EPH/Ephrin Signaling in Normal Hematopoiesis and Hematologic Malignancies: Deciphering Their Intricate Role and Unraveling Possible New Therapeutic Targets
Abstract
:Simple Summary
Abstract
1. Introduction
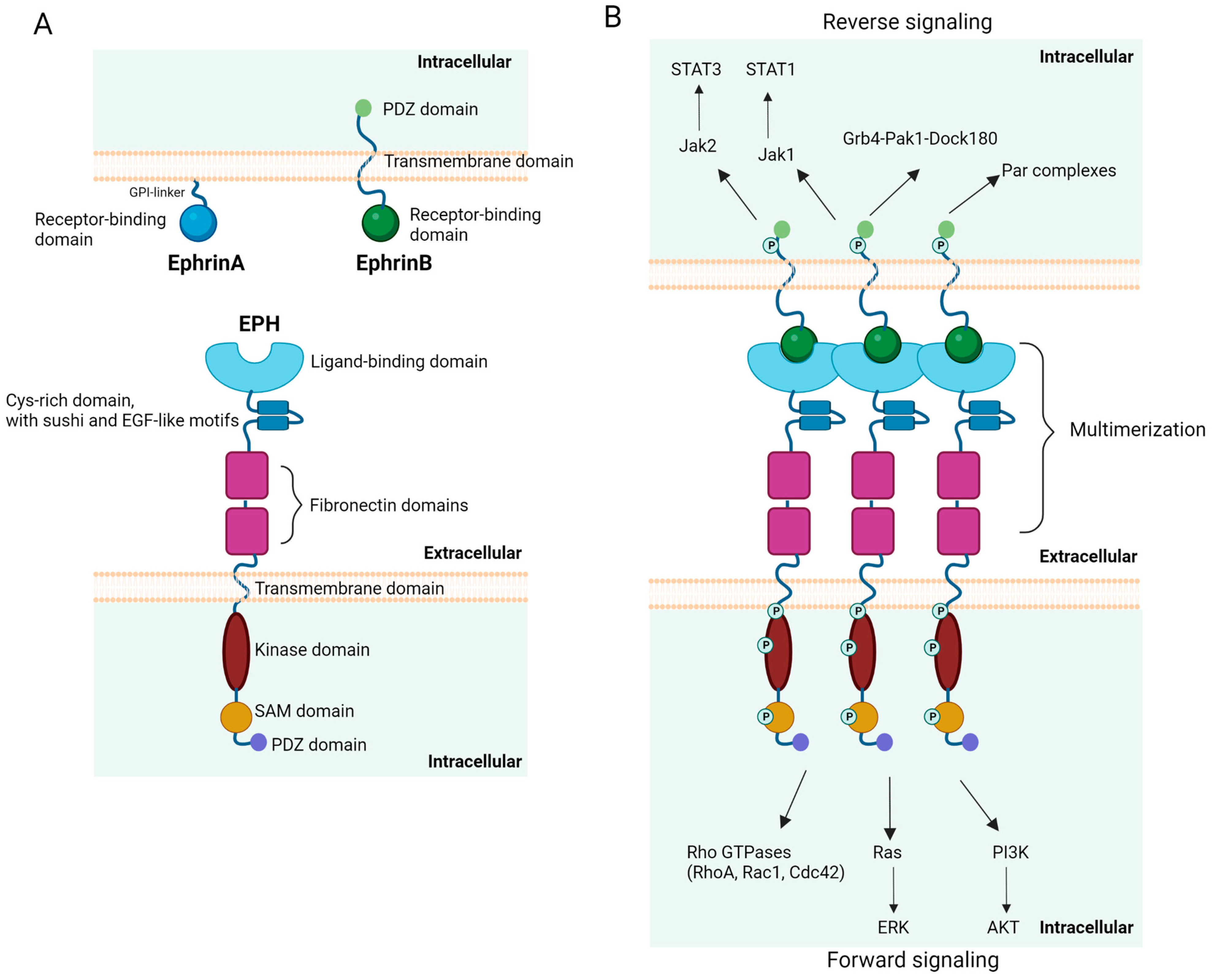
2. Ephrins and EPHs: Structure and Signaling Pathways
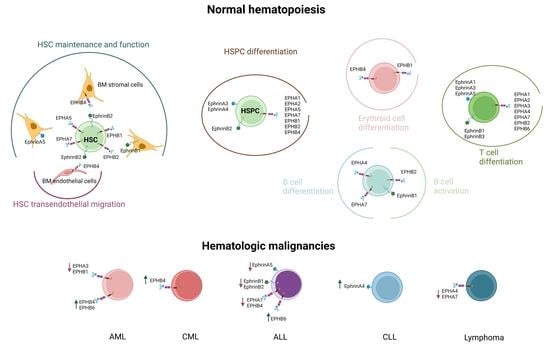
3. EPH/Ephrin Expression in Hematopoiesis
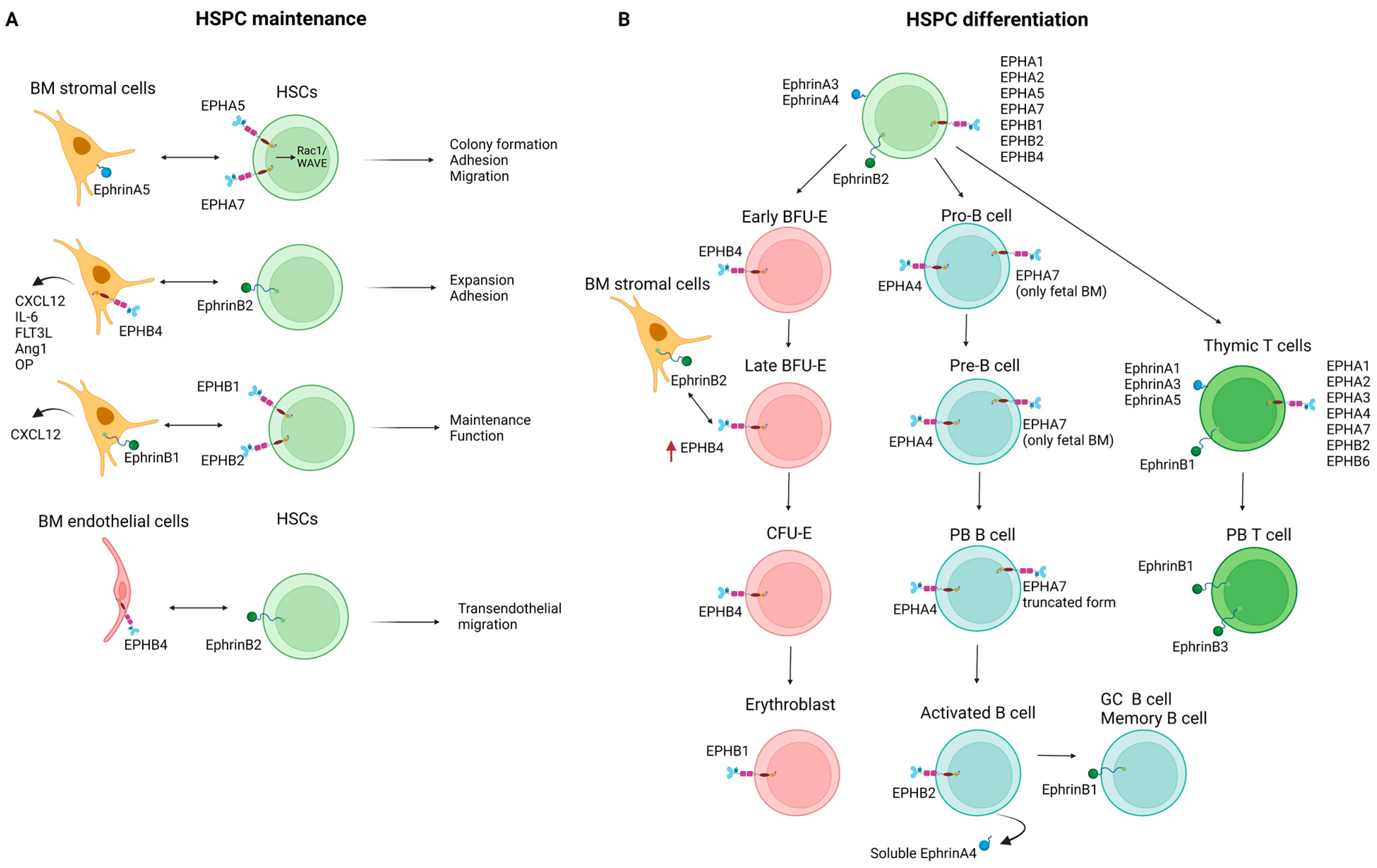
3.1. Hematopoietic Stem and Progenitor Cells
3.2. Erythropoiesis
3.3. Lymphopoiesis
3.4. Other Hematopoietic and Immune Cells
4. EPH/Ephrin Expression in the Setting of Hematologic Neoplasia
4.1. EPH/Ephrin Expression in Leukemia/Lymphoma Cell Lines
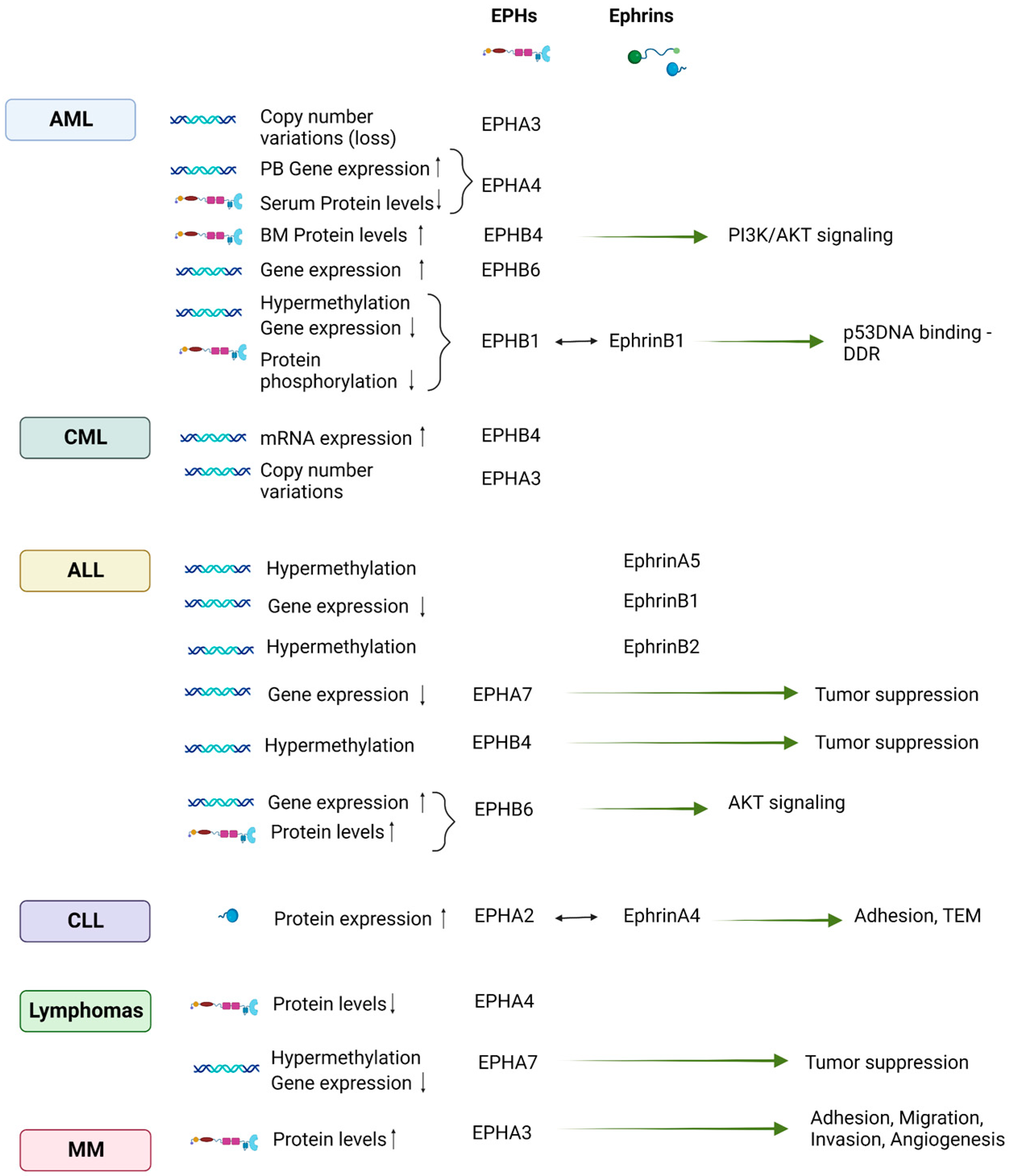
4.2. EPH/Ephrin Expression in Hematologic Malignancies
4.2.1. Leukemias of Myeloid Origin
4.2.2. Leukemias of Lymphoid Origin
4.2.3. Lymphomas
4.2.4. Multiple Myeloma (MM)
5. EPH/Ephrin Therapeutic Targeting in Hematologic Malignancies
6. Discussion
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hirai, H.; Maru, Y.; Hagiwara, K.; Nishida, J.; Takaku, F. A Novel Putative Tyrosine Kinase Receptor Encoded by the eph Gene. Science 1987, 238, 1717–1720. [Google Scholar] [CrossRef]
- Gale, N.W.; Holland, S.J.; Valenzuela, D.M.; Flenniken, A.; Pan, L.; Ryan, T.E.; Henkemeyer, M.; Strebhardt, K.; Hirai, H.; Wilkinson, D.G.; et al. Eph receptors and ligands comprise two major specificity subclasses and are reciprocally compartmentalized during embryogenesis. Neuron 1996, 17, 9–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holder, N.; Klein, R. Eph receptors and ephrins: Effectors of morphogenesis. Development 1999, 126, 2033–2044. [Google Scholar] [CrossRef] [PubMed]
- Janes, P.W.; Vail, M.E.; Ernst, M.; Scott, A.M. Eph Receptors in the Immunosuppressive Tumor Microenvironment. Cancer Res. 2021, 81, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Irie, F.Y.Y. Eph Receptor Signaling and Spine Morphology; Academic Press: Oxford, UK, 2009. [Google Scholar]
- Brantley-Sieders, D.M.; Chen, J. Eph receptor tyrosine kinases in angiogenesis: From development to disease. Angiogenesis 2004, 7, 17–28. [Google Scholar] [CrossRef]
- Kim, Y.H.; Hu, H.; Guevara-Gallardo, S.; Lam, M.T.; Fong, S.Y.; Wang, R.A. Artery and vein size is balanced by Notch and ephrin B2/EphB4 during angiogenesis. Development 2008, 135, 3755–3764. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.U.; Chen, Z.F.; Anderson, D.J. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell 1998, 93, 741–753. [Google Scholar] [CrossRef] [Green Version]
- Boyd, A.W.; Bartlett, P.F.; Lackmann, M. Therapeutic targeting of EPH receptors and their ligands. Nat. Rev. Drug Discov. 2014, 13, 39–62. [Google Scholar] [CrossRef]
- Pasquale, E.B. Eph receptors and ephrins in cancer: Bidirectional signalling and beyond. Nat. Rev. Cancer 2010, 10, 165–180. [Google Scholar] [CrossRef] [Green Version]
- Shiuan, E.; Chen, J. Eph Receptor Tyrosine Kinases in Tumor Immunity. Cancer Res. 2016, 76, 6452–6457. [Google Scholar] [CrossRef] [Green Version]
- Darling, T.K.; Lamb, T.J. Emerging Roles for Eph Receptors and Ephrin Ligands in Immunity. Front. Immunol. 2019, 10, 1473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, A.; Sugamoto, Y.; Tokunaga, Y.; Yoshimuta, T.; Hayashi, K.; Konno, T.; Kawashiri, M.A.; Takeda, Y.; Yamagishi, M. Expression profiling of the ephrin (EFN) and Eph receptor (EPH) family of genes in atherosclerosis-related human cells. J. Int. Med. Res. 2011, 39, 522–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vreeken, D.; Zhang, H.; van Zonneveld, A.J.; van Gils, J.M. Ephs and Ephrins in Adult Endothelial Biology. Int. J. Mol. Sci. 2020, 21, 5623. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Rockel, J.S.; Lagares, D.; Kapoor, M. Ephrins and Eph Receptor Signaling in Tissue Repair and Fibrosis. Curr. Rheumatol. Rep. 2019, 21, 23. [Google Scholar] [CrossRef]
- Arora, S.; Scott, A.M.; Janes, P.W. Eph Receptors in Cancer. Biomedicines 2023, 11, 315. [Google Scholar] [CrossRef] [PubMed]
- Giaginis, C.; Tsourouflis, G.; Zizi-Serbetzoglou, A.; Kouraklis, G.; Chatzopoulou, E.; Dimakopoulou, K.; Theocharis, S.E. Clinical significance of ephrin (eph)-A1, -A2, -a4, -a5 and -a7 receptors in pancreatic ductal adenocarcinoma. Pathol. Oncol. Res. 2010, 16, 267–276. [Google Scholar] [CrossRef]
- Nikas, I.; Giaginis, C.; Petrouska, K.; Alexandrou, P.; Michail, A.; Sarantis, P.; Tsourouflis, G.; Danas, E.; Pergaris, A.; Politis, P.K.; et al. EPHA2, EPHA4, and EPHA7 Expression in Triple-Negative Breast Cancer. Diagnostics 2022, 12, 366. [Google Scholar] [CrossRef]
- Pergaris, A.; Danas, E.; Goutas, D.; Sykaras, A.G.; Soranidis, A.; Theocharis, S. The Clinical Impact of the EPH/Ephrin System in Cancer: Unwinding the Thread. Int. J. Mol. Sci. 2021, 22, 8412. [Google Scholar] [CrossRef]
- Papadakos, S.P.; Dedes, N.; Gkolemi, N.; Machairas, N.; Theocharis, S. The EPH/Ephrin System in Pancreatic Ductal Adenocarcinoma (PDAC): From Pathogenesis to Treatment. Int. J. Mol. Sci. 2023, 24, 3015. [Google Scholar] [CrossRef]
- Papadakos, S.P.; Petrogiannopoulos, L.; Pergaris, A.; Theocharis, S. The EPH/Ephrin System in Colorectal Cancer. Int. J. Mol. Sci. 2022, 23, 2761. [Google Scholar] [CrossRef]
- Psilopatis, I.; Karniadakis, I.; Danos, K.S.; Vrettou, K.; Michaelidou, K.; Mavridis, K.; Agelaki, S.; Theocharis, S. May EPH/Ephrin Targeting Revolutionize Lung Cancer Treatment? Int. J. Mol. Sci. 2022, 24, 93. [Google Scholar] [CrossRef] [PubMed]
- Psilopatis, I.; Souferi-Chronopoulou, E.; Vrettou, K.; Troungos, C.; Theocharis, S. EPH/Ephrin-Targeting Treatment in Breast Cancer: A New Chapter in Breast Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 15275. [Google Scholar] [CrossRef] [PubMed]
- Hadjimichael, A.C.; Pergaris, A.; Kaspiris, A.; Foukas, A.F.; Kokkali, S.; Tsourouflis, G.; Theocharis, S. The EPH/Ephrin System in Bone and Soft Tissue Sarcomas’ Pathogenesis and Therapy: New Advancements and a Literature Review. Int. J. Mol. Sci. 2022, 23, 5171. [Google Scholar] [CrossRef] [PubMed]
- Pergaris, A.; Danas, E.; Gajdzis, P.; Levidou, G.; Gajdzis, M.; Cassoux, N.; Gardrat, S.; Donizy, P.; Korkolopoulou, P.; Kavantzas, N.; et al. EPHA2, EPHA4, and EPHA6 Expression in Uveal Melanomas: Searching for the Culprits of Neoplasia. Diagnostics 2022, 12, 1025. [Google Scholar] [CrossRef] [PubMed]
- Psilopatis, I.; Pergaris, A.; Vrettou, K.; Tsourouflis, G.; Theocharis, S. The EPH/Ephrin System in Gynecological Cancers: Focusing on the Roots of Carcinogenesis for Better Patient Management. Int. J. Mol. Sci. 2022, 23, 3249. [Google Scholar] [CrossRef]
- Masaoutis, C.; Georgantzoglou, N.; Sarantis, P.; Theochari, I.; Tsoukalas, N.; Bobos, M.; Alexandrou, P.; Pergaris, A.; Rontogianni, D.; Theocharis, S. Ephrin Receptors (Ephs) Expression in Thymic Epithelial Tumors: Prognostic Implications and Future Therapeutic Approaches. Diagnostics 2021, 11, 2265. [Google Scholar] [CrossRef] [PubMed]
- Goutas, D.; Pergaris, A.; Goutas, N.; Theocharis, S. Utilizing Exosomal-EPHs/Ephrins as Biomarkers and as a Potential Platform for Targeted Delivery of Therapeutic Exosomes. Int. J. Mol. Sci. 2022, 23, 3551. [Google Scholar] [CrossRef]
- Brantley-Sieders, D.M.; Fang, W.B.; Hicks, D.J.; Zhuang, G.; Shyr, Y.; Chen, J. Impaired tumor microenvironment in EphA2-deficient mice inhibits tumor angiogenesis and metastatic progression. FASEB J. 2005, 19, 1884–1886. [Google Scholar] [CrossRef]
- Brantley-Sieders, D.M.; Jiang, A.; Sarma, K.; Badu-Nkansah, A.; Walter, D.L.; Shyr, Y.; Chen, J. Eph/ephrin profiling in human breast cancer reveals significant associations between expression level and clinical outcome. PLoS ONE 2011, 6, e24426. [Google Scholar] [CrossRef] [Green Version]
- Markosyan, N.; Li, J.; Sun, Y.H.; Richman, L.P.; Lin, J.H.; Yan, F.; Quinones, L.; Sela, Y.; Yamazoe, T.; Gordon, N.; et al. Tumor cell-intrinsic EPHA2 suppresses anti-tumor immunity by regulating PTGS2 (COX-2). J. Clin. Investig. 2019, 129, 3594–3609. [Google Scholar] [CrossRef] [Green Version]
- Vail, M.E.; Murone, C.; Tan, A.; Hii, L.; Abebe, D.; Janes, P.W.; Lee, F.-T.; Baer, M.; Palath, V.; Bebbington, C.; et al. Targeting EphA3 Inhibits Cancer Growth by Disrupting the Tumor Stromal Microenvironment. Cancer Res. 2014, 74, 4470–4481. [Google Scholar] [CrossRef] [Green Version]
- Herath, N.I.; Doecke, J.; Spanevello, M.D.; Leggett, B.A.; Boyd, A.W. Epigenetic silencing of EphA1 expression in colorectal cancer is correlated with poor survival. Br. J. Cancer 2009, 100, 1095–1102. [Google Scholar] [CrossRef] [PubMed]
- Day, B.W.; Stringer, B.W.; Al-Ejeh, F.; Ting, M.J.; Wilson, J.; Ensbey, K.S.; Jamieson, P.R.; Bruce, Z.C.; Lim, Y.C.; Offenhäuser, C.; et al. EphA3 maintains tumorigenicity and is a therapeutic target in glioblastoma multiforme. Cancer Cell 2013, 23, 238–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saintigny, P.; Peng, S.; Zhang, L.; Sen, B.; Wistuba, I.I.; Lippman, S.M.; Girard, L.; Minna, J.D.; Heymach, J.V.; Johnson, F.M. Global evaluation of Eph receptors and ephrins in lung adenocarcinomas identifies EphA4 as an inhibitor of cell migration and invasion. Mol. Cancer Ther. 2012, 11, 2021–2032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, J.H.; Chan, L.C.; Wang, Y.N.; Chu, Y.Y.; Wang, C.H.; Lee, H.H.; Xia, W.; Shyu, W.C.; Liu, S.P.; Yao, J.; et al. Ephrin receptor A10 monoclonal antibodies and the derived chimeric antigen receptor T cells exert an antitumor response in mouse models of triple-negative breast cancer. J. Biol. Chem. 2022, 298, 101817. [Google Scholar] [CrossRef]
- Xuan, Z.; Huang, J.; Gao, L.; Wang, Y.; Wang, J.; Sun, Y. Receptor Tyrosine Kinase EphB3: A Prognostic Indicator in Colorectal Carcinoma. Pathol. Oncol. Res. 2020, 26, 541–549. [Google Scholar] [CrossRef]
- Magic, Z.; Sandström, J.; Perez-Tenorio, G. Ephrin-B2 inhibits cell proliferation and motility in vitro and predicts longer metastasis-free survival in breast cancer. Int. J. Oncol. 2019, 55, 1275–1286. [Google Scholar] [CrossRef] [Green Version]
- Himanen, J.P.; Nikolov, D.B. Eph signaling: A structural view. Trends Neurosci. 2003, 26, 46–51. [Google Scholar] [CrossRef]
- Pasquale, E.B. Eph receptor signalling casts a wide net on cell behaviour. Nat. Rev. Mol. Cell Biol. 2005, 6, 462–475. [Google Scholar] [CrossRef]
- Pasquale, E.B. Eph–ephrin promiscuity is now crystal clear. Nat. Neurosci. 2004, 7, 417–418. [Google Scholar] [CrossRef]
- Mosch, B.; Reissenweber, B.; Neuber, C.; Pietzsch, J. Eph Receptors and Ephrin Ligands: Important Players in Angiogenesis and Tumor Angiogenesis. J. Oncol. 2010, 2010, 135285. [Google Scholar] [CrossRef]
- Kania, A.; Klein, R. Mechanisms of ephrin-Eph signalling in development, physiology and disease. Nat. Rev. Mol. Cell Biol. 2016, 17, 240–256. [Google Scholar] [CrossRef] [PubMed]
- Coulthard, M.G.; Lickliter, J.D.; Subanesan, N.; Chen, K.; Webb, G.C.; Lowry, A.J.; Koblar, S.; Bottema, C.D.; Boyd, A.W. Characterization of the Epha1 receptor tyrosine kinase: Expression in epithelial tissues. Growth Factors 2001, 18, 303–317. [Google Scholar] [CrossRef]
- Lackmann, M.; Mann, R.J.; Kravets, L.; Smith, F.M.; Bucci, T.A.; Maxwell, K.F.; Howlett, G.J.; Olsson, J.E.; Vanden Bos, T.; Cerretti, D.P.; et al. Ligand for EPH-related kinase (LERK) 7 is the preferred high affinity ligand for the HEK receptor. J. Biol. Chem. 1997, 272, 16521–16530. [Google Scholar] [CrossRef] [Green Version]
- Blits-Huizinga, C.T.; Nelersa, C.M.; Malhotra, A.; Liebl, D.J. Ephrins and their receptors: Binding versus biology. IUBMB Life 2004, 56, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, F.; Amode, M.R.; Barrell, D.; Beal, K.; Billis, K.; Brent, S.; Carvalho-Silva, D.; Clapham, P.; Coates, G.; Fitzgerald, S.; et al. Ensembl 2015. Nucleic Acids Res. 2015, 43, D662–D669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alford, S.C.; Bazowski, J.; Lorimer, H.; Elowe, S.; Howard, P.L. Tissue transglutaminase clusters soluble A-type ephrins into functionally active high molecular weight oligomers. Exp. Cell Res. 2007, 313, 4170–4179. [Google Scholar] [CrossRef]
- Wykosky, J.; Palma, E.; Gibo, D.M.; Ringler, S.; Turner, C.P.; Debinski, W. Soluble monomeric EphrinA1 is released from tumor cells and is a functional ligand for the EphA2 receptor. Oncogene 2008, 27, 7260–7273. [Google Scholar] [CrossRef] [Green Version]
- Himanen, J.P.; Yermekbayeva, L.; Janes, P.W.; Walker, J.R.; Xu, K.; Atapattu, L.; Rajashankar, K.R.; Mensinga, A.; Lackmann, M.; Nikolov, D.B.; et al. Architecture of Eph receptor clusters. Proc. Natl. Acad. Sci. USA 2010, 107, 10860–10865. [Google Scholar] [CrossRef]
- Surawska, H.; Ma, P.C.; Salgia, R. The role of ephrins and Eph receptors in cancer. Cytokine Growth Factor Rev. 2004, 15, 419–433. [Google Scholar] [CrossRef]
- Egea, J.; Nissen, U.V.; Dufour, A.; Sahin, M.; Greer, P.; Kullander, K.; Mrsic-Flogel, T.D.; Greenberg, M.E.; Kiehn, O.; Vanderhaeghen, P.; et al. Regulation of EphA 4 kinase activity is required for a subset of axon guidance decisions suggesting a key role for receptor clustering in Eph function. Neuron 2005, 47, 515–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stein, E.; Lane, A.A.; Cerretti, D.P.; Schoecklmann, H.O.; Schroff, A.D.; Van Etten, R.L.; Daniel, T.O. Eph receptors discriminate specific ligand oligomers to determine alternative signaling complexes, attachment, and assembly responses. Genes Dev. 1998, 12, 667–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holland, S.J.; Gale, N.W.; Mbamalu, G.; Yancopoulos, G.D.; Henkemeyer, M.; Pawson, T. Bidirectional signalling through the EPH-family receptor Nuk and its transmembrane ligands. Nature 1996, 383, 722–725. [Google Scholar] [CrossRef] [PubMed]
- Pasquale, E.B. Eph-ephrin bidirectional signaling in physiology and disease. Cell 2008, 133, 38–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kullander, K.; Mather, N.K.; Diella, F.; Dottori, M.; Boyd, A.W.; Klein, R. Kinase-dependent and kinase-independent functions of EphA4 receptors in major axon tract formation in vivo. Neuron 2001, 29, 73–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brückner, K.; Pasquale, E.B.; Klein, R. Tyrosine phosphorylation of transmembrane ligands for Eph receptors. Science 1997, 275, 1640–1643. [Google Scholar] [CrossRef] [Green Version]
- Palmer, A.; Zimmer, M.; Erdmann, K.S.; Eulenburg, V.; Porthin, A.; Heumann, R.; Deutsch, U.; Klein, R. EphrinB phosphorylation and reverse signaling: Regulation by Src kinases and PTP-BL phosphatase. Mol. Cell 2002, 9, 725–737. [Google Scholar] [CrossRef]
- Dravis, C.; Henkemeyer, M. Ephrin-B reverse signaling controls septation events at the embryonic midline through separate tyrosine phosphorylation-independent signaling avenues. Dev. Biol. 2011, 355, 138–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kullander, K.; Klein, R. Mechanisms and functions of Eph and ephrin signalling. Nat. Rev. Mol. Cell Biol. 2002, 3, 475–486. [Google Scholar] [CrossRef]
- Wagner, M.J.; Stacey, M.M.; Liu, B.A.; Pawson, T. Molecular mechanisms of SH2- and PTB-domain-containing proteins in receptor tyrosine kinase signaling. Cold Spring Harb. Perspect. Biol. 2013, 5, a008987. [Google Scholar] [CrossRef] [PubMed]
- Lisabeth, E.M.; Falivelli, G.; Pasquale, E.B. Eph receptor signaling and ephrins. Cold Spring Harb. Perspect. Biol. 2013, 5, a009159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Picco, V.; Hudson, C.; Yasuo, H. Ephrin-Eph signalling drives the asymmetric division of notochord/neural precursors in Ciona embryos. Development 2007, 134, 1491–1497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, W.; Levine, M. Ephrin signaling establishes asymmetric cell fates in an endomesoderm lineage of the Ciona embryo. Development 2008, 135, 931–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minami, M.; Koyama, T.; Wakayama, Y.; Fukuhara, S.; Mochizuki, N. EphrinA/EphA signal facilitates insulin-like growth factor-I-induced myogenic differentiation through suppression of the Ras/extracellular signal-regulated kinase 1/2 cascade in myoblast cell lines. Mol. Biol. Cell 2011, 22, 3508–3519. [Google Scholar] [CrossRef]
- Meier, C.; Anastasiadou, S.; Knöll, B. Ephrin-A5 suppresses neurotrophin evoked neuronal motility, ERK activation and gene expression. PLoS ONE 2011, 6, e26089. [Google Scholar] [CrossRef] [Green Version]
- Bush, J.O.; Soriano, P. Ephrin-B1 forward signaling regulates craniofacial morphogenesis by controlling cell proliferation across Eph-ephrin boundaries. Genes Dev. 2010, 24, 2068–2080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vindis, C.; Cerretti, D.P.; Daniel, T.O.; Huynh-Do, U. EphB1 recruits c-Src and p52Shc to activate MAPK/ERK and promote chemotaxis. J. Cell Biol. 2003, 162, 661–671. [Google Scholar] [CrossRef]
- Xiao, Z.; Carrasco, R.; Kinneer, K.; Sabol, D.; Jallal, B.; Coats, S.; Tice, D.A. EphB4 promotes or suppresses Ras/MEK/ERK pathway in a context-dependent manner: Implications for EphB4 as a cancer target. Cancer Biol. Ther. 2012, 13, 630–637. [Google Scholar] [CrossRef] [Green Version]
- Poliakov, A.; Cotrina, M.L.; Pasini, A.; Wilkinson, D.G. Regulation of EphB2 activation and cell repulsion by feedback control of the MAPK pathway. J. Cell Biol. 2008, 183, 933–947. [Google Scholar] [CrossRef]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef] [Green Version]
- Chang, Q.; Jorgensen, C.; Pawson, T.; Hedley, D.W. Effects of dasatinib on EphA2 receptor tyrosine kinase activity and downstream signalling in pancreatic cancer. Br. J. Cancer 2008, 99, 1074–1082. [Google Scholar] [CrossRef] [Green Version]
- Maddigan, A.; Truitt, L.; Arsenault, R.; Freywald, T.; Allonby, O.; Dean, J.; Narendran, A.; Xiang, J.; Weng, A.; Napper, S.; et al. EphB receptors trigger Akt activation and suppress Fas receptor-induced apoptosis in malignant T lymphocytes. J. Immunol. 2011, 187, 5983–5994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menges, C.W.; McCance, D.J. Constitutive activation of the Raf-MAPK pathway causes negative feedback inhibition of Ras-PI3K-AKT and cellular arrest through the EphA2 receptor. Oncogene 2008, 27, 2934–2940. [Google Scholar] [CrossRef]
- Miao, H.; Li, D.Q.; Mukherjee, A.; Guo, H.; Petty, A.; Cutter, J.; Basilion, J.P.; Sedor, J.; Wu, J.; Danielpour, D.; et al. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell 2009, 16, 9–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, N.Y.; Fernandez, C.; Richter, M.; Xiao, Z.; Valencia, F.; Tice, D.A.; Pasquale, E.B. Crosstalk of the EphA2 receptor with a serine/threonine phosphatase suppresses the Akt-mTORC1 pathway in cancer cells. Cell. Signal. 2011, 23, 201–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Ji, X.D.; Gao, H.; Zhao, J.S.; Xu, J.F.; Sun, Z.J.; Deng, Y.Z.; Shi, S.; Feng, Y.X.; Zhu, Y.Q.; et al. EphB3 suppresses non-small-cell lung cancer metastasis via a PP2A/RACK1/Akt signalling complex. Nat. Commun. 2012, 3, 667. [Google Scholar] [CrossRef] [Green Version]
- Bong, Y.S.; Lee, H.S.; Carim-Todd, L.; Mood, K.; Nishanian, T.G.; Tessarollo, L.; Daar, I.O. ephrinB1 signals from the cell surface to the nucleus by recruitment of STAT3. Proc. Natl. Acad. Sci. USA 2007, 104, 17305–17310. [Google Scholar] [CrossRef] [PubMed]
- Salvucci, O.; Ohnuki, H.; Maric, D.; Hou, X.; Li, X.; Yoon, S.O.; Segarra, M.; Eberhart, C.G.; Acker-Palmer, A.; Tosato, G. EphrinB2 controls vessel pruning through STAT1-JNK3 signalling. Nat. Commun. 2015, 6, 6576. [Google Scholar] [CrossRef] [Green Version]
- Bong, Y.-S.; Park, Y.-H.; Lee, H.-S.; Mood, K.; Ishimura, A.; Daar, I.O. Tyr-298 in ephrinB1 is critical for an interaction with the Grb4 adaptor protein. Biochem. J. 2004, 377, 499–507. [Google Scholar] [CrossRef]
- Cowan, C.A.; Henkemeyer, M. The SH2/SH3 adaptor Grb4 transduces B-ephrin reverse signals. Nature 2001, 413, 174–179. [Google Scholar] [CrossRef]
- Lee, H.S.; Nishanian, T.G.; Mood, K.; Bong, Y.S.; Daar, I.O. EphrinB1 controls cell-cell junctions through the Par polarity complex. Nat. Cell Biol. 2008, 10, 979–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baron, M.H. Embryonic origins of mammalian hematopoiesis. Exp. Hematol. 2003, 31, 1160–1169. [Google Scholar] [CrossRef] [PubMed]
- Adams, R.H.; Wilkinson, G.A.; Weiss, C.; Diella, F.; Gale, N.W.; Deutsch, U.; Risau, W.; Klein, R. Roles of ephrinB ligands and EphB receptors in cardiovascular development: Demarcation of arterial/venous domains, vascular morphogenesis, and sprouting angiogenesis. Genes Dev. 1999, 13, 295–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuijper, S.; Turner, C.J.; Adams, R.H. Regulation of angiogenesis by Eph-ephrin interactions. Trends Cardiovasc. Med. 2007, 17, 145–151. [Google Scholar] [CrossRef]
- Ivanova, N.B.; Dimos, J.T.; Schaniel, C.; Hackney, J.A.; Moore, K.A.; Lemischka, I.R. A stem cell molecular signature. Science 2002, 298, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Steidl, U.; Bork, S.; Schaub, S.; Selbach, O.; Seres, J.; Aivado, M.; Schroeder, T.; Rohr, U.-P.; Fenk, R.; Kliszewski, S.; et al. Primary human CD34+ hematopoietic stem and progenitor cells express functionally active receptors of neuromediators. Blood 2004, 104, 81–88. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, J.J.; Alonso, C.L.; Sacedón, R.; Crompton, T.; Vicente, A.; Jiménez, E.; Varas, A.; Zapata, A.G. Expression and function of the Eph A receptors and their ligands ephrins A in the rat thymus. J. Immunol. 2002, 169, 177–184. [Google Scholar] [CrossRef] [Green Version]
- Lazarova, P.; Wu, Q.; Kvalheim, G.; Suo, Z.; Haakenstad, K.W.; Metodiev, K.; Nesland, J.M. Growth factor receptors in hematopoietic stem cells: EPH family expression in CD34+ and CD133+ cell populations from mobilized peripheral blood. Int. J. Immunopathol. Pharmacol. 2006, 19, 49–56. [Google Scholar] [CrossRef] [Green Version]
- Ting, M.J.; Day, B.W.; Spanevello, M.D.; Boyd, A.W. Activation of ephrin A proteins influences hematopoietic stem cell adhesion and trafficking patterns. Exp. Hematol. 2010, 38, 1087–1098. [Google Scholar] [CrossRef]
- Prévost, N.; Woulfe, D.S.; Jiang, H.; Stalker, T.J.; Marchese, P.; Ruggeri, Z.M.; Brass, L.F. Eph kinases and ephrins support thrombus growth and stability by regulating integrin outside-in signaling in platelets. Proc. Natl. Acad. Sci. USA 2005, 102, 9820–9825. [Google Scholar] [CrossRef]
- Prevost, N.; Woulfe, D.; Tanaka, T.; Brass, L.F. Interactions between Eph kinases and ephrins provide a mechanism to support platelet aggregation once cell-to-cell contact has occurred. Proc. Natl. Acad. Sci. USA 2002, 99, 9219–9224. [Google Scholar] [CrossRef] [PubMed]
- Aasheim, H.-C.; Terstappen, L.W.M.M.; Logtenberg, T. Regulated Expression of the Eph-Related Receptor Tyrosine Kinase Hek11 in Early Human B Lymphopoiesis. Blood 1997, 90, 3613–3622. [Google Scholar] [CrossRef] [PubMed]
- Dawson, D.W.; Hong, J.S.; Shen, R.R.; French, S.W.; Troke, J.J.; Wu, Y.Z.; Chen, S.S.; Gui, D.; Regelson, M.; Marahrens, Y.; et al. Global DNA methylation profiling reveals silencing of a secreted form of Epha7 in mouse and human germinal center B-cell lymphomas. Oncogene 2007, 26, 4243–4252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Cohen, K.; Shao, Y.; Mole, P.; Dombkowski, D.; Scadden, D.T. Ephrin receptor, EphB4, regulates ES cell differentiation of primitive mammalian hemangioblasts, blood, cardiomyocytes, and blood vessels. Blood 2004, 103, 100–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suenobu, S.; Takakura, N.; Inada, T.; Yamada, Y.; Yuasa, H.; Zhang, X.Q.; Sakano, S.; Oike, Y.; Suda, T. A role of EphB4 receptor and its ligand, ephrin-B2, in erythropoiesis. Biochem. Biophys. Res. Commun. 2002, 293, 1124–1131. [Google Scholar] [CrossRef] [PubMed]
- Okubo, T.; Yanai, N.; Obinata, M. Stromal cells modulate ephrinB2 expression and transmigration of hematopoietic cells. Exp. Hematol. 2006, 34, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, M.; Matsuoka, H.; Nagata, A.; Iwata, N.; Tamekane, A.; Okamura, A.; Gomyo, H.; Ito, M.; Jishage, K.; Kamada, N.; et al. Developmental expression of EphB6 in the thymus: Lessons from EphB6 knockout mice. Biochem. Biophys. Res. Commun. 2002, 298, 87–94. [Google Scholar] [CrossRef]
- Yu, G.; Mao, J.; Wu, Y.; Luo, H.; Wu, J. Ephrin-B1 is critical in T-cell development. J. Biol. Chem. 2006, 281, 10222–10229. [Google Scholar] [CrossRef] [Green Version]
- Bennett, B.D.; Wang, Z.; Kuang, W.J.; Wang, A.; Groopman, J.E.; Goeddel, D.V.; Scadden, D.T. Cloning and characterization of HTK, a novel transmembrane tyrosine kinase of the EPH subfamily. J. Biol. Chem. 1994, 269, 14211–14218. [Google Scholar] [CrossRef]
- Suda, T.; Iwama, A.; Hashiyama, M.; Sakano, S.; Ohno, M. Receptor tyrosine kinases involved in hematopoietic progenitor cells. Leukemia 1997, 11 (Suppl. 3), 451–453. [Google Scholar]
- Guder, C.; Gravius, S.; Burger, C.; Wirtz, D.C.; Schildberg, F.A. Osteoimmunology: A Current Update of the Interplay Between Bone and the Immune System. Front. Immunol. 2020, 11, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukasaki, M.; Takayanagi, H. Osteoimmunology: Evolving concepts in bone-immune interactions in health and disease. Nat. Rev. Immunol. 2019, 19, 626–642. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.M.; Arthur, A.; Gronthos, S. The role of Eph/ephrin molecules in stromal–hematopoietic interactions. Int. J. Hematol. 2016, 103, 145–154. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Arthur, A.; Zannettino, A.C.; Gronthos, S. EphA5 and EphA7 forward signaling enhances human hematopoietic stem and progenitor cell maintenance, migration, and adhesion via Rac1 activation. Exp. Hematol. 2017, 48, 72–78. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.M.; Arthur, A.; Panagopoulos, R.; Paton, S.; Hayball, J.D.; Zannettino, A.C.W.; Purton, L.E.; Matsuo, K.; Gronthos, S. EphB4 Expressing Stromal Cells Exhibit an Enhanced Capacity for Hematopoietic Stem Cell Maintenance. Stem Cells 2015, 33, 2838–2849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arthur, A.; Nguyen, T.M.; Paton, S.; Zannettino, A.C.W.; Gronthos, S. Loss of EfnB1 in the osteogenic lineage compromises their capacity to support hematopoietic stem/progenitor cell maintenance. Exp. Hematol. 2019, 69, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Kwak, H.; Salvucci, O.; Weigert, R.; Martinez-Torrecuadrada, J.L.; Henkemeyer, M.; Poulos, M.G.; Butler, J.M.; Tosato, G. Sinusoidal ephrin receptor EPHB4 controls hematopoietic progenitor cell mobilization from bone marrow. J. Clin. Investig. 2016, 126, 4554–4568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakano, S.; Serizawa, R.; Inada, T.; Iwama, A.; Itoh, A.; Kato, C.; Shimizu, Y.; Shinkai, F.; Shimizu, R.; Kondo, S.; et al. Characterization of a ligand for receptor protein-tyrosine kinase HTK expressed in immature hematopoietic cells. Oncogene 1996, 13, 813–822. [Google Scholar]
- Inada, T.; Iwama, A.; Sakano, S.; Ohno, M.; Sawada, K.; Suda, T. Selective expression of the receptor tyrosine kinase, HTK, on human erythroid progenitor cells. Blood 1997, 89, 2757–2765. [Google Scholar] [CrossRef]
- Wang, Z.; Miura, N.; Bonelli, A.; Mole, P.; Carlesso, N.; Olson, D.P.; Scadden, D.T. Receptor tyrosine kinase, EphB4 (HTK), accelerates differentiation of select human hematopoietic cells. Blood 2002, 99, 2740–2747. [Google Scholar] [CrossRef] [Green Version]
- Anselmo, A.; Lauranzano, E.; Soldani, C.; Ploia, C.; Angioni, R.; D’Amico, G.; Sarukhan, A.; Mazzon, C.; Viola, A. Identification of a novel agrin-dependent pathway in cell signaling and adhesion within the erythroid niche. Cell Death Differ. 2016, 23, 1322–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aasheim, H.-C.; Munthe, E.; Funderud, S.; Smeland, E.B.; Beiske, K.; Logtenberg, T. A splice variant of human ephrin-A4 encodes a soluble molecule that is secreted by activated human B lymphocytes. Blood 2000, 95, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Liang, W.; Wen, S.; Zhao, T.; Zhu, M.X.; Li, H.H.; Long, Q.; Wang, M.; Cheng, X.; Liao, Y.H.; et al. EphB2 contributes to human naive B-cell activation and is regulated by miR-185. FASEB J. 2014, 28, 3609–3617. [Google Scholar] [CrossRef] [PubMed]
- Laidlaw, B.J.; Schmidt, T.H.; Green, J.A.; Allen, C.D.; Okada, T.; Cyster, J.G. The Eph-related tyrosine kinase ligand Ephrin-B1 marks germinal center and memory precursor B cells. J. Exp. Med. 2017, 214, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Lu, P.; Shih, C.; Qi, H. Ephrin B1-mediated repulsion and signaling control germinal center T cell territoriality and function. Science 2017, 356, aai9264. [Google Scholar] [CrossRef]
- García-Ceca, J.; Alfaro, D.; Montero-Herradón, S.; Tobajas, E.; Muñoz, J.J.; Zapata, A.G. Eph/Ephrins-Mediated Thymocyte-Thymic Epithelial Cell Interactions Control Numerous Processes of Thymus Biology. Front. Immunol. 2015, 6, 333. [Google Scholar] [CrossRef] [Green Version]
- Cejalvo, T.; Munoz, J.J.; Tobajas, E.; Fanlo, L.; Alfaro, D.; García-Ceca, J.; Zapata, A. Ephrin-B-dependent thymic epithelial cell-thymocyte interactions are necessary for correct T cell differentiation and thymus histology organization: Relevance for thymic cortex development. J. Immunol. 2013, 190, 2670–2681. [Google Scholar] [CrossRef] [Green Version]
- Aasheim, H.C.; Delabie, J.; Finne, E.F. Ephrin-A1 binding to CD4+ T lymphocytes stimulates migration and induces tyrosine phosphorylation of PYK2. Blood 2005, 105, 2869–2876. [Google Scholar] [CrossRef] [Green Version]
- Sharfe, N.; Freywald, A.; Toro, A.; Dadi, H.; Roifman, C. Ephrin stimulation modulates T cell chemotaxis. Eur. J. Immunol. 2002, 32, 3745–3755. [Google Scholar] [CrossRef]
- Yu, G.; Luo, H.; Wu, Y.; Wu, J. Mouse ephrinB3 augments T-cell signaling and responses to T-cell receptor ligation. J. Biol. Chem. 2003, 278, 47209–47216. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Luo, H.; Wu, Y.; Wu, J. EphrinB1 is essential in T-cell-T-cell co-operation during T-cell activation. J. Biol. Chem. 2004, 279, 55531–55539. [Google Scholar] [CrossRef] [Green Version]
- Sharfe, N.; Nikolic, M.; Cimpeon, L.; Van De Kratts, A.; Freywald, A.; Roifman, C.M. EphA and ephrin-A proteins regulate integrin-mediated T lymphocyte interactions. Mol. Immunol. 2008, 45, 1208–1220. [Google Scholar] [CrossRef]
- Prévost, N.; Woulfe, D.S.; Tognolini, M.; Tanaka, T.; Jian, W.; Fortna, R.R.; Jiang, H.; Brass, L.F. Signaling by ephrinB1 and Eph kinases in platelets promotes Rap1 activation, platelet adhesion, and aggregation via effector pathways that do not require phosphorylation of ephrinB1. Blood 2004, 103, 1348–1355. [Google Scholar] [CrossRef]
- Vaiyapuri, S.; Sage, T.; Rana, R.H.; Schenk, M.P.; Ali, M.S.; Unsworth, A.J.; Jones, C.I.; Stainer, A.R.; Kriek, N.; Moraes, L.A.; et al. EphB2 regulates contact-dependent and contact-independent signaling to control platelet function. Blood 2015, 125, 720–730. [Google Scholar] [CrossRef] [Green Version]
- Berrou, E.; Soukaseum, C.; Favier, R.; Adam, F.; Elaib, Z.; Kauskot, A.; Bordet, J.C.; Ballerini, P.; Loyau, S.; Feng, M.; et al. A mutation of the human EPHB2 gene leads to a major platelet functional defect. Blood 2018, 132, 2067–2077. [Google Scholar] [CrossRef] [Green Version]
- Mukai, M.; Suruga, N.; Saeki, N.; Ogawa, K. EphA receptors and ephrin-A ligands are upregulated by monocytic differentiation/maturation and promote cell adhesion and protrusion formation in HL60 monocytes. BMC Cell Biol. 2017, 18, 28. [Google Scholar] [CrossRef] [Green Version]
- Rissoan, M.C.; Duhen, T.; Bridon, J.M.; Bendriss-Vermare, N.; Péronne, C.; de Saint Vis, B.; Brière, F.; Bates, E.E. Subtractive hybridization reveals the expression of immunoglobulin-like transcript 7, Eph-B1, granzyme B, and 3 novel transcripts in human plasmacytoid dendritic cells. Blood 2002, 100, 3295–3303. [Google Scholar] [CrossRef] [Green Version]
- de Saint-Vis, B.; Bouchet, C.; Gautier, G.g.; Valladeau, J.; Caux, C.; Garrone, P. Human dendritic cells express neuronal Eph receptor tyrosine kinases: Role of EphA2 in regulating adhesion to fibronectin. Blood 2003, 102, 4431–4440. [Google Scholar] [CrossRef] [Green Version]
- Mimche, P.N.; Brady, L.M.; Keeton, S.; Fenne, D.S.; King, T.P.; Quicke, K.M.; Hudson, L.E.; Lamb, T.J. Expression of the Receptor Tyrosine Kinase EphB2 on Dendritic Cells Is Modulated by Toll-Like Receptor Ligation but Is Not Required for T Cell Activation. PLoS ONE 2015, 10, e0138835. [Google Scholar] [CrossRef]
- Kuang, S.Q.; Bai, H.; Fang, Z.H.; Lopez, G.; Yang, H.; Tong, W.; Wang, Z.Z.; Garcia-Manero, G. Aberrant DNA methylation and epigenetic inactivation of Eph receptor tyrosine kinases and ephrin ligands in acute lymphoblastic leukemia. Blood 2010, 115, 2412–2419. [Google Scholar] [CrossRef] [Green Version]
- Boyd, A.W.; Ward, L.D.; Wicks, I.P.; Simpson, R.J.; Salvaris, E.; Wilks, A.; Welch, K.; Loudovaris, M.; Rockman, S.; Busmanis, I. Isolation and characterization of a novel receptor-type protein tyrosine kinase (hek) from a human pre-B cell line. J. Biol. Chem. 1992, 267, 3262–3267. [Google Scholar] [CrossRef]
- Wicks, I.P.; Wilkinson, D.; Salvaris, E.; Boyd, A.W. Molecular cloning of HEK, the gene encoding a receptor tyrosine kinase expressed by human lymphoid tumor cell lines. Proc. Natl. Acad. Sci. USA 1992, 89, 1611–1615. [Google Scholar] [CrossRef]
- Smith, L.M.; Walsh, P.T.; Rüdiger, T.; Cotter, T.G.; Mc Carthy, T.V.; Marx, A.; O’Connor, R. EphA3 is induced by CD28 and IGF-1 and regulates cell adhesion. Exp. Cell Res. 2004, 292, 295–303. [Google Scholar] [CrossRef]
- Ashton, J.M.; Balys, M.; Neering, S.J.; Hassane, D.C.; Cowley, G.; Root, D.E.; Miller, P.G.; Ebert, B.L.; McMurray, H.R.; Land, H.; et al. Gene Sets Identified with Oncogene Cooperativity Analysis Regulate In Vivo Growth and Survival of Leukemia Stem Cells. Cell Stem Cell 2012, 11, 359–372. [Google Scholar] [CrossRef] [Green Version]
- Guan, M.; Liu, L.; Zhao, X.; Wu, Q.; Yu, B.; Shao, Y.; Yang, H.; Fu, X.; Wan, J.; Zhang, W. Copy Number Variations of EphA3 Are Associated With Multiple Types of Hematologic Malignancies. Clin. Lymphoma Myeloma Leuk. 2011, 11, 50–53. [Google Scholar] [CrossRef] [Green Version]
- Walter, M.J.; Payton, J.E.; Ries, R.E.; Shannon, W.D.; Deshmukh, H.; Zhao, Y.; Baty, J.; Heath, S.; Westervelt, P.; Watson, M.A.; et al. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc. Natl. Acad. Sci. USA 2009, 106, 12950–12955. [Google Scholar] [CrossRef]
- Caivano, A.; La Rocca, F.; Laurenzana, I.; Annese, T.; Tamma, R.; Famigliari, U.; Simeon, V.; Trino, S.; De Luca, L.; Villani, O.; et al. Epha3 acts as proangiogenic factor in multiple myeloma. Oncotarget 2017, 8, 34298–34309. [Google Scholar] [CrossRef] [Green Version]
- Nakanishi, H.; Nakamura, T.; Canaani, E.; Croce, C.M. ALL1 fusion proteins induce deregulation of EphA7 and ERK phosphorylation in human acute leukemias. Proc. Natl. Acad. Sci. USA 2007, 104, 14442–14447. [Google Scholar] [CrossRef]
- López-Nieva, P.; Vaquero, C.; Fernández-Navarro, P.; González-Sánchez, L.; Villa-Morales, M.; Santos, J.; Esteller, M.; Fernández-Piqueras, J. EPHA7, a new target gene for 6q deletion in T-cell lymphoblastic lymphomas. Carcinogenesis 2012, 33, 452–458. [Google Scholar] [CrossRef]
- Oricchio, E.; Nanjangud, G.; Wolfe, A.L.; Schatz, J.H.; Mavrakis, K.J.; Jiang, M.; Liu, X.; Bruno, J.; Heguy, A.; Olshen, A.B.; et al. The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell 2011, 147, 554–564. [Google Scholar] [CrossRef] [Green Version]
- Kampen, K.R.; Scherpen, F.J.; Garcia-Manero, G.; Yang, H.; Kaspers, G.J.; Cloos, J.; Zwaan, C.M.; van den Heuvel-Eibrink, M.M.; Kornblau, S.M.; De Bont, E.S. EphB1 Suppression in Acute Myelogenous Leukemia: Regulating the DNA Damage Control System. Mol. Cancer Res. 2015, 13, 982–992. [Google Scholar] [CrossRef] [Green Version]
- Steube, K.G.; Meyer, C.; Habig, S.; Uphoff, C.C.; Drexler, H.G. Expression of receptor tyrosine kinase HTK (hepatoma transmembrane kinase) and HTK ligand by human leukemia-lymphoma cell lines. Leuk. Lymphoma 1999, 33, 371–376. [Google Scholar] [CrossRef]
- Li, L.; Xu, N.; Zhang, J.F.; Xu, L.L.; Zhou, X.; Huang, B.T.; Li, Y.L.; Liu, X.L. EphB4/ephrinB2 Contributes to Imatinib Resistance in Chronic Myeloid Leukemia Involved in Cytoskeletal Proteins. Int. J. Med. Sci. 2016, 13, 365–373. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.; Abe, A.; Imagama, S.; Nomura, Y.; Tanizaki, R.; Minami, Y.; Hayakawa, F.; Ito, Y.; Katsumi, A.; Yamamoto, K.; et al. BCR-ABL-independent and RAS/MAPK pathway-dependent form of imatinib resistance in Ph-positive acute lymphoblastic leukemia cell line with activation of EphB4. Eur. J. Haematol. 2010, 84, 229–238. [Google Scholar] [CrossRef]
- Merchant, A.A.; Jorapur, A.; McManus, A.; Liu, R.; Krasnoperov, V.; Chaudhry, P.; Singh, M.; Harton, L.; Agajanian, M.; Kim, M.; et al. EPHB4 is a therapeutic target in AML and promotes leukemia cell survival via AKT. Blood Adv. 2017, 1, 1635–1644. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, H.; Chen, X.; Mai, H.; Li, C.; Wen, F. Aberrant EPHB4 gene methylation and childhood acute lymphoblastic leukemia. Oncol. Lett. 2017, 14, 4433–4440. [Google Scholar] [CrossRef]
- Huang, Y.C.; Lin, S.J.; Lin, K.M.; Chou, Y.C.; Lin, C.W.; Yu, S.C.; Chen, C.L.; Shen, T.L.; Chen, C.K.; Lu, J.; et al. Regulation of EBV LMP1-triggered EphA4 downregulation in EBV-associated B lymphoma and its impact on patients’ survival. Blood 2016, 128, 1578–1589. [Google Scholar] [CrossRef]
- Müller-Tidow, C.; Schwäble, J.; Steffen, B.R.; Tidow, N.; Brandt, B.; Becker, K.; Schulze-Bahr, E.; Halfter, H.; Vogt, U.; Metzger, R.; et al. High-Throughput Analysis of Genome-Wide Receptor Tyrosine Kinase Expression in Human Cancers Identifies Potential Novel Drug Targets. Clin. Cancer Res. 2004, 10, 1241–1249. [Google Scholar] [CrossRef] [Green Version]
- Shimoyama, M.; Matsuoka, H.; Tamekane, A.; Ito, M.; Iwata, N.; Inoue, R.; Chihara, K.; Furuya, A.; Hanai, N.; Matsui, T. T-cell-specific expression of kinase-defective Eph-family receptor protein, EphB6 in normal as well as transformed hematopoietic cells. Growth Factors 2000, 18, 63–78. [Google Scholar] [CrossRef]
- El Zawily, A.; McEwen, E.; Toosi, B.; Vizeacoumar, F.S.; Freywald, T.; Vizeacoumar, F.J.; Freywald, A. The EphB6 receptor is overexpressed in pediatric T cell acute lymphoblastic leukemia and increases its sensitivity to doxorubicin treatment. Sci. Rep. 2017, 7, 14767. [Google Scholar] [CrossRef] [Green Version]
- Alonso, C.L.; Trinidad, E.M.; de Garcillan, B.; Ballesteros, M.; Castellanos, M.; Cotillo, I.; Muñoz, J.J.; Zapata, A.G. Expression profile of Eph receptors and ephrin ligands in healthy human B lymphocytes and chronic lymphocytic leukemia B-cells. Leuk. Res. 2009, 33, 395–406. [Google Scholar] [CrossRef]
- Trinidad, E.M.; Ballesteros, M.; Zuloaga, J.; Zapata, A.; Alonso-Colmenar, L.M. An impaired transendothelial migration potential of chronic lymphocytic leukemia (CLL) cells can be linked to ephrin-A4 expression. Blood 2009, 114, 5081–5090. [Google Scholar] [CrossRef] [Green Version]
- Jiang, G.; Freywald, T.; Webster, J.; Kozan, D.; Geyer, R.; DeCoteau, J.; Narendran, A.; Freywald, A. In human leukemia cells ephrin-B-induced invasive activity is supported by Lck and is associated with reassembling of lipid raft signaling complexes. Mol. Cancer Res. 2008, 6, 291–305. [Google Scholar] [CrossRef]
- Xiang, C.; Wu, J.; Yu, L. Construction of three-gene-based prognostic signature and analysis of immune cells infiltration in children and young adults with B-acute lymphoblastic leukemia. Mol. Genet. Genomic. Med. 2022, 10, e1964. [Google Scholar] [CrossRef]
- Takahashi, Y.; Itoh, M.A.I.; Nara, N.; Tohda, S. Effect of EPH–Ephrin Signaling on the Growth of Human Leukemia Cells. Anticancer Res. 2014, 34, 2913. [Google Scholar]
- Wimmer-Kleikamp, S.H.; Nievergall, E.; Gegenbauer, K.; Adikari, S.; Mansour, M.; Yeadon, T.; Boyd, A.W.; Patani, N.R.; Lackmann, M. Elevated protein tyrosine phosphatase activity provokes Eph/ephrin-facilitated adhesion of pre-B leukemia cells. Blood 2008, 112, 721–732. [Google Scholar] [CrossRef] [Green Version]
- Holen, H.L.; Shadidi, M.; Narvhus, K.; Kjøsnes, O.; Tierens, A.; Aasheim, H.C. Signaling through ephrin-A ligand leads to activation of Src-family kinases, Akt phosphorylation, and inhibition of antigen receptor-induced apoptosis. J. Leukoc. Biol. 2008, 84, 1183–1191. [Google Scholar] [CrossRef]
- Cimino, G.; Rapanotti, M.C.; Sprovieri, T.; Elia, L. ALL1 gene alterations in acute leukemia: Biological and clinical aspects. Haematologica 1998, 83, 350–357. [Google Scholar]
- Gu, Y.; Nakamura, T.; Alder, H.; Prasad, R.; Canaani, O.; Cimino, G.; Croce, C.M.; Canaani, E. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 1992, 71, 701–708. [Google Scholar] [CrossRef]
- Winters, A.C.; Bernt, K.M. MLL-Rearranged Leukemias-An Update on Science and Clinical Approaches. Front. Pediatr. 2017, 5, 4. [Google Scholar] [CrossRef] [Green Version]
- Ebrahim, E.K.; Assem, M.M.; Amin, A.I.; Kamel, M.M.; El Meligui, Y.M.; Metwally, A.M. FLT3 Internal Tandem Duplication Mutation, cMPL and CD34 Expressions Predict Low Survival in Acute Myeloid Leukemia Patients. Ann. Clin. Lab. Sci. 2016, 46, 592–600. [Google Scholar]
- El-Sisi, M.G.; Radwan, S.M.; Saeed, A.M.; El-Mesallamy, H.O. Serum levels of FAK and some of its effectors in adult AML: Correlation with prognostic factors and survival. Mol. Cell Biochem. 2021, 476, 1949–1963. [Google Scholar] [CrossRef]
- McMurray, H.R.; Sampson, E.R.; Compitello, G.; Kinsey, C.; Newman, L.; Smith, B.; Chen, S.R.; Klebanov, L.; Salzman, P.; Yakovlev, A.; et al. Synergistic response to oncogenic mutations defines gene class critical to cancer phenotype. Nature 2008, 453, 1112–1116. [Google Scholar] [CrossRef] [Green Version]
- Kong, L.; Zhang, X.; Li, C.; Zhou, L. Potential therapeutic targets and small molecular drugs for pediatric B-precursor acute lymphoblastic leukemia treatment based on microarray data. Oncol. Lett. 2017, 14, 1543–1549. [Google Scholar] [CrossRef]
- Trinidad, E.M.; Zapata, A.G.; Alonso-Colmenar, L.M. Eph-ephrin bidirectional signaling comes into the context of lymphocyte transendothelial migration. Cell Adh. Migr. 2010, 4, 363–367. [Google Scholar] [CrossRef]
- Flores, M.A.; Fortea, P.; Trinidad, E.M.; García, D.; Soler, G.; Ortuño, F.J.; Zapata, A.G.; Alonso-Colmenar, L.M. EphrinA4 plays a critical role in α4 and αL mediated survival of human CLL cells during extravasation. Oncotarget 2016, 7, 48481–48500. [Google Scholar] [CrossRef]
- Charmsaz, S.; Beckett, K.; Smith, F.M.; Bruedigam, C.; Moore, A.S.; Al-Ejeh, F.; Lane, S.W.; Boyd, A.W. EphA2 Is a Therapy Target in EphA2-Positive Leukemias but Is Not Essential for Normal Hematopoiesis or Leukemia. PLoS ONE 2015, 10, e0130692. [Google Scholar] [CrossRef] [Green Version]
- Charmsaz, S.; Al-Ejeh, F.; Yeadon, T.M.; Miller, K.J.; Smith, F.M.; Stringer, B.W.; Moore, A.S.; Lee, F.T.; Cooper, L.T.; Stylianou, C.; et al. EphA3 as a target for antibody immunotherapy in acute lymphoblastic leukemia. Leukemia 2017, 31, 1779–1787. [Google Scholar] [CrossRef] [Green Version]
- Swords, R.T.; Greenberg, P.L.; Wei, A.H.; Durrant, S.; Advani, A.S.; Hertzberg, M.S.; Jonas, B.A.; Lewis, I.D.; Rivera, G.; Gratzinger, D.; et al. KB004, a first in class monoclonal antibody targeting the receptor tyrosine kinase EphA3, in patients with advanced hematologic malignancies: Results from a phase 1 study. Leuk. Res. 2016, 50, 123–131. [Google Scholar] [CrossRef]
- Wang, C.; Dong, J.; Zhang, Y.; Wang, F.; Gao, H.; Li, P.; Wang, S.; Zhang, J. Design, synthesis and biological evaluation of biphenyl urea derivatives as novel VEGFR-2 inhibitors. MedChemComm 2013, 4, 1434–1438. [Google Scholar] [CrossRef]
- Sawamiphak, S.; Seidel, S.; Essmann, C.L.; Wilkinson, G.A.; Pitulescu, M.E.; Acker, T.; Acker-Palmer, A. Ephrin-B2 regulates VEGFR2 function in developmental and tumour angiogenesis. Nature 2010, 465, 487–491. [Google Scholar] [CrossRef]
- Jabbarzadeh Kaboli, P.; Rahmat, A.; Ismail, P.; Ling, K.H. Targets and mechanisms of berberine, a natural drug with potential to treat cancer with special focus on breast cancer. Eur. J. Pharmacol. 2014, 740, 584–595. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhu, M.; Zhang, D.; Yang, L.; Yang, T.; Li, X.; Zhang, Y. Berberine inhibits the proliferation and migration of breast cancer ZR-75-30 cells by targeting Ephrin-B2. Phytomedicine 2017, 25, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhu, M.; Yang, L.; Yang, T.; Zhang, Y. Synergistic Effect of TPD7 and Berberine against Leukemia Jurkat Cell Growth through Regulating Ephrin-B2 Signaling. Phytother. Res. 2017, 31, 1392–1399. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Gao, X.; Yu, X.; Liu, X.; Du, Q. Ephrin type-B receptor 4 is an essential mediator in drug-resistance of acute myeloid leukemia cells to Adriamycin. J. Exp. Ther. Oncol. 2018, 12, 249–259. [Google Scholar]
- Ma, W.; Zhu, M.; Wang, B.; Gong, Z.; Du, X.; Yang, T.; Shi, X.; Dai, B.; Zhan, Y.; Zhang, D.; et al. Vandetanib drives growth arrest and promotes sensitivity to imatinib in chronic myeloid leukemia by targeting ephrin type-B receptor 4. Mol. Oncol. 2022, 16, 2747–2765. [Google Scholar] [CrossRef]
| EPHA1 | Human CD34+ HSCs [86,87] Rat T lymphocytes—during thymic development [88] |
| EPHA2 | Human CD34+ HSCs [89] Human CD133+ HSCs [89] Mouse Lin-ckit + sca1+ HSCs [90] Rat T lymphocytes—during thymic development [88] |
| EPHA3 | Mouse Lin-ckit + sca1+ HSCs [90] Rat T lymphocytes—during thymic development [88] |
| EPHA4 | Human platelets [91,92] Rat T lymphocytes—during thymic development [88] Human pro- and pre-B cells [93] |
| EPHA6 | Mouse Lin-ckit + sca1+ HSCs [90] |
| EPHA7 | Rat T lymphocytes—during thymic development [88] Human pro- and pre-B cells [93] Normal lymphocytes [94] |
| EPHA8 | Mouse Lin-ckit + sca1+ HSCs [90] |
| EPHB1 | Human platelets [92] |
| EPHB2 | Human CD133+ HSCs [89] Human CD34+ HSCs—partially expressed [89] |
| EPHB4 | Bone marrow CD34+ cells [95] Erythroid progenitor cells [96] HSPCs [97] |
| EPHB6 | Mouse thymic T cells [98] |
| EphrinA1 | Rat T lymphocytes—during thymic development [88] |
| EphrinA3 | Human CD34+ HSPCs [87] Rat T lymphocytes—during thymic development [88] |
| EphrinA4 | Human CD34+ HSPCs [87] Mouse Lin-ckit + sca1+ HSCs [90] |
| EphrinA5 | Mouse Lin-ckit + sca1+ HSCs [90] Rat T lymphocytes—during thymic development [88] |
| EphrinB1 | Human platelets [92] Mouse T lymphocytes—during thymic development [99] |
| Leukemia/Lymphoma Cell Lines | Hematologic Malignancies | |
|---|---|---|
| EPHA2 | ALL: EPHA2 hypermethylation [131] | |
| EPHA3 | LK63 pre-B ALL cell line [132] T-cell leukemia cell lines (Jurkat, JM, HSB-2) [133] EPHA3 identified as a CD28-responsive gene in Jurkat cells [134] | Mouse model of CML blast crisis: EPHA3 identified as a common CRG [135] AML: EPHA3 CNVs [136,137] ALL: EPHA3 CNVs [136] CLL: EPHA3 CNVs [136] CML: EPHA3 CNVs [136] MDS: EPHA3 CNVs [136] MM: EPHA3 highly expressed in endothelial BM cells [138] |
| EPHA4 | ALL: EPHA4 hypermethylation [131] | |
| EPHA5 | ALL: EPHA5 hypermethylation [131] | |
| EPHA6 | ALL: EPHA6 hypermethylation [131] | |
| EPHA7 | Leukemic cell lines with ALL1 gene translocations: transcriptional up-regulation of the EPHA7 [139] | ALL: EPHA7 hypermethylation [131] T-ALL/lymphoma with 6q deletion: EPHA7 as a tumor suppressor [140] GC B cell NHL: EPHA7 hypermethylation and repression [94] FL: EPHA7 proposed as a tumor suppressor [141] |
| EPHA10 | ALL: EPHA10 hypermethylation [131] | |
| EPHB1 | pediatric AML: decreased EPHB1 peptide phosphorylation and mRNA expression compared to healthy controls, hypermethylation of the EPHB1 promoter [142] ALL: EPHB1 hypermethylation [131] | |
| EPHB2 | ALL: EPHB2 hypermethylation [131] | |
| EPHB3 | T-ALL cell lines H9 and E6.1 [73] | ALL: EPHB3 hypermethylation [131] |
| EPHB4 | Human leukemia/lymphoma cell lines: EPHB4 mRNA expression in 68 of the 70 studied cell lines [143] K562-R cell line (human imatinib-resistant CML cell line): EPHB4 overexpression [144] Philadelphia chromosome-positive ALL cell lines: imatinib resistance mediated by EPHB4 activation [145] | AML: 28% (7/25) of newly diagnosed AML BM samples over-expressed EPHB4 protein [146] CML: EPHB4 mRNA levels in BM cells significantly increased according to clinical stages (increased EPHB4 expression in blast crisis compared to chronic phase) [144] Childhood ALL: prevalent methylation of the EPHB4 [147] ALL: EPHB4 hypermethylation [131] PTLDs: EPHB4 expression suppressed in EBV+ PTLDs [148] |
| EPHB6 | T-ALL cell lines H9 and E6.1 [73] | AML: Increased EPHB6 expression [149] T-cell leukemia/lymphoma: increased EPHB6 expression [150,151] T-ALL: EPHB6 expression confers increased sensitivity to doxorubicin [151] CLL: EPHB6 expression correlated with a high content of ZAP-70 mRNA and a poor prognosis [152] |
| EphrinA1 | ALL: EFNA1 hypermethylation [131] | |
| EphrinA3 | ALL: EFNA3 hypermethylation [131] | |
| EphrinA4 | CLL: High serum levels of a soluble ephrinA4 isoform positively correlated with increasing PB lymphocyte counts and lymphadenopathy [152] CLL: ephrinA4 expression on PB CLL cells inversely correlated with lymphadenopathy [153] | |
| EphrinA5 | ALL: EFNA5 hypermethylation [131] | |
| EphrinB1 | Jurkat cells: EphrinB1 enhanced their metastatic potential [154] | Pediatric B precursor ALL: EFNB1 identified among downregulated DEGs [155] B-ALL: EFNB1 as an independent prognostic factor for B-ALL; EFNB1 mRNA levels significantly lower in B-ALL patients compared to controls [155] ALL: EFNB1 hypermethylation [131] |
| EphrinB2 | Human leukemia/lymphoma cell lines: EphrinB2 mRNA expression in 58 of the 70 studied cell lines [143] Erythroid leukemia-derived cell line: Induction of proliferation and colony formation, upregulation of growth-related gene expression [156] | ALL: EFNB2 hypermethylation [131] |
| Target | Type of Hematologic Malignancy | Drug | Type of Study | Outcome | Reference |
|---|---|---|---|---|---|
| EPHA2 | ALL1/AF9 leukemias | EPHA2 mAb (IF7) radiolabeled with Lutetium-177 | Preclinical: Mouse model | Survival advantage | [168] |
| EPHA3 | Pre-B-ALL | EPHA3 with α-particle-emitting Bismuth-213 payload | In vitro: EPHA3-expressing leukemic xenografts | Antitumor effect | [169] |
| AML, MDS/MPN, MDS, DLBCL, MF | EPHA3 mAb (KB004) | Clinical: phase I | Responses in AML, MF, MDS/MPN, and MDS patients | [170] | |
| EPHA7 | Leukemias with ALL1 translocations | ERK inhibitor (indirect EPHA7 signaling inhibition via its downstream target ERK) | In vitro: K562 cells | Induction of apoptotic cell death | [139] |
| Lymphomas | EPHA7 delivered by anti-CD20 mAb (Rituximab) | Preclinical: xenografted human lymphomas | Inhibition of ERK and SRC activity; induction of cell death | [141] | |
| EPHB4 | Imatinib-resistant CML | Vandetanib | Preclinical: K562 cells | Growth arrest; overcoming of imatinib resistance | [177] |
| EPHB4 | AML cases with high EPHB4 expression | EPHB4 mAb (MAb131) | Preclinical: myeloid leukemia cell lines and human myeloid leukemia xenograft models | Effective against AML in vitro and in vivo | [139] |
| EphrinB2 | T-ALL | Combination of TPD7 and berberine (TAB) | Preclinical: Jurkat cells | Decrease in the levels and phosphorylation of ephrinB2; downregulation of PDZ domain-interacting proteins (syntenin-1 and PICK1); decreased phosphorylation of VEGFR2; inhibition of Rac1; upregulation of PTEN | [175] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stergiou, I.E.; Papadakos, S.P.; Karyda, A.; Tsitsilonis, O.E.; Dimopoulos, M.-A.; Theocharis, S. EPH/Ephrin Signaling in Normal Hematopoiesis and Hematologic Malignancies: Deciphering Their Intricate Role and Unraveling Possible New Therapeutic Targets. Cancers 2023, 15, 3963. https://doi.org/10.3390/cancers15153963
Stergiou IE, Papadakos SP, Karyda A, Tsitsilonis OE, Dimopoulos M-A, Theocharis S. EPH/Ephrin Signaling in Normal Hematopoiesis and Hematologic Malignancies: Deciphering Their Intricate Role and Unraveling Possible New Therapeutic Targets. Cancers. 2023; 15(15):3963. https://doi.org/10.3390/cancers15153963
Chicago/Turabian StyleStergiou, Ioanna E., Stavros P. Papadakos, Anna Karyda, Ourania E. Tsitsilonis, Meletios-Athanasios Dimopoulos, and Stamatios Theocharis. 2023. "EPH/Ephrin Signaling in Normal Hematopoiesis and Hematologic Malignancies: Deciphering Their Intricate Role and Unraveling Possible New Therapeutic Targets" Cancers 15, no. 15: 3963. https://doi.org/10.3390/cancers15153963
APA StyleStergiou, I. E., Papadakos, S. P., Karyda, A., Tsitsilonis, O. E., Dimopoulos, M.-A., & Theocharis, S. (2023). EPH/Ephrin Signaling in Normal Hematopoiesis and Hematologic Malignancies: Deciphering Their Intricate Role and Unraveling Possible New Therapeutic Targets. Cancers, 15(15), 3963. https://doi.org/10.3390/cancers15153963