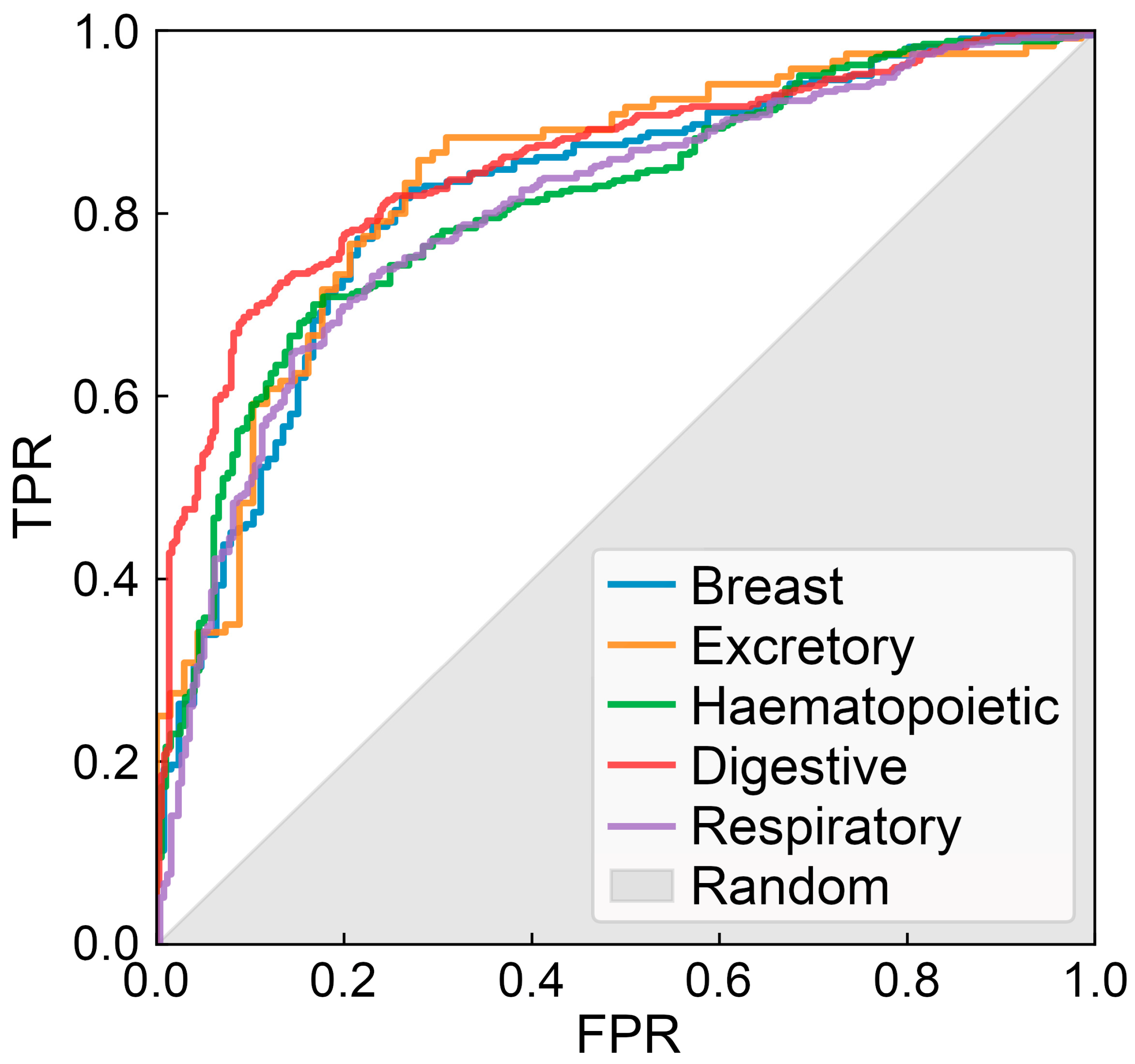
3.1. Architecture and Large-Scale Benchmarks of CancerOmicsNet
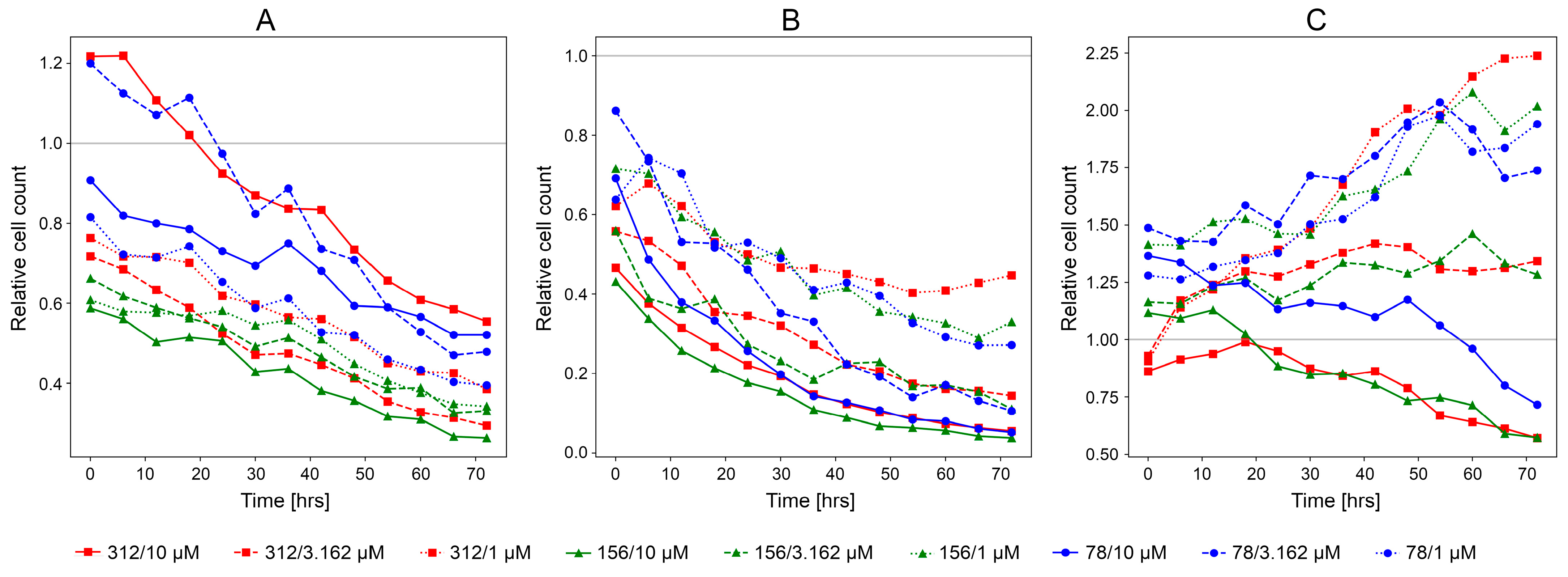
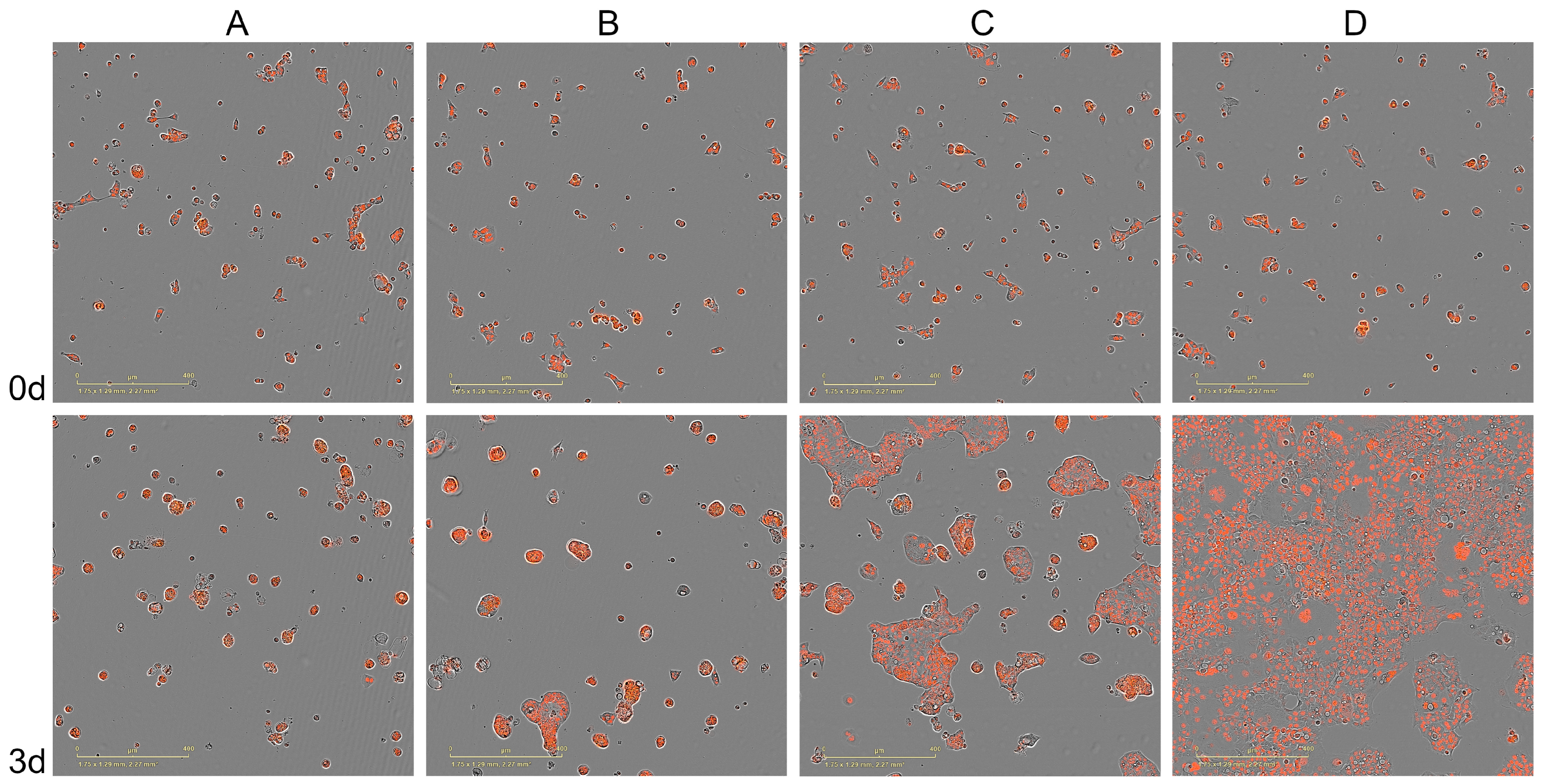
3.2. Experimental Validation of CancerOmicsNet
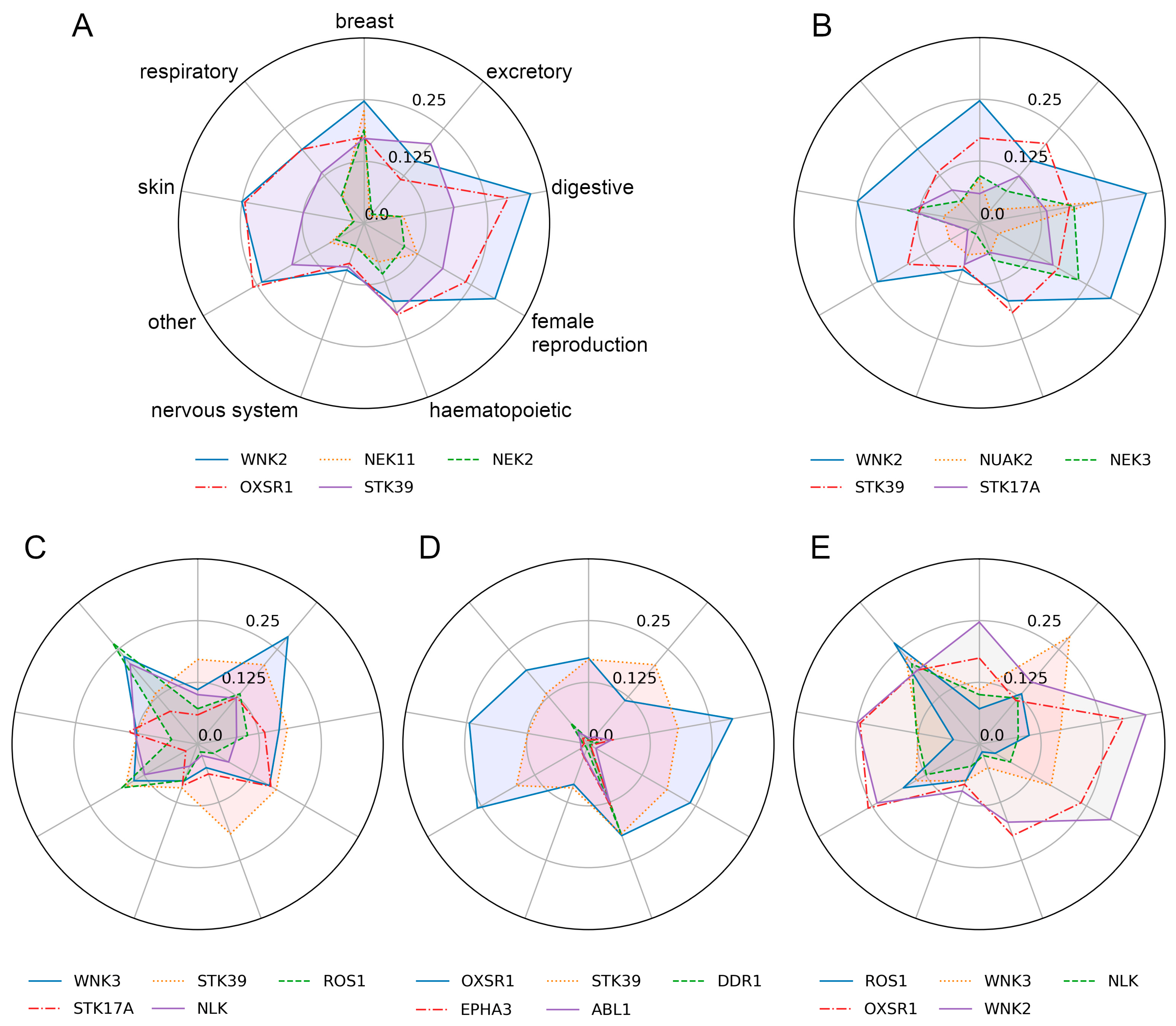
3.3. Explanation for the Decision-Making Process of CancerOmicsNet
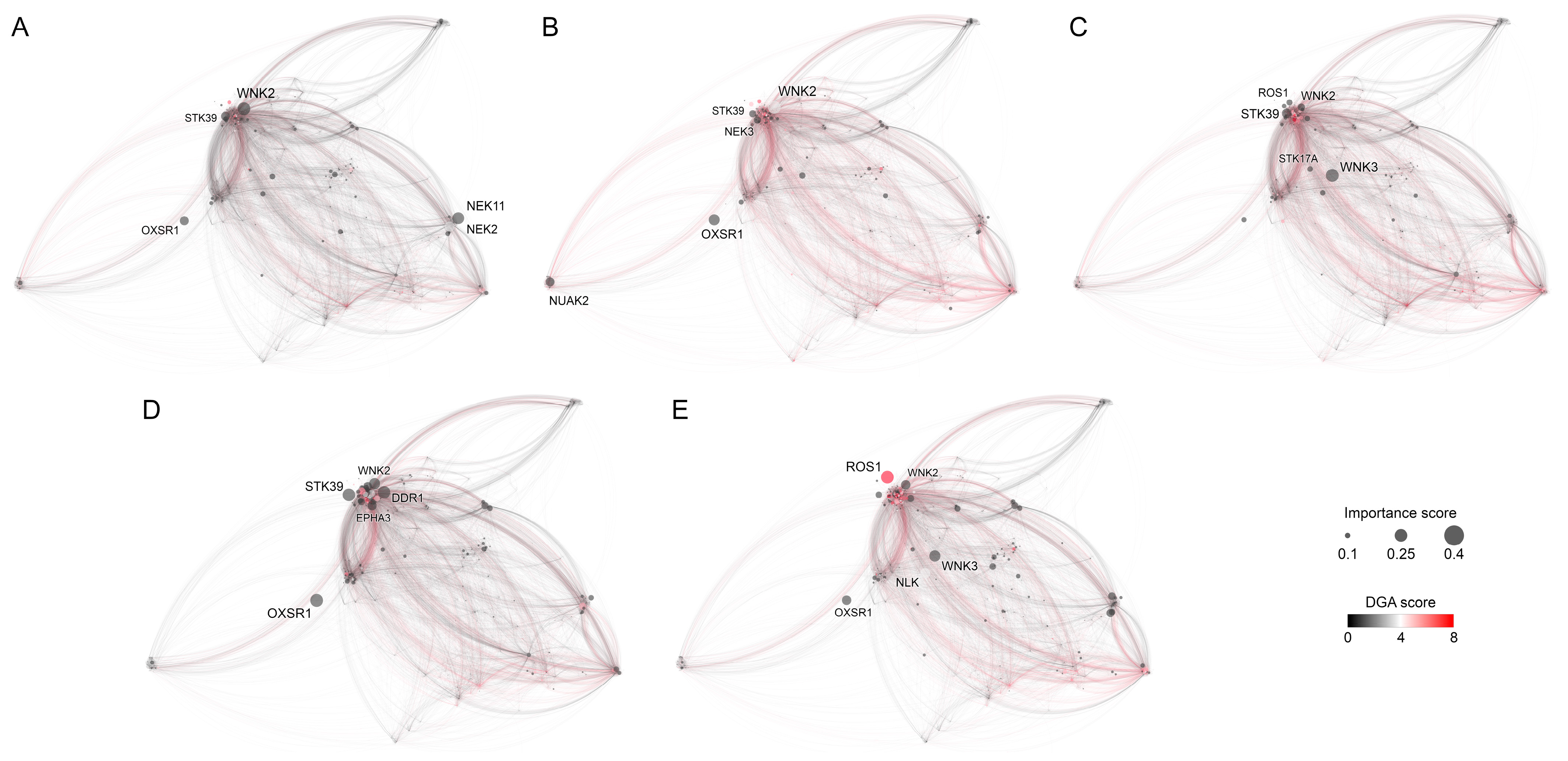
3.3.1. Breast Tissue
3.3.2. Digestive System
3.3.3. Excretory Tissue
3.3.4. Haematopoietic and Lymphoid Tissue
3.3.5. Respiratory Tissue
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cicenas, J.; Zalyte, E.; Bairoch, A.; Gaudet, P. Kinases and cancer. Cancers 2018, 10, 63. [Google Scholar] [CrossRef]
- Paul, M.K.; Mukhopadhyay, A.K. Tyrosine kinase—Role and significance in Cancer. Int. J. Med. Sci. 2004, 1, 101–115. [Google Scholar] [CrossRef]
- Hunter, T. Signaling-2000 and beyond. Cell 2000, 100, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Schlessinger, J. Cell Signaling by Receptor Tyrosine Kinases. Cell 2000, 103, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Blume-Jensen, P.; Hunter, T. Oncogenic kinase signalling. Nature 2001, 411, 355–365. [Google Scholar] [CrossRef]
- Coussens, L.; Parker, P.J.; Rhee, L.; Yang-Feng, T.L.; Chen, E.; Waterfield, M.D.; Francke, U.; Ullrich, A. Multiple, distinct forms of bovine and human protein kinase C suggest diversity in cellular signaling pathways. Science 1986, 233, 859–866. [Google Scholar] [CrossRef]
- Futreal, P.A.; Coin, L.; Marshall, M.; Down, T.; Hubbard, T.; Wooster, R.; Rahman, N.; Stratton, M.R. A census of human cancer genes. Nat. Rev. Cancer 2004, 4, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Chong, Z.Z.; Shang, Y.C.; Wang, S.; Maiese, K. A critical kinase cascade in neurological disorders: PI 3-K, Akt, and mTOR. Future Neurol. 2012, 7, 733–748. [Google Scholar] [CrossRef]
- Mueller, B.K.; Mack, H.; Teusch, N. Rho kinase, a promising drug target for neurological disorders. Nat. Rev. Drug Discov. 2005, 4, 387–398. [Google Scholar] [CrossRef]
- Sato, S.; Sanjo, H.; Takeda, K.; Ninomiya-Tsuji, J.; Yamamoto, M.; Kawai, T.; Matsumoto, K.; Takeuchi, O.; Akira, S. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat. Immunol. 2005, 6, 1087–1095. [Google Scholar] [CrossRef]
- Tabit, C.E.; Shenouda, S.M.; Holbrook, M.; Frame, A.A.; Kluge, M.A.; Duess, M.-A.; Kim, B.H.; Levit, A.D.; Held, A.; Rosenzweig, J.L.; et al. Protein kinase-C beta activation contributes to impaired endothelial insulin signaling in humans with diabetes mellitus. J. Am. Coll. Cardiol. 2012, 59, E2133. [Google Scholar] [CrossRef][Green Version]
- Shibuya, M.; Suzuki, Y. Treatment of cerebral vasospasm by a protein kinase inhibitor AT 877. No Shinkei= Brain Nerve 1993, 45, 819–824. [Google Scholar] [PubMed]
- Bardelli, A.; Parsons, D.W.; Silliman, N.; Ptak, J.; Szabo, S.; Saha, S.; Markowitz, S.; Willson, J.K.V.; Parmigiani, G.; Kinzler, K.W.; et al. Mutational Analysis of the Tyrosine Kinome in Colorectal Cancers. Science 2003, 300, 949. [Google Scholar] [CrossRef] [PubMed]
- Chalhoub, N.; Baker, S.J. PTEN and the PI3-Kinase Pathway in Cancer. Annu. Rev. Pathol. Mech. Dis. 2009, 4, 127–150. [Google Scholar] [CrossRef]
- Grego-Bessa, J.; Bloomekatz, J.; Castel, P.; Omelchenko, T.; Baselga, J.; Anderson, K.V. The tumor suppressor PTEN and the PDK1 kinase regulate formation of the columnar neural epithelium. Elife 2016, 5, e12034. [Google Scholar] [CrossRef]
- Davies, S.P.; Reddy, H.; Caivano, M.; Cohen, P. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 2000, 351 Pt 1, 95–105. [Google Scholar] [CrossRef]
- Druker, B.J.; Guilhot, F.; O’Brien, S.G.; Gathmann, I.; Kantarjian, H.; Gattermann, N.; Deininger, M.W.; Silver, R.T.; Goldman, J.M.; Stone, R.M.; et al. Five-Year Follow-up of Patients Receiving Imatinib for Chronic Myeloid Leukemia. New Engl. J. Med. 2006, 355, 2408–2417. [Google Scholar] [CrossRef]
- Lombardo, L.J.; Lee, F.Y.; Chen, P.; Norris, D.; Barrish, J.C.; Behnia, K.; Castaneda, S.; Cornelius, L.A.M.; Das, J.; Doweyko, A.M.; et al. Discovery of N-(2-chloro-6-methyl- phenyl)-2-(6-(4-(2-hydroxyethyl)- piperazin-1-yl)-2-methylpyrimidin-4- ylamino)thiazole-5-carboxamide (BMS-354825), a dual Src/Abl kinase inhibitor with potent antitumor activity in preclinical assays. J. Med. Chem. 2004, 47, 6658–6661. [Google Scholar] [CrossRef]
- Shah, N.P.; Tran, C.; Lee, F.Y.; Chen, P.; Norris, D.; Sawyers, C.L. Overriding Imatinib Resistance with a Novel ABL Kinase Inhibitor. Science 2004, 305, 399–401. [Google Scholar] [CrossRef]
- Fabian, M.A.; Biggs, W.H., III; Treiber, D.K.; Atteridge, C.E.; Azimioara, M.D.; Benedetti, M.G.; Carter, T.A.; Ciceri, P.; Edeen, P.T.; Floyd, M.; et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 2005, 23, 329–336. [Google Scholar] [CrossRef]
- Köstler, W.J.; Zielinski, C.C. Targeting Receptor Tyrosine Kinases in Cancer, in Receptor Tyrosine Kinases: Structure, Functions and Role in Human Disease; Wheeler, D.L., Yarden, Y., Eds.; Springer: New York, NY, USA, 2015; pp. 225–278. [Google Scholar]
- Karaman, M.W.; Herrgard, S.; Treiber, D.K.; Gallant, P.; Atteridge, C.E.; Campbell, B.T.; Chan, K.W.; Ciceri, P.; Davis, M.I.; Edeen, P.T.; et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 2008, 26, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.; Robey, R.W.; Bates, S.E.; Ambudkar, S.V. Sunitinib (Sutent, SU11248), a Small-Molecule Receptor Tyrosine Kinase Inhibitor, Blocks Function of the ATP-Binding Cassette (ABC) Transporters P-Glycoprotein (ABCB1) and ABCG2. Drug Metab. Dispos. 2009, 37, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Kirkland, L.O.; McInnes, C. Non-ATP competitive protein kinase inhibitors as anti-tumor therapeutics. Biochem. Pharmacol. 2009, 77, 1561–1571. [Google Scholar] [CrossRef] [PubMed]
- Essegian, D.; Khurana, R.; Stathias, V.; Schürer, S.C. The Clinical Kinase Index: A Method to Prioritize Understudied Kinases as Drug Targets for the Treatment of Cancer. Cell Rep. Med. 2020, 1, 100128. [Google Scholar] [CrossRef]
- Richardson, C.J.; Gao, Q.; Mitsopoulous, C.; Zvelebil, M.; Pearl, L.H.; Pearl, F.M. MoKCa database-mutations of kinases in cancer. Nucleic Acids Res. 2009, 37, D824–D831. [Google Scholar] [CrossRef][Green Version]
- Dixit, A.; Verkhivker, G.M. Structure-Functional Prediction and Analysis of Cancer Mutation Effects in Protein Kinases. Comput. Math. Methods Med. 2014, 2014, 653487. [Google Scholar] [CrossRef]
- Yuan, H.; Yu, H.; Gui, S.; Ji, S. Explainability in Graph Neural Networks: A Taxonomic Survey. arXiv 2020, arXiv:2012.15445. [Google Scholar] [CrossRef]
- Zeiler, M.D.; Krishnan, D.; Taylor, G.W.; Fergus, R. Deconvolutional networks. In Proceedings of the 2010 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, San Francisco, CA, USA, 13–18 June 2010. [Google Scholar]
- Pu, L.; Singha, M.; Ramanujam, J.; Brylinski, M. CancerOmicsNet: A multi-omics network-based approach to anti-cancer drug profiling. Oncotarget 2022, 13, 695–706. [Google Scholar] [CrossRef]
- Pu, L.; Singha, M.; Wu, H.-C.; Busch, C.; Ramanujam, J.; Brylinski, M. An integrated network representation of multiple cancer-specific data for graph-based machine learning. NPJ Syst. Biol. Appl. 2022, 8, 1–8. [Google Scholar] [CrossRef]
- Singha, M.; Pu, L.; Stanfield, B.A.; Uche, I.K.; Rider, P.J.F.; Kousoulas, K.G.; Ramanujam, J.; Brylinski, M. Artificial intelligence to guide precision anticancer therapy with multitargeted kinase inhibitors. BMC Cancer 2022, 22, 1211. [Google Scholar] [CrossRef]
- Sharma, G.; Jurie, F.; Schmid, C. Discriminative spatial saliency for image classification. In Proceedings of the 2012 IEEE Conference on Computer Vision and Pattern Recognition, Providence, RI, USA, 16–21 June 2012. [Google Scholar]
- Simonyan, K.; Vedaldi, A.; Zisserman, A. Deep inside convolutional networks: Visualising image classification models and saliency maps. arXiv 2013, arXiv:1312.6034. [Google Scholar]
- Klinke, D.J., 2nd. Signal transduction networks in cancer: Quantitative parameters influence network topology. Cancer Res. 2010, 70, 1773–1782. [Google Scholar] [CrossRef]
- Xiang, X.; Wang, Z.; Zhao, Z.; Su, F. Multiple Saliency and Channel Sensitivity Network for Aggregated Convolutional Feature. Proc. Conf. AAAI Artif. Intell. 2019, 33, 9013–9020. [Google Scholar] [CrossRef]
- Rajaraman, A.; Ullman, J.D. Data Mining, in Mining of Massive Datasets; Rajaraman, A., Ullman, J.D., Eds.; Cambridge University Press: Cambridge, UK, 2011; pp. 1–17. [Google Scholar]
- Jia, Y.; Wang, Y.; Zhang, C.; Chen, M.Y. Upregulated CBX8 promotes cancer metastasis via the WNK2/MMP2 pathway. Mol. Ther. Oncolytics. 2020, 19, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Liu, X. microRNA-370 Promotes Cell Growth by Targeting WNK2 in Breast Cancer. DNA Cell Biol. 2019, 38, 501–509. [Google Scholar] [CrossRef]
- Gao, W.-L.; Niu, L.; Chen, W.-L.; Zhang, Y.-Q.; Huang, W.-H. Integrative Analysis of the Expression Levels and Prognostic Values for NEK Family Members in Breast Cancer. Front. Genet. 2022, 13, 798170. [Google Scholar] [CrossRef]
- Anuraga, G.; Wang, W.-J.; Phan, N.N.; An Ton, N.T.A.; Ta, H.D.K.; Berenice Prayugo, F.; Minh Xuan, D.T.; Ku, S.-C.; Wu, Y.-F.; Andriani, V.; et al. Potential Prognostic Biomarkers of NIMA (Never in Mitosis, Gene A)-Related Kinase (NEK) Family Members in Breast Cancer. J. Pers. Med. 2021, 11, 1089. [Google Scholar] [CrossRef]
- Li, Y.; Li, L.; Qin, J.; Wu, J.; Dai, X.; Xu, J. OSR1 phosphorylates the Smad2/3 linker region and induces TGF-beta1 autocrine to promote EMT and metastasis in breast cancer. Oncogene 2021, 40, 68–84. [Google Scholar] [CrossRef]
- Li, Y.; Qin, J.; Wu, J.; Dai, X.; Xu, J. High expression of OSR1 as a predictive biomarker for poor prognosis and lymph node metastasis in breast cancer. Breast Cancer Res. Treat. 2020, 182, 35–46. [Google Scholar] [CrossRef]
- Qiu, Z.; Dong, B.; Guo, W.; Piotr, R.; Longmore, G.; Yang, X.; Yu, Z.; Deng, J.; Evers, B.M.; Wu, Y. STK39 promotes breast cancer invasion and metastasis by increasing SNAI1 activity upon phosphorylation. Theranostics 2021, 11, 7658–7670. [Google Scholar] [CrossRef]
- Zhou, S.-L.; Zhou, Z.-J.; Hu, Z.-Q.; Song, C.; Luo, Y.-J.; Luo, C.-B.; Xin, H.-Y.; Yang, X.-R.; Shi, Y.-H.; Wang, Z.; et al. Genomic sequencing identifies WNK2 as a driver in hepatocellular carcinoma and a risk factor for early recurrence. J. Hepatol. 2019, 71, 1152–1163. [Google Scholar] [CrossRef] [PubMed]
- Moon, H.; Cho, K.; Shin, S.; Kim, D.Y.; Han, K.-H.; Ro, S.W. High Risk of Hepatocellular Carcinoma Development in Fibrotic Liver: Role of the Hippo-YAP/TAZ Signaling Pathway. Int. J. Mol. Sci. 2019, 20, 581. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.-C.; Pepe-Mooney, B.; Galli, G.G.; Dill, M.T.; Huang, H.-T.; Hao, M.; Wang, Y.; Liang, H.; Calogero, R.A.; Camargo, F.D. NUAK2 is a critical YAP target in liver cancer. Nat. Commun. 2018, 9, 4834. [Google Scholar] [CrossRef]
- Cao, Y.; Song, J.; Chen, J.; Xiao, J.; Ni, J.; Wu, C. Overexpression of NEK3 is associated with poor prognosis in patients with gastric cancer. Medicine 2018, 97, e9630. [Google Scholar] [CrossRef] [PubMed]
- Panchal, N.K.; Prince, S.E. The NEK family of serine/threonine kinases as a biomarker for cancer. Clin. Exp. Med. 2023, 23, 17–30. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, X.; Fang, D.; Xu, P.; Mo, X.; Hu, C.; Xia, H. STK39 is a novel kinase contributing to the progression of hepatocellular carcinoma by the PLK1/ERK signaling pathway. Theranostics 2021, 11, 2108–2122. [Google Scholar] [CrossRef] [PubMed]
- Short, S.P.; Thompson, J.J.; Bilotta, A.J.; Chen, X.; Revetta, F.L.; Washington, M.K.; Williams, C.S. Serine Threonine Kinase 17A Maintains the Epithelial State in Colorectal Cancer Cells. Mol. Cancer Res. 2019, 17, 882–894. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Miller, Z.; Musich, P.R.; Thomas, A.E.; Yao, Z.Q.; Xie, Q.; Howe, P.H.; Jiang, Y. DSTYK Promotes Metastasis and Chemoresistance via EMT in Colorectal Cancer. Front. Pharmacol. 2020, 11, 2108. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Dai, F.; Yuan, M.; Wang, F.; Wu, N.; Xu, M.; Bai, Y.; Liu, Y. A construction and comprehensive analysis of ceRNA networks and infiltrating immune cells in papillary renal cell carcinoma. Cancer Med. 2021, 10, 8192–8209. [Google Scholar] [CrossRef]
- Zhao, Q.; Zhu, Y.; Liu, L.; Wang, H.; Jiang, S.; Hu, X.; Guo, J. STK39 blockage by RNA interference inhibits the proliferation and induces the apoptosis of renal cell carcinoma. OncoTargets Ther. 2018, 11, 1511–1519. [Google Scholar] [CrossRef]
- Luo, L.-X.; Fan, X.-X.; Li, Y.; Peng, X.; Ji, Y.-C.; Hsiao, W.W.-L.; Liu, L.; Leung, E.L.-H.; Yao, X.-J. Identification of mitoxantrone as a new inhibitor of ROS1 fusion protein in non-small cell lung cancer cells. MedChemComm 2017, 8, 621–624. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Monroe, J.; Zhang, Y.; George, D.; Bremer, E.; Li, H. Proteasome inhibition induces both pro- and anti-cell death pathways in prostate cancer cells. Cancer Lett. 2006, 243, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Emami, K.H.; Brown, L.G.; Pitts, T.E.; Sun, X.; Vessella, R.L.; Corey, E. Nemo-like kinase induces apoptosis and inhibits androgen receptor signaling in prostate cancer cells. Prostate 2009, 69, 1481–1492. [Google Scholar] [CrossRef] [PubMed]
- Zong, L.; Sun, Y. OSR1 suppresses acute myeloid leukaemia cell proliferation by inhibiting LGR5-mediated JNK signalling. Autoimmunity 2021, 54, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Balatoni, C.E.; Dawson, D.W.; Suh, J.; Sherman, M.H.; Sanders, G.; Hong, J.S.; Frank, M.J.; Malone, C.S.; Said, J.W.; Teitell, M.A. Epigenetic Silencing of Stk39 in B-Cell Lymphoma Inhibits Apoptosis from Genotoxic Stress. Am. J. Pathol. 2009, 175, 1653–1661. [Google Scholar] [CrossRef]
- Barisione, G.; Fabbi, M.; Cutrona, G.; De Cecco, L.; Zupo, S.; Leitinger, B.; Gentile, M.; Manzoni, M.; Neri, A.; Morabito, F.; et al. Heterogeneous expression of the collagen receptor DDR1 in chronic lymphocytic leukaemia and correlation with progression. Blood Cancer J. 2017, 7, e513. [Google Scholar] [CrossRef][Green Version]
- Caivano, A.; La Rocca, F.; Laurenzana, I.; Annese, T.; Tamma, R.; Famigliari, U.; Simeon, V.; Trino, S.; De Luca, L.; Villani, O.; et al. Epha3 acts as proangiogenic factor in multiple myeloma. Oncotarget 2017, 8, 34298–34309. [Google Scholar] [CrossRef]
- Dasgupta, Y.; Koptyra, M.; Hoser, G.; Kantekure, K.; Roy, D.; Gornicka, B.; Nieborowska-Skorska, M.; Bolton-Gillespie, E.; Cerny-Reiterer, S.; Müschen, M.; et al. Normal ABL1 is a tumor suppressor and therapeutic target in human and mouse leukemias expressing oncogenic ABL1 kinases. Blood 2016, 127, 2131–2143. [Google Scholar] [CrossRef]
- Katayama, R.; Gong, B.O.; Togashi, N.; Miyamoto, M.; Kiga, M.; Iwasaki, S.; Isoyama, T. The new-generation selective ROS1/NTRK inhibitor DS-6051b overcomes crizotinib resistant ROS1-G2032R mutation in preclinical models. Nat. Commun. 2019, 10, 3604. [Google Scholar] [CrossRef]
- Roys, A.; Chang, X.; Liu, Y.; Xu, X.; Wu, Y.; Zuo, D. Resistance mechanisms and potent-targeted therapies of ROS1-positive lung cancer. Cancer Chemother. Pharmacol. 2019, 84, 679–688. [Google Scholar] [CrossRef]
- Zhao, X.; Jin, X.; Zhang, Q.; Liu, R.; Luo, H.; Yang, Z.; Geng, Y.; Feng, S.; Li, C.; Wang, L.; et al. Silencing of the lncRNA H19 enhances sensitivity to X-ray and carbon-ions through the miR-130a-3p /WNK3 signaling axis in NSCLC cells. Cancer Cell Int. 2021, 21, 644. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Wang, Y.; Zheng, Y.-W.; Fei, L.-R.; Shen, H.-Y.; Li, Z.-H.; Huang, W.-J.; Yu, J.-H.; Xu, H.-T. Overexpression of Nemo-like Kinase Promotes the Proliferation and Invasion of Lung Cancer Cells and Indicates Poor Prognosis. Curr. Cancer Drug Targets 2019, 19, 674–680. [Google Scholar] [CrossRef]
- Tahmasbpour, E.; Ghanei, M.; Qazvini, A.; Vahedi, E.; Panahi, Y. Gene expression profile of oxidative stress and antioxidant defense in lung tissue of patients exposed to sulfur mustard. Mutat. Res. Toxicol. Environ. Mutagen. 2016, 800–801, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Gashaw, I.; Ellinghaus, P.; Sommer, A.; Asadullah, K. What makes a good drug target? Drug Discov. Today 2012, 17, S24–S30. [Google Scholar] [CrossRef] [PubMed]
- Plenge, R.M.; Scolnick, E.M.; Altshuler, D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 2013, 12, 581–594. [Google Scholar] [CrossRef]
- Consortium, G.T. The Genotype-Tissue Expression (GTEx) project. Nat. Genet 2013, 45, 580–585. [Google Scholar]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singha, M.; Pu, L.; Srivastava, G.; Ni, X.; Stanfield, B.A.; Uche, I.K.; Rider, P.J.F.; Kousoulas, K.G.; Ramanujam, J.; Brylinski, M. Unlocking the Potential of Kinase Targets in Cancer: Insights from CancerOmicsNet, an AI-Driven Approach to Drug Response Prediction in Cancer. Cancers 2023, 15, 4050. https://doi.org/10.3390/cancers15164050
Singha M, Pu L, Srivastava G, Ni X, Stanfield BA, Uche IK, Rider PJF, Kousoulas KG, Ramanujam J, Brylinski M. Unlocking the Potential of Kinase Targets in Cancer: Insights from CancerOmicsNet, an AI-Driven Approach to Drug Response Prediction in Cancer. Cancers. 2023; 15(16):4050. https://doi.org/10.3390/cancers15164050
Chicago/Turabian StyleSingha, Manali, Limeng Pu, Gopal Srivastava, Xialong Ni, Brent A. Stanfield, Ifeanyi K. Uche, Paul J. F. Rider, Konstantin G. Kousoulas, J. Ramanujam, and Michal Brylinski. 2023. "Unlocking the Potential of Kinase Targets in Cancer: Insights from CancerOmicsNet, an AI-Driven Approach to Drug Response Prediction in Cancer" Cancers 15, no. 16: 4050. https://doi.org/10.3390/cancers15164050
APA StyleSingha, M., Pu, L., Srivastava, G., Ni, X., Stanfield, B. A., Uche, I. K., Rider, P. J. F., Kousoulas, K. G., Ramanujam, J., & Brylinski, M. (2023). Unlocking the Potential of Kinase Targets in Cancer: Insights from CancerOmicsNet, an AI-Driven Approach to Drug Response Prediction in Cancer. Cancers, 15(16), 4050. https://doi.org/10.3390/cancers15164050






