Interruption of p53-MDM2 Interaction by Nutlin-3a in Human Lymphoma Cell Models Initiates a Cell-Dependent Global Effect on Transcriptome and Proteome Level
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Statistical Rationale
2.2. Chemical and Biochemical Reagents
2.3. Cell Lines, Drug Treatment and Chemical Reagents
2.4. Cell Viability, Proliferation, Apoptosis Assays
2.5. Cell Cycle Analysis
2.6. Cell Lysis and Western Blotting
2.7. Mitochondrial Labelling Assay—Measurement of ROS Generation via High Content Imaging
2.8. Transmission Electron Microscopy Imaging of the Biological Samples
2.9. Affymetrix Microarray Analysis
2.10. Isotope Coded Protein Labeling, Protein Fractionation by SDS-PAGE and in Gel Tryptic Digestion
2.11. Filter Aided Sample Preparation and in-Solution Tryptic Digestion
2.12. Mass Spectrometric Analysis by nLC ESI-MS/MS
2.13. Mass Spectrometric Data Processing, Protein Identification, and Relative Quantitation
2.14. Comparison of Transcriptome and Proteome Analysis
- Τhe altered transcripts and proteins between treated and untreated lymphoma samples in each cell type and in all lymphoma groups were compared, to identify the overlapping and unique genes/proteins between the two levels of analyses. The comparison was performed on pooled lymphoma data and on paired control-treated cells per cell line that derived separately from transcriptomics and proteomics analysis (proteomic experiments consisting of three independent biological replicates), to lessen the variability between the different samples.
- The analysis of the deregulated GO terms and functional pathways in transcriptome and proteome data was performed according to Section 2.15. The main objective of this analysis was to identify the common and unique functional contexts extracted from the integrated omics analysis on the level of protein interaction networks.
2.15. Functional and Pathway Analysis of Deregulated Proteins
2.16. Protein–Protein Interaction Network Analysis and Construction
3. Results
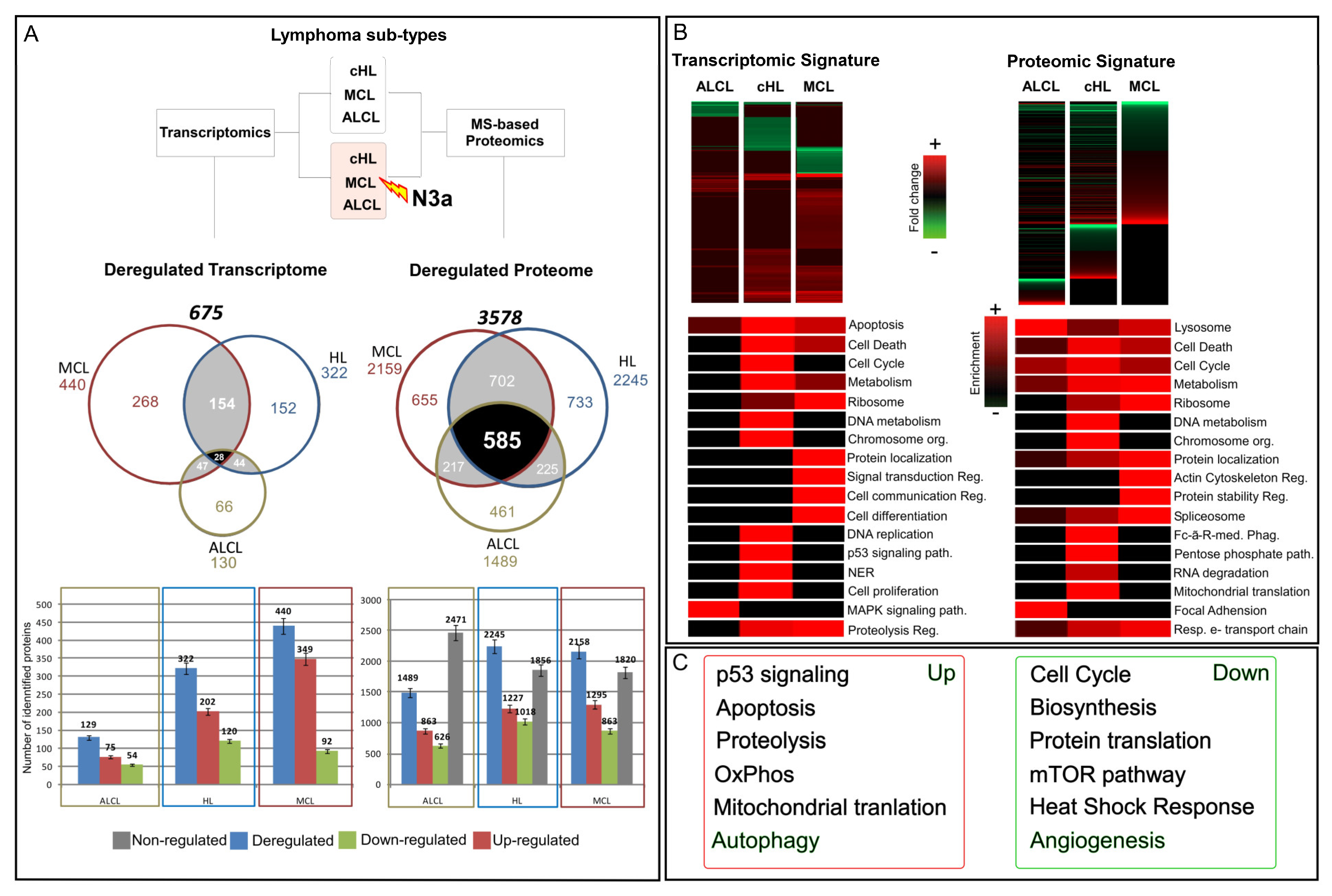
3.1. N3a-Induced Transcriptome and Proteome Profile of the Three Lymphoma Types
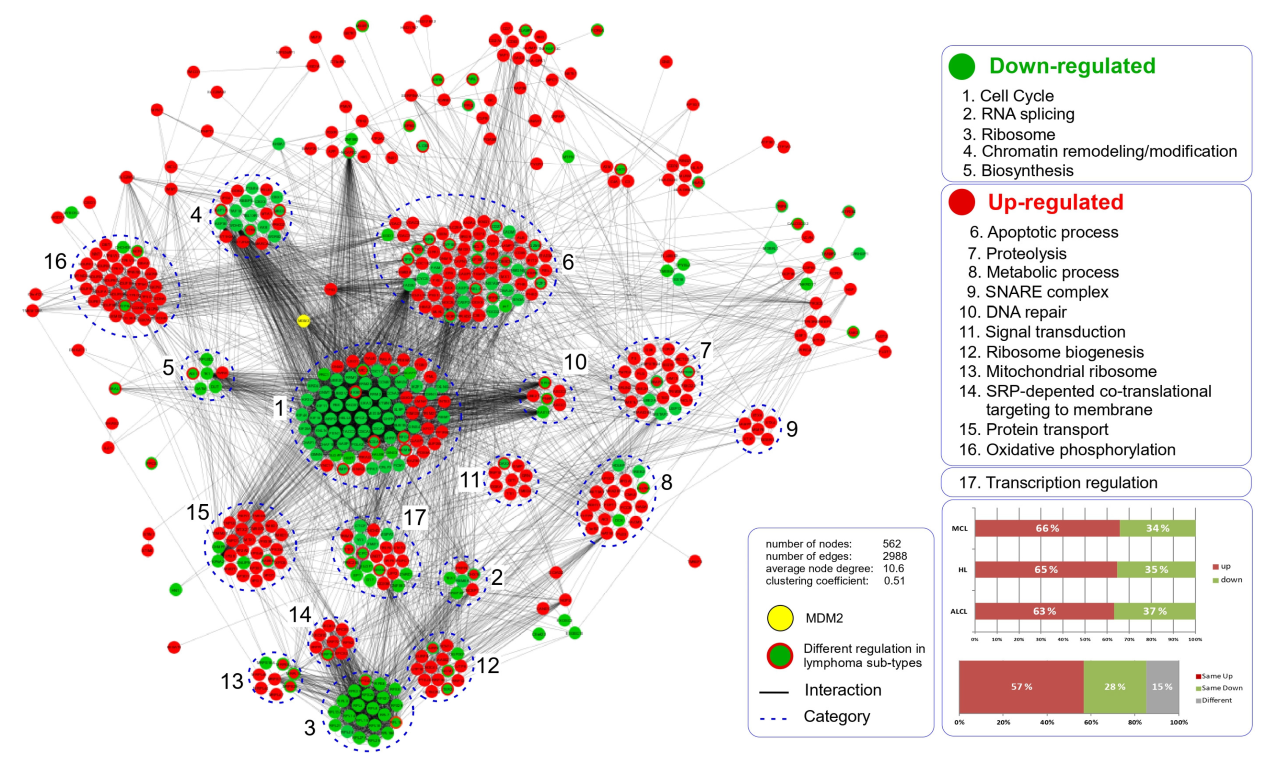
3.2. Gene Ontology Terms and Pathway Enrichment Analysis of the N3a-Affected Transcripts and Proteins
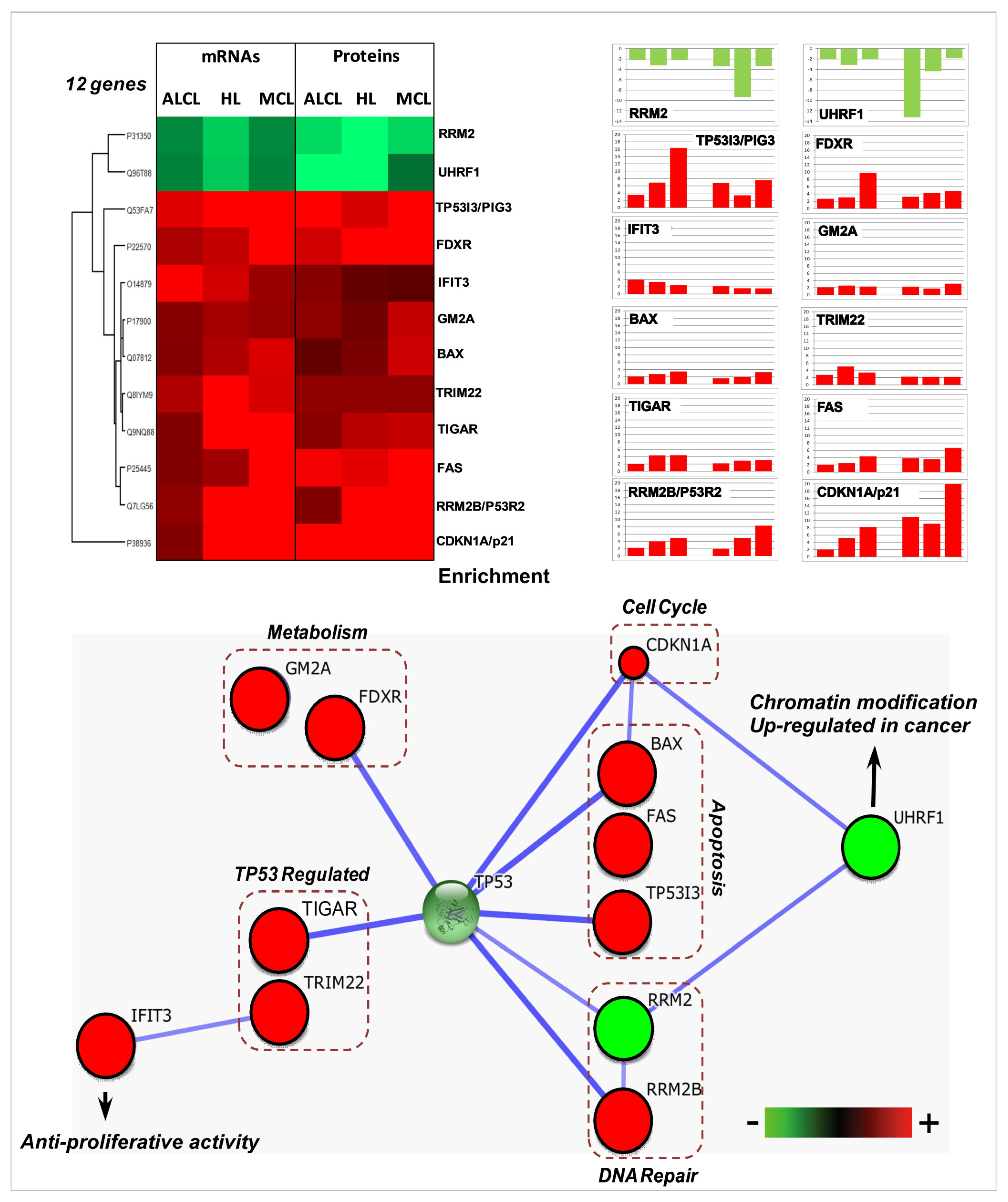
3.3. Common Molecular Signatures after N3a Treatment in All Three Lymphoma Types
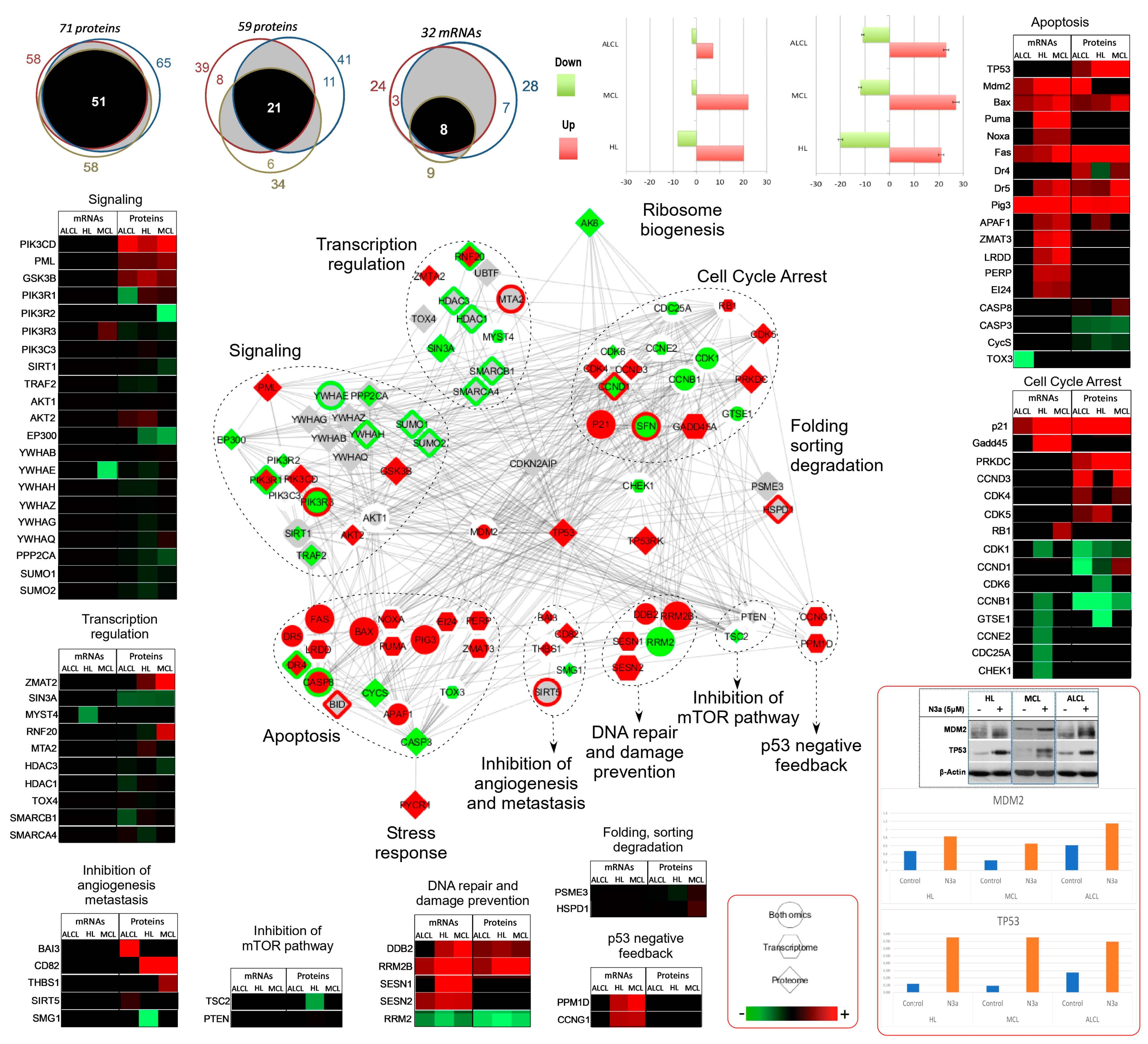
3.4. Unique Molecular Signatures of the N3a Effect in the Three Lymphoma Types
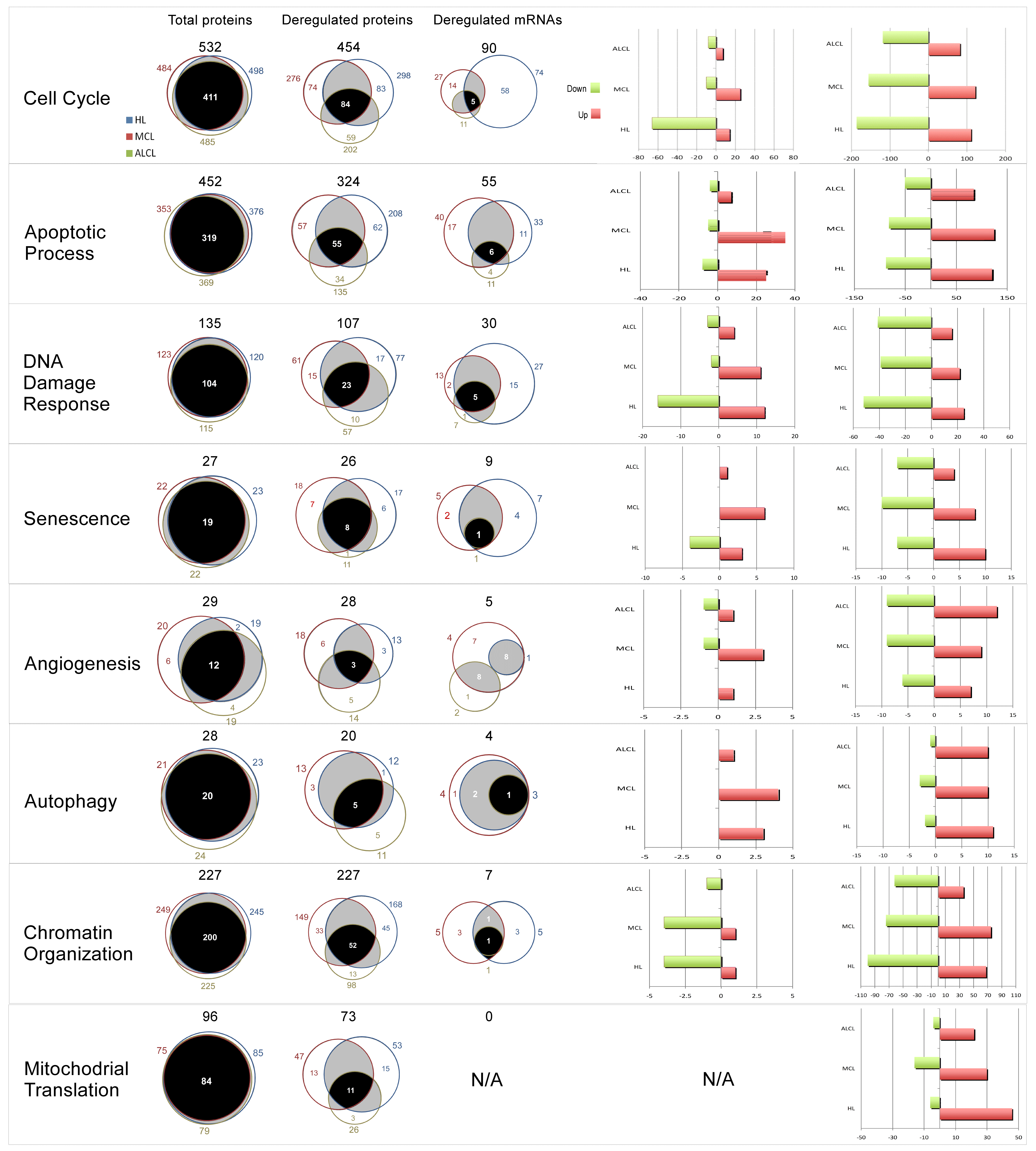
3.5. Cellular Pathways and Biological Processes Affected by N3a in the Three Lymphoma Types
3.5.1. Activation of p53 Signaling Pathway by N3a
3.5.2. N3a-Induced Effect on Cell-Cycle Pathway
3.5.3. N3a-Induced Deregulation of Proteins Involved in Apoptotic Process
3.5.4. Deregulation of Proteins Involved in DNA Damage Response
3.5.5. p53 Activation Affects Angiogenesis, Autophagy, Metabolism, and Chromatin Organization in Lymphoma
3.6. The Effect of p53 Activation on Mitochondrial Translation
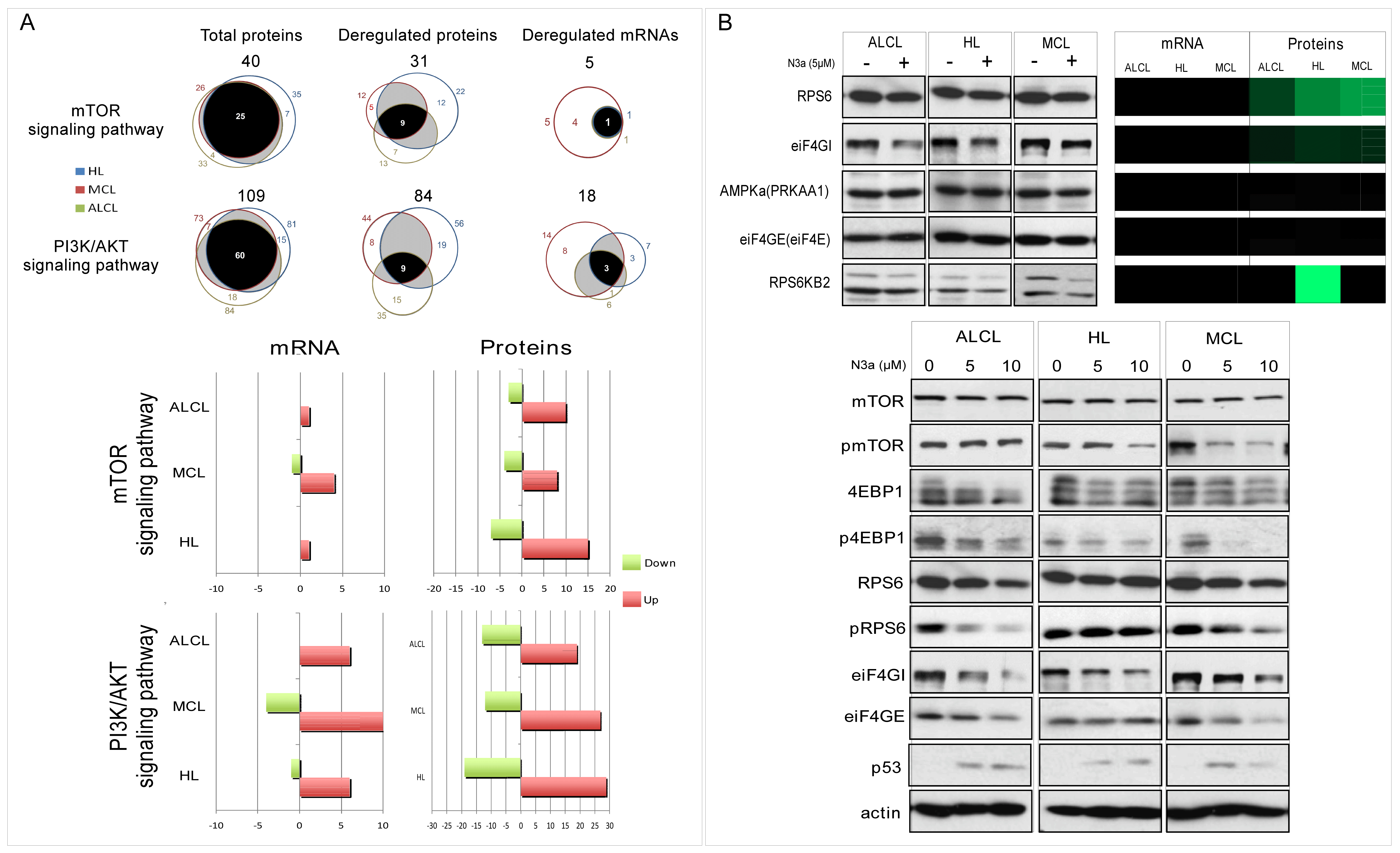
3.7. Oncogenic Pathways Are Significantly Affected by p53 Activation
3.8. The PI3K/mTOR Pathway
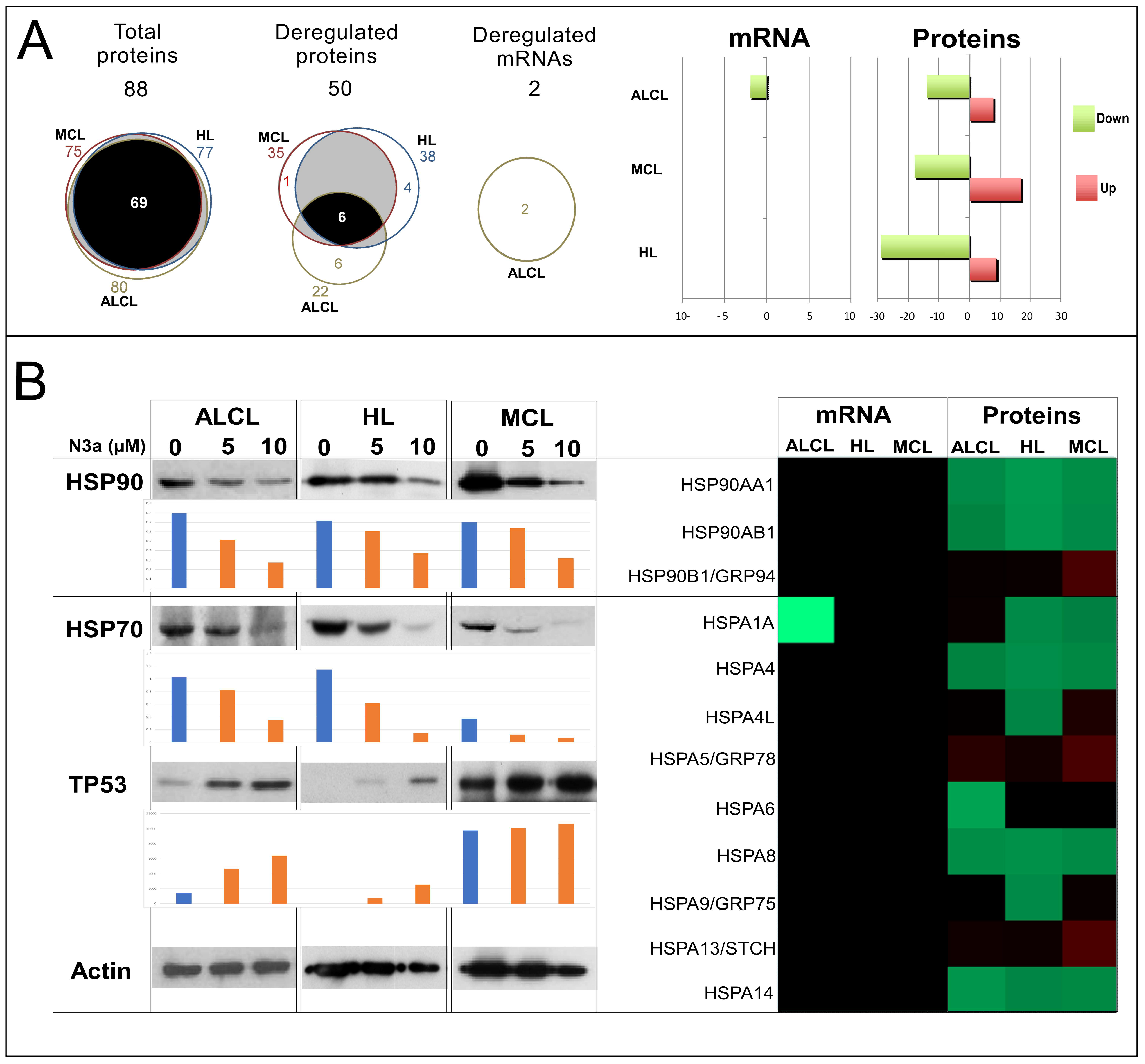
3.9. Deregulation of Heat Shock Response (HSR) by N3a
3.10. Synergistic N3a/Hsp90 Inhibition Effect on Lymphoma Cells
3.11. Enhanced N3a-Effect on Lymphoma Cells in Combination to PI3K/mTOR Inhibition
3.12. N3a Promotes Metabolic Switch to Oxphos in Lymphoma Cells
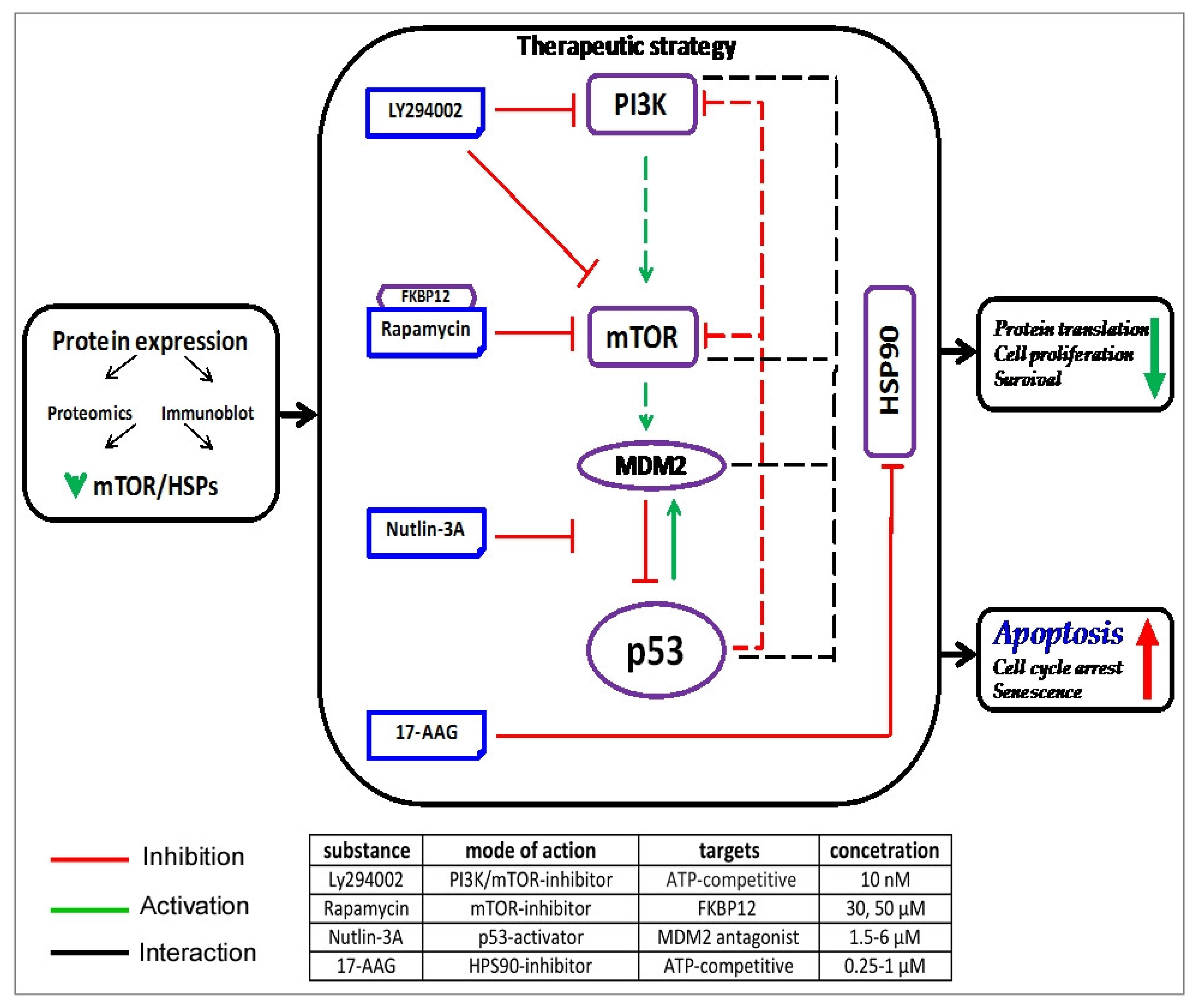
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xu-Monette, Z.Y.; Medeiros, L.J.; Li, Y.; Orlowski, R.Z.; Andreeff, M.; Bueso-Ramos, C.E.; Greiner, T.C.; McDonnell, T.J.; Young, K.H. Dysfunction of the TP53 tumor suppressor gene in lymphoid malignancies. Blood 2012, 119, 3668–3683. [Google Scholar] [CrossRef]
- Saha, M.N.; Qiu, L.; Chang, H. Targeting p53 by small molecules in hematological malignancies. J. Hematol. Oncol. 2013, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Brooks, C.L.; Gu, W. p53 Ubiquitination: Mdm2 and Beyond. Mol. Cell 2006, 21, 307–315. [Google Scholar] [CrossRef]
- Tovar, C.; Rosinski, J.; Filipovic, Z.; Higgins, B.; Kolinsky, K.; Hilton, H.; Zhao, X.; Vu, B.T.; Qing, W.; Packman, K.; et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: Implications for therapy. Proc. Natl. Acad. Sci. USA 2006, 103, 1888–1893. [Google Scholar] [CrossRef]
- Nicholson, J.; Neelagandan, K.; Huart, A.-S.; Ball, K.; Molloy, M.; Hupp, T. An iTRAQ Proteomics Screen Reveals the Effects of the MDM2 Binding Ligand Nutlin-3 on Cellular Proteostasis. J. Proteome Res. 2012, 11, 5464–5478. [Google Scholar] [CrossRef] [PubMed]
- Haaland, I.; Opsahl, J.A.; Berven, F.S.; Reikvam, H.; Fredly, H.K.; Haugse, R.; Thiede, B.; McCormack, E.; Lain, S.; Bruserud, Ø.; et al. Molecular mechanisms of nutlin-3 involve acetylation of p53, histones and heat shock proteins in acute myeloid leukemia. Mol. Cancer 2014, 13, 116. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.S. Proteomic Profiling and Target Identification in Lymphoma. In Neoplastic Hematopathology; Springer: Berlin/Heidelberg, Germany, 2010; pp. 573–582. [Google Scholar] [CrossRef]
- Psatha, K.; Kollipara, L.; Voutyraki, C.; Divanach, P.; Sickmann, A.; Rassidakis, G.Z.; Drakos, E.; Aivaliotis, M. Deciphering lymphoma pathogenesis via state-of-the-art mass spectrometry-based quantitative proteomics. J. Chromatogr. B Analyt. Technol. Biomed Life Sci. 2017, 1047, 2–14. [Google Scholar] [CrossRef] [PubMed]
- Drakos, E.; Atsaves, V.; Li, J.; Leventaki, V.; Andreeff, M.; Medeiros, L.J.; Rassidakis, G.Z. Stabilization and activation of p53 downregulates mTOR signaling through AMPK in mantle cell lymphoma. Leukemia 2009, 23, 784–790. [Google Scholar] [CrossRef]
- Drakos, E.; Singh, R.R.; Rassidakis, G.Z.; Schlette, E.; Li, J.; Claret, F.X.; Ford, R.J.; Vega, F.; Medeiros, L.J. Activation of the p53 pathway by the MDM2 inhibitor nutlin-3a overcomes BCL2 overexpression in a preclinical model of diffuse large B-cell lymphoma associated with t(14;18)(q32;q21). Leukemia 2011, 25, 856–867. [Google Scholar] [CrossRef]
- Drakos, E.; Atsaves, V.; Schlette, E.; Li, J.; Papanastasi, I.; Rassidakis, G.Z.; Medeiros, L.J. The therapeutic potential of p53 reactivation by nutlin-3a in ALK+ anaplastic large cell lymphoma with wild-type or mutated p53. Leukemia 2009, 23, 2290–2299. [Google Scholar] [CrossRef]
- Drakos, E.; Thomaides, A.; Medeiros, L.J.; Li, J.; Leventaki, V.; Konopleva, M.; Andreeff, M.; Rassidakis, G.Z. Inhibition of p53-Murine Double Minute 2 Interaction by Nutlin-3A Stabilizes p53 and Induces Cell Cycle Arrest and Apoptosis in Hodgkin Lymphoma. Clin. Cancer Res. 2007, 13, 3380–3387. [Google Scholar] [CrossRef] [PubMed]
- Esposti, M.D.; Hatzinisiriou, I.; McLennan, H.; Ralph, S. Bcl-2 and mitochondrial oxygen radicals. New approaches with reactive oxygen species-sensitive probes. J. Biol. Chem. 1999, 274, 29831–29837. [Google Scholar] [CrossRef] [PubMed]
- Tebbe, A.; Schmidt, A.; Konstantinidis, K.; Falb, M.; Bisle, B.; Klein, C.; Aivaliotis, M.; Kellermann, J.; Siedler, F.; Pfeiffer, F.; et al. Life-style changes of a halophilic archaeon analyzed by quantitative proteomics. Proteomics 2009, 9, 3843–3855. [Google Scholar] [CrossRef]
- Schmidt, A.; Kellermann, J.; Lottspeich, F. A novel strategy for quantitative proteomics using isotope-coded protein labels. Proteomics 2005, 5, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Chahrour, O.; Cobice, D.; Malone, J. Stable isotope labelling methods in mass spectrometry-based quantitative proteomics. J. Pharm. Biomed. Anal. 2015, 113, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Candiano, G.; Bruschi, M.; Musante, L.; Santucci, L.; Ghiggeri, G.M.; Carnemolla, B.; Orecchia, P.; Zardi, L.; Righetti, P.G. Blue silver: A very sensitive colloidal Coomassie G-250 staining for proteome analysis. Electrophoresis 2004, 25, 1327–1333. [Google Scholar] [CrossRef]
- Shevchenko, A.; Wilm, M.; Vorm, O.; Mann, M. Mass Spectrometric Sequencing of Proteins from Silver-Stained Polyacrylamide Gels. Anal. Chem. 1996, 68, 850–858. [Google Scholar] [CrossRef]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
- Aivaliotis, M.; Macek, B.; Gnad, F.; Reichelt, P.; Mann, M.; Oesterhelt, D. Ser/Thr/Tyr Protein Phosphorylation in the Archaeon Halobacterium salinarum—A Representative of the Third Domain of Life. PLoS ONE 2009, 4, e4777. [Google Scholar] [CrossRef]
- UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010, 38, D142–D148. [Google Scholar] [CrossRef]
- Vaudel, M.; Barsnes, H.; Berven, F.S.; Sickmann, A.; Martens, L. SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X!Tandem searches. Proteomics 2011, 11, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Vaudel, M.; Burkhart, J.M.; Zahedi, R.; Oveland, E.; Berven, F.S.; Sickmann, A.; Martens, L.; Barsnes, H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015, 33, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015, 44, D457–D462. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. Gene Ontology Consortium: Going forward. Nucleic Acids Res. 2015, 43, D1049–D1056. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; Von Mering, C.; et al. STRING v9.1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef]
- Franz, M.; Lopes, C.T.; Fong, D.; Kucera, M.; Cheung, M.; Siper, M.C.; Huck, G.; Dong, Y.; Sumer, O.; Bader, G.D. Cytoscape.js 2023 update: A graph theory library for visualization and analysis. Bioinformatics 2023, 39, btad031. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.; Tan, M.; Yang, J.; Inuzuka, H.; Dai, X.; Wu, T.; Liu, J.; Shaik, S.; Chen, G.; Deng, J.; et al. APC(Cdc20) suppresses apoptosis through targeting Bim for ubiquitination and destruction. Dev Cell 2014, 29, 377–391. [Google Scholar] [CrossRef] [PubMed]
- Dusek, R.L.; Attardi, L.D. Desmosomes: New perpetrators in tumour suppression. Nat. Rev. Cancer 2011, 11, 317–323. [Google Scholar] [CrossRef]
- Davis, R.E.; Ngo, V.N.; Lenz, G.; Tolar, P.; Young, R.M.; Romesser, P.B.; Kohlhammer, H.; Lamy, L.; Zhao, H.; Yang, Y.; et al. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 2009, 463, 88–92. [Google Scholar] [CrossRef]
- Widlak, P.; Garrard, W.T. Roles of the Major Apoptotic Nuclease-DNA Fragmentation Factor-in Biology and Disease. Cell. Mol. Life Sci. 2009, 66, 263–274. [Google Scholar] [CrossRef] [PubMed]
- Krivtsov, A.V.; Armstrong, S.A. MLL translocations, histone modifications and leukaemia stem-cell development. Nat. Rev. Cancer 2007, 7, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Arias-Lopez, C.; Lazaro-Trueba, I.; Kerr, P.; Lord, C.J.; Dexter, T.; Iravani, M.; Ashworth, A.; Silva, A. p53 modulates homologous recombination by transcriptional regulation of the RAD51 gene. EMBO Rep. 2006, 7, 219–224. [Google Scholar] [CrossRef]
- Kim, T.W.; Kim, B.; Kim, J.H.; Kang, S.; Park, S.-B.; Jeong, G.; Kang, H.-S.; Kim, S.J. Nuclear-encoded mitochondrial MTO1 and MRPL41 are regulated in an opposite epigenetic mode based on estrogen receptor status in breast cancer. BMC Cancer 2013, 13, 502. [Google Scholar] [CrossRef]
- Vaseva, A.V.; Yallowitz, A.R.; Marchenko, N.D.; Xu, S.; Moll, U.M. Blockade of Hsp90 by 17AAG antagonizes MDMX and synergizes with Nutlin to induce p53-mediated apoptosis in solid tumors. Cell Death Dis. 2011, 2, e156. [Google Scholar] [CrossRef]
- Manfe, V.; Biskup, E.; Rosbjerg, A.; Kamstrup, M.; Skov, A.G.; Lerche, C.M.; Lauenborg, B.T.; Ødum, N.; Gniadecki, R. miR-122 regulates p53/Akt signalling and the chemotherapy-induced apoptosis in cutaneous T-cell lymphoma. PLoS ONE 2012, 7, e29541. [Google Scholar] [CrossRef]
- Lin, Z.; Crockett, D.K.; Jenson, S.D.; Lim, M.S.; Elenitoba-Johnson, K.S.J. Quantitative Proteomic and Transcriptional Analysis of the Response to the p38 Mitogen-activated Protein Kinase Inhibitor SB203580 in Transformed Follicular Lymphoma Cells. Mol. Cell. Proteom. 2004, 3, 820–833. [Google Scholar] [CrossRef]
- Díez, P.; Droste, C.; Dégano, R.M.; González-Muñoz, M.; Ibarrola, N.; Pérez-Andrés, M.; Garin-Muga, A.; Segura, V.; Marko-Varga, G.; LaBaer, J.; et al. Integration of Proteomics and Transcriptomics Data Sets for the Analysis of a Lymphoma B-Cell Line in the Context of the Chromosome-Centric Human Proteome Project. J. Proteome Res. 2015, 14, 3530–3540. [Google Scholar] [CrossRef] [PubMed]
- Weinkauf, M.; Christopeit, M.; Hiddemann, W.; Dreyling, M. Proteome- and microarray-based expression analysis of lymphoma cell lines identifies a p53-centered cluster of differentially expressed proteins in mantle cell and follicular lymphoma. Electrophoresis 2007, 28, 4416–4426. [Google Scholar] [CrossRef] [PubMed]
- Manfé, V.; Biskup, E.; Johansen, P.; Kamstrup, M.R.; Krejsgaard, T.F.; Morling, N.; Wulf, H.C.; Gniadecki, R. MDM2 Inhibitor Nutlin-3a Induces Apoptosis and Senescence in Cutaneous T-Cell Lymphoma: Role of p53. J. Investig. Dermatol. 2012, 132, 1487–1496. [Google Scholar] [CrossRef]
- Patterson, D.M.; Gao, D.; Trahan, D.N.; Johnson, B.A.; Ludwig, A.; Barbieri, E.; Chen, Z.; Diaz-Miron, J.; Vassilev, L.; Shohet, J.M.; et al. Effect of MDM2 and vascular endothelial growth factor inhibition on tumor angiogenesis and metastasis in neuroblastoma. Angiogenesis 2011, 14, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.P.; Bartek, J. The DNA-damage response in human biology and disease. Nature 2009, 461, 1071–1078. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Rigatti, M.J.; Belinsky, G.S.; Godman, C.A.; Giardina, C. Faculty Opinions recommendation of DNA damage response to the Mdm2 inhibitor nutlin-3. Biochem. Pharmacol. 2010, 79, 565–574. [Google Scholar] [CrossRef]
- Valentine, J.M.; Kumar, S.; Moumen, A. A p53-independent role for the MDM2 antagonist Nutlin-3 in DNA damage response initiation. BMC Cancer 2011, 11, 79. [Google Scholar] [CrossRef] [PubMed]
- Linke, S.; Sengupta, S.; Khabie, N.; Jeffries, B.A.; Buchhop, S.; Miska, S.; Henning, W.; Pedeux, R.; Wang, X.W.; Hofseth, L.J.; et al. p53 interacts with hRAD51 and hRAD54, and directly modulates homologous recombination. Cancer Res. 2003, 63, 2596–2605. [Google Scholar]
- Romanova, L.Y.; Willers, H.; Blagosklonny, M.V.; Powell, S.N. The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene 2004, 23, 9025–9033. [Google Scholar] [CrossRef] [PubMed]
- Aylon, Y.; Liefshitz, B.; Kupiec, M. The CDK regulates repair of double-strand breaks by homologous recombination during the cell cycle. EMBO J. 2004, 23, 4868–4875. [Google Scholar] [CrossRef]
- Kumamoto, K.; Spillare, E.A.; Fujita, K.; Horikawa, I.; Yamashita, T.; Appella, E.; Nagashima, M.; Takenoshita, S.; Yokota, J.; Harris, C.C.; et al. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008, 68, 3193–3203. [Google Scholar] [CrossRef]
- Laptenko, O.; Beckerman, R.; Freulich, E.; Prives, C. p53 binding to nucleosomes within the p21 promoter in vivo leads to nucleosome loss and transcriptional activation. Proc. Natl. Acad. Sci. USA 2011, 108, 10385–10390. [Google Scholar] [CrossRef]
- Sidhu, H.; Capalash, N. UHRF1: The key regulator of epigenetics and molecular target for cancer therapeutics. Tumor Biol. 2017, 39, 1010428317692205. [Google Scholar] [CrossRef]
- Robaina, M.C.; Mazzoccoli, L.; Arruda, V.O.; Reis, F.R.D.S.; Apa, A.G.; de Rezende, L.M.M.; Klumb, C.E. Deregulation of DNMT1, DNMT3B and miR-29s in Burkitt lymphoma suggests novel contribution for disease pathogenesis. Exp. Mol. Pathol. 2015, 98, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Radulović, V.; de Haan, G.; Klauke, K. Polycomb-group proteins in hematopoietic stem cell regulation and hematopoietic neoplasms. Leukemia 2013, 27, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Scorrano, L.; Petronilli, V.; Colonna, R.; di Lisa, F.; Bernardi, P. Interactions of chloromethyltetramethylrosamine (Mitotracker Orange) with isolated mitochondria and intact cells. Ann. N. Y. Acad. Sci. 1999, 893, 391–395. [Google Scholar] [CrossRef] [PubMed]
- Bolisetty, S.; Jaimes, E.A. Mitochondria and Reactive Oxygen Species: Physiology and Pathophysiology. Int. J. Mol. Sci. 2013, 14, 6306–6344. [Google Scholar] [CrossRef]
- Kruiswijk, F.; Labuschagne, C.F.; Vousden, K.H. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 2015, 16, 393–405. [Google Scholar] [CrossRef]
- Duan, L.; Perez, R.E.; Davaadelger, B.; Dedkova, E.N.; Blatter, L.A.; Maki, C.G. p53-regulated autophagy is controlled by glycolysis and determines cell fate. Oncotarget 2015, 6, 23135–23156. [Google Scholar] [CrossRef]
- Duan, L.; Perez, R.E.; Chen, L.; Blatter, L.A.; Maki, C.G. p53 promotes AKT and SP1-dependent metabolism through the pentose phosphate pathway that inhibits apoptosis in response to Nutlin-3a. J. Mol. Cell Biol. 2018, 10, 331–340. [Google Scholar] [CrossRef]
- Westin, J.R. Status of PI3K/Akt/mTOR Pathway Inhibitors in Lymphoma. Clin. Lymphoma Myeloma Leuk. 2014, 14, 335–342. [Google Scholar] [CrossRef]
- Efeyan, A.; Sabatini, D.M. mTOR and cancer: Many loops in one pathway. Curr. Opin. Cell Biol. 2010, 22, 169–176. [Google Scholar] [CrossRef]
- Budanov, A.V.; Karin, M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 2008, 134, 451–460. [Google Scholar] [CrossRef]
- Constantinou, C.; Clemens, M.J. Regulation of the phosphorylation and integrity of protein synthesis initiation factor eIF4GI and the translational repressor 4E-BP1 by p53. Oncogene 2005, 24, 4839–4850. [Google Scholar] [CrossRef]
- Brady, C.A.; Jiang, D.; Mello, S.S.; Johnson, T.M.; Jarvis, L.A.; Kozak, M.M.; Broz, D.K.; Basak, S.; Park, E.J.; McLaughlin, M.; et al. Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 2011, 145, 571–583. [Google Scholar] [CrossRef]
- Sung, C.O.; Kim, S.C.; Karnan, S.; Karube, K.; Shin, H.J.; Nam, D.-H.; Suh, Y.-L.; Kim, S.-H.; Kim, J.Y.; Kim, W.S.; et al. Genomic profiling combined with gene expression profiling in primary central nervous system lymphoma. Blood 2011, 117, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Chipoy, C.; Brounais, B.; Trichet, V.; Battaglia, S.; Berreur, M.; Oliver, L.; Juin, P.; Rédini, F.; Heymann, D.; Blanchard, F. Sensitization of osteosarcoma cells to apoptosis by oncostatin M depends on STAT5 and p53. Oncogene 2007, 26, 6653–6664. [Google Scholar] [CrossRef]
- Nencioni, A.; Cea, M.; Montecucco, F.; Longo, V.D.; Patrone, F.; Carella, A.M.; Holyoake, T.L.; Helgason, G.V. Autophagy in blood cancers: Biological role and therapeutic implications. Haematologica 2013, 98, 1335–1343. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Abrams, J. p53: The Janus of autophagy? Nat. Cell Biol. 2008, 10, 637–639. [Google Scholar] [CrossRef]
- Leu, J.I.-J.; Pimkina, J.; Frank, A.; Murphy, M.E.; George, D.L. A Small Molecule Inhibitor of Inducible Heat Shock Protein 70. Mol. Cell 2009, 36, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Rosich, L.; Xargay-Torrent, S.; López-Guerra, M.; Campo, E.; Colomer, D.; Roué, G. Counteracting autophagy overcomes resistance to everolimus in mantle cell lymphoma. Clin. Cancer Res. 2012, 18, 5278–5289. [Google Scholar] [CrossRef]
- Mc Cormack, E.; Haaland, I.; Venås, G.; Forthun, R.B.; Huseby, S.; Gausdal, G.; Knappskog, S.; Micklem, D.R.; Lorens, J.B.; Bruserud, Ø.; et al. Synergistic induction of p53 mediated apoptosis by valproic acid and nutlin-3 in acute myeloid leukemia. Leukemia 2012, 26, 910–917. [Google Scholar] [CrossRef]
- Borthakur, G.; Duvvuri, S.; Ruvolo, V.; Tripathi, D.; Piya, S.; Burks, J.; Jacamo, R.; Kojima, K.; Ruvolo, P.; Fueyo-Margareto, J.; et al. MDM2 Inhibitor, Nutlin 3a, Induces p53 Dependent Autophagy in Acute Leukemia by AMP Kinase Activation. PLoS ONE 2015, 10, e0139254. [Google Scholar] [CrossRef]
- Lee, J.; Giordano, S.; Zhang, J. Autophagy, mitochondria and oxidative stress: Cross-talk and redox signalling. Biochem. J. 2012, 441, 523–540. [Google Scholar] [CrossRef]
- Chen, Y.; Azad, M.B.; Gibson, S.B. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ. 2009, 16, 1040–1052. [Google Scholar] [CrossRef] [PubMed]
- Santón, A.; García-Cosío, M.; Cristóbal, E.; Pascual, A.; Muriel, A.; García-Laraña, J. Expression of heat shock proteins in classical Hodgkin lymphoma: Correlation with apoptotic pathways and prognostic significance. Histopathology 2011, 58, 1072–1080. [Google Scholar] [CrossRef] [PubMed]
- Valbuena, J.R.; Rassidakis, G.Z.; Lin, P.; Atwell, C.; Georgakis, G.V.; Younes, A.; Jones, D.; Medeiros, L.J. Expression of heat-shock protein-90 in non-Hodgkin’s lymphomas. Mod. Pathol. 2005, 18, 1343–1349. [Google Scholar] [CrossRef]
- Calderwood, S.K.; Khaleque, A.; Sawyer, D.B.; Ciocca, D.R. Heat shock proteins in cancer: Chaperones of tumorigenesis. Trends Biochem. Sci. 2006, 31, 164–172. [Google Scholar] [CrossRef] [PubMed]
- Dai, C.; Whitesell, L.; Rogers, A.B.; Lindquist, S. Heat Shock Factor 1 Is a Powerful Multifaceted Modifier of Carcinogenesis. Cell 2007, 130, 1005–1018. [Google Scholar] [CrossRef] [PubMed]
- Min, J.-N.; Huang, L.; Zimonjic, D.B.; Moskophidis, D.; Mivechi, N.F. Selective suppression of lymphomas by functional loss of Hsf1 in a p53-deficient mouse model for spontaneous tumors. Oncogene 2007, 26, 5086–5097. [Google Scholar] [CrossRef]
- Logan, I.R.; McNeill, H.V.; Cook, S.; Lu, X.; Meek, D.W.; Fuller-Pace, F.V.; Lunec, J.; Robson, C.N. Heat shock factor-1 modulates p53 activity in the transcriptional response to DNA damage. Nucleic Acids Res. 2009, 37, 2962–2973. [Google Scholar] [CrossRef]
- Chauhan, D.; Li, G.; Hideshima, T.; Podar, K.; Mitsiades, C.; Mitsiades, N.; Catley, L.; Tai, Y.T.; Hayashi, T.; Shringarpure, R.; et al. Hsp27 inhibits release of mitochondrial protein Smac in multiple myeloma cells and confers dexamethasone resistance. Blood 2003, 102, 3379–3386. [Google Scholar] [CrossRef]
- Palani, C.D.; Beck, J.F.; Sonnemann, J. Histone deacetylase inhibitors enhance the anticancer activity of nutlin-3 and induce p53 hyperacetylation and downregulation of MDM2 and MDM4 gene expression. Investig. New Drugs 2012, 30, 25–36. [Google Scholar] [CrossRef]
- Yang, Y.; Rao, R.; Shen, J.; Tang, Y.; Fiskus, W.; Nechtman, J.; Atadja, P.; Bhalla, K. Role of Acetylation and Extracellular Location of Heat Shock Protein 90α in Tumor Cell Invasion. Cancer Res. 2008, 68, 4833–4842. [Google Scholar] [CrossRef] [PubMed]
- Georgakis, G.V.; Li, Y.; Rassidakis, G.Z.; Medeiros, L.J.; Younes, A. The HSP90 inhibitor 17-AAG synergizes with doxorubicin and U0126 in anaplastic large cell lymphoma irrespective of ALK expression. Exp. Hematol. 2006, 34, 1670–1679. [Google Scholar] [CrossRef] [PubMed]
- Georgakis, G.V.; Li, Y.; Rassidakis, G.Z.; Martinez-Valdez, H.; Medeiros, L.J.; Younes, A. Inhibition of heat shock protein 90 function by 17-allylamino-17-demethoxy-geldanamycin in Hodgkin’s lymphoma cells down-regulates Akt kinase, dephosphorylates extracellular signal-regulated kinase, and induces cell cycle arrest and cell death. Clin. Cancer Res. 2006, 12, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Georgakis, G.V.; Li, Y.; Younes, A. The heat shock protein 90 inhibitor 17-AAG induces cell cycle arrest and apoptosis in mantle cell lymphoma cell lines by depleting cyclin D1, Akt, Bid and activating caspase 9. Br. J. Haematol. 2006, 135, 68–71. [Google Scholar] [CrossRef]
- Cheng, T.H.; Cohen, S.N. Human MDM2 isoforms translated differentially on constitutive versus p53-regulated transcripts have distinct functions in the p53/MDM2 and TSG101/MDM2 feedback control loops. Mol. Cell. Biol. 2007, 27, 111–119. [Google Scholar] [CrossRef]
- Chou, T.-C. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010, 70, 440–446. [Google Scholar] [CrossRef]
- Kollipara, L.; Zahedi, R.P. Protein carbamylation: In vivo modification or in vitro artefact? Proteomics 2013, 13, 941–944. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, J.M.; Schumbrutzki, C.; Wortelkamp, S.; Sickmann, A.; Zahedi, R.P. Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. J. Proteom. 2012, 75, 1454–1462. [Google Scholar] [CrossRef] [PubMed]
- Aivaliotis, M.; Gevaert, K.; Falb, M.; Tebbe, A.; Konstantinidis, K.; Bisle, B.; Klein, C.; Martens, L.; Staes, A.; Timmerman, E.; et al. Large-Scale Identification of N-Terminal Peptides in the Halophilic Archaea Halobacterium salinarum and Natronomonas pharaonis. J. Proteome Res. 2007, 6, 2195–2204. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Ryder, U.; Lamond, A.I.; Mann, M. Large-Scale Proteomic Analysis of the Human Spliceosome. Genome Res. 2002, 12, 1231–1245. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. 1D and 2D annotation enrichment: A statistical method integrating quantitative proteomics with complementary high-throughput data. BMC Bioinform. 2012, 13, S12. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Smoot, M.E.; Ono, K.; Ruscheinski, J.; Wang, P.-L.; Ideker, T. Cytoscape 2.8: New features for data integration and network visualization. Bioinformatics 2011, 27, 431–432. [Google Scholar] [CrossRef] [PubMed]
- Chou, T.-C. Theoretical Basis, Experimental Design, and Computerized Simulation of Synergism and Antagonism in Drug Combination Studies. Pharmacol. Rev. 2006, 58, 621–681. [Google Scholar] [CrossRef]
- Martens, L.; Hermjakob, H.; Jones, P.; Adamski, M.; Taylor, C.; States, D.; Gevaert, K.; Vandekerckhove, J.; Apweiler, R. PRIDE: The proteomics identifications database. Proteomics 2005, 5, 3537–3545. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Psatha, K.; Kollipara, L.; Drakos, E.; Deligianni, E.; Brintakis, K.; Patsouris, E.; Sickmann, A.; Rassidakis, G.Z.; Aivaliotis, M. Interruption of p53-MDM2 Interaction by Nutlin-3a in Human Lymphoma Cell Models Initiates a Cell-Dependent Global Effect on Transcriptome and Proteome Level. Cancers 2023, 15, 3903. https://doi.org/10.3390/cancers15153903
Psatha K, Kollipara L, Drakos E, Deligianni E, Brintakis K, Patsouris E, Sickmann A, Rassidakis GZ, Aivaliotis M. Interruption of p53-MDM2 Interaction by Nutlin-3a in Human Lymphoma Cell Models Initiates a Cell-Dependent Global Effect on Transcriptome and Proteome Level. Cancers. 2023; 15(15):3903. https://doi.org/10.3390/cancers15153903
Chicago/Turabian StylePsatha, Konstantina, Laxmikanth Kollipara, Elias Drakos, Elena Deligianni, Konstantinos Brintakis, Eustratios Patsouris, Albert Sickmann, George Z. Rassidakis, and Michalis Aivaliotis. 2023. "Interruption of p53-MDM2 Interaction by Nutlin-3a in Human Lymphoma Cell Models Initiates a Cell-Dependent Global Effect on Transcriptome and Proteome Level" Cancers 15, no. 15: 3903. https://doi.org/10.3390/cancers15153903
APA StylePsatha, K., Kollipara, L., Drakos, E., Deligianni, E., Brintakis, K., Patsouris, E., Sickmann, A., Rassidakis, G. Z., & Aivaliotis, M. (2023). Interruption of p53-MDM2 Interaction by Nutlin-3a in Human Lymphoma Cell Models Initiates a Cell-Dependent Global Effect on Transcriptome and Proteome Level. Cancers, 15(15), 3903. https://doi.org/10.3390/cancers15153903









