Machine Learning Identifies a Signature of Nine Exosomal RNAs That Predicts Hepatocellular Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
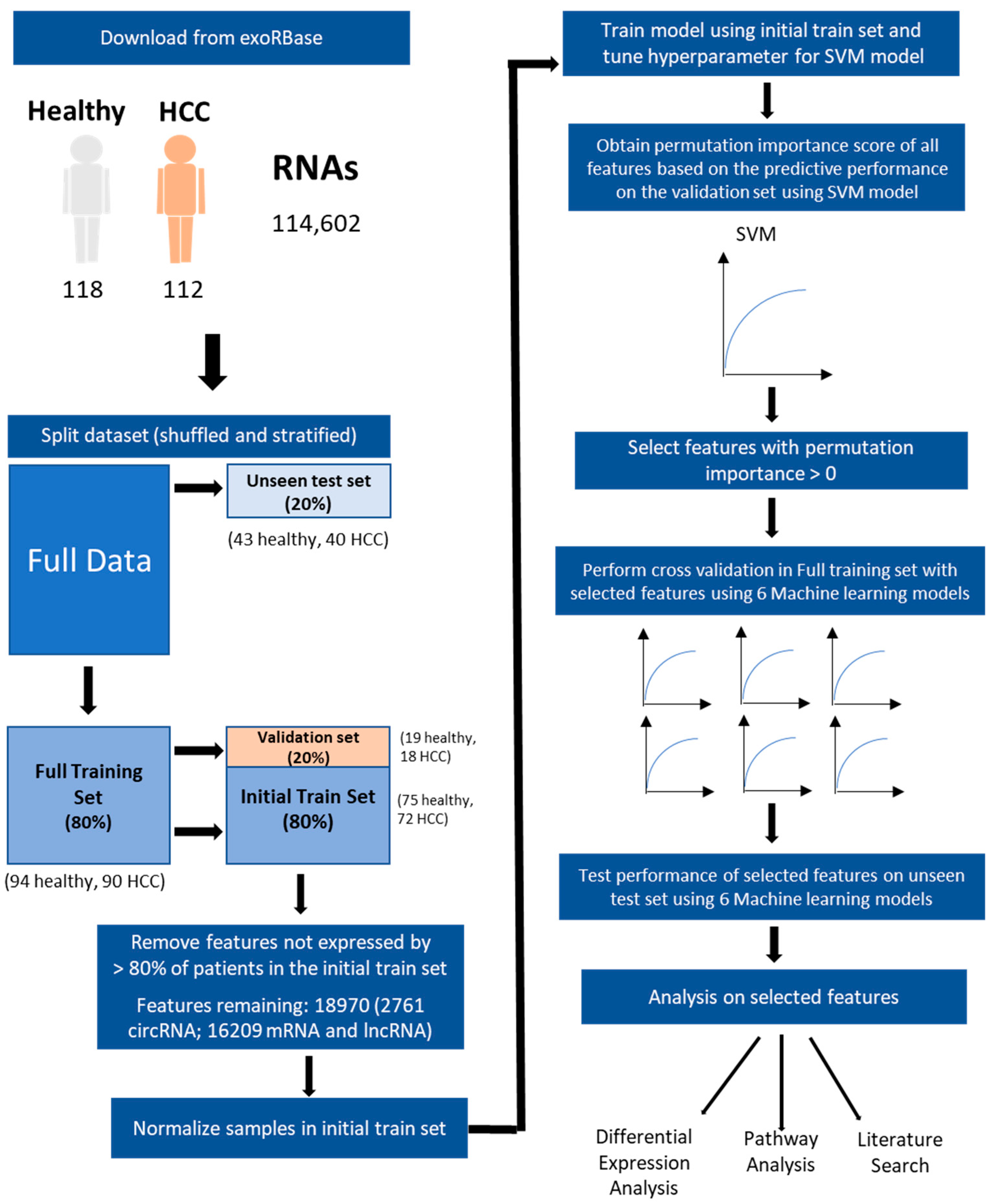
2. Materials and Methods
2.1. Exosomal RNA Expression Data
2.2. Splitting and Processing of Data
2.3. Model Training and Feature Selection by Permutation Importance
2.4. Evaluating the Predictiveness of Selected Features
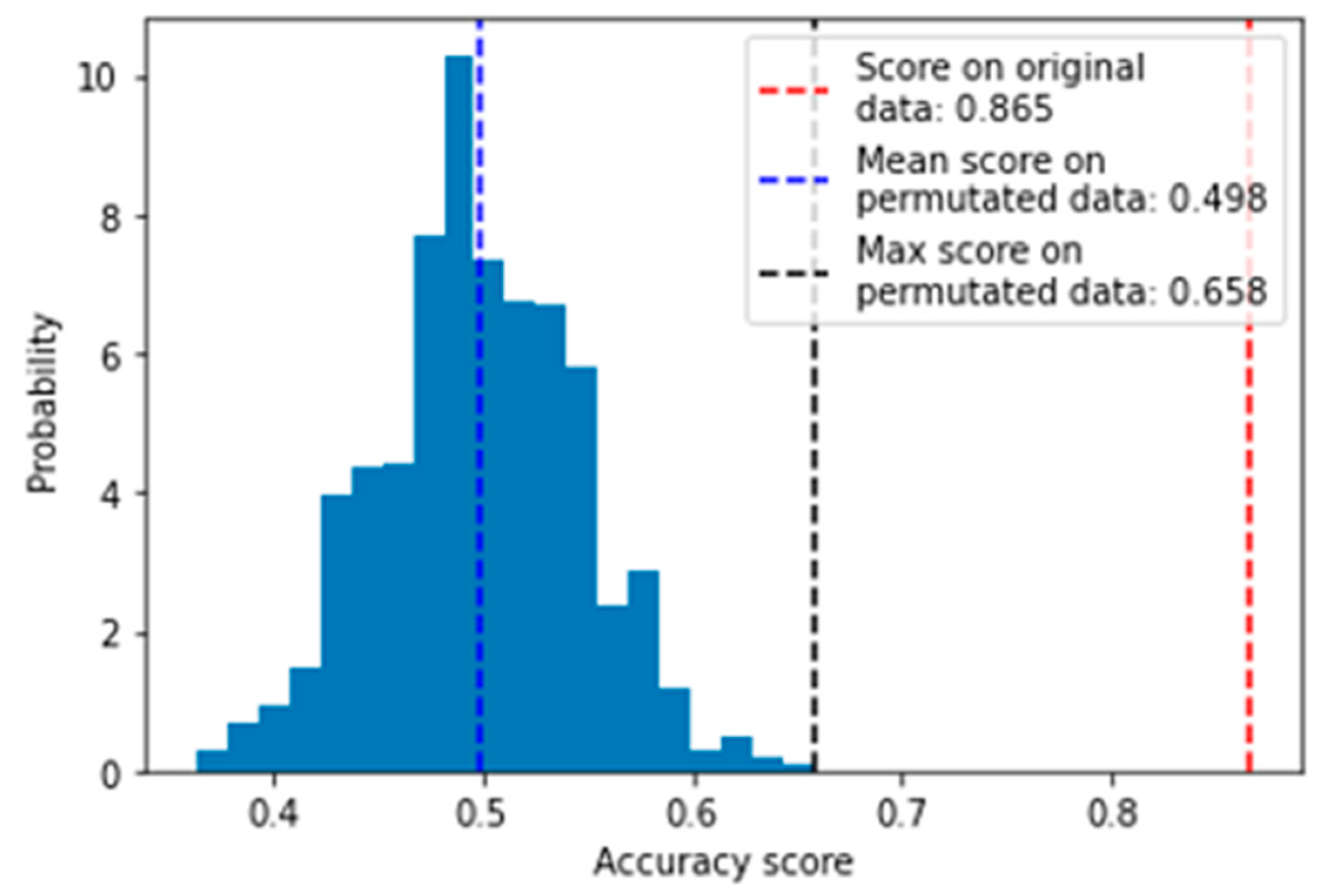
2.4.1. Evaluation with Permutation Test
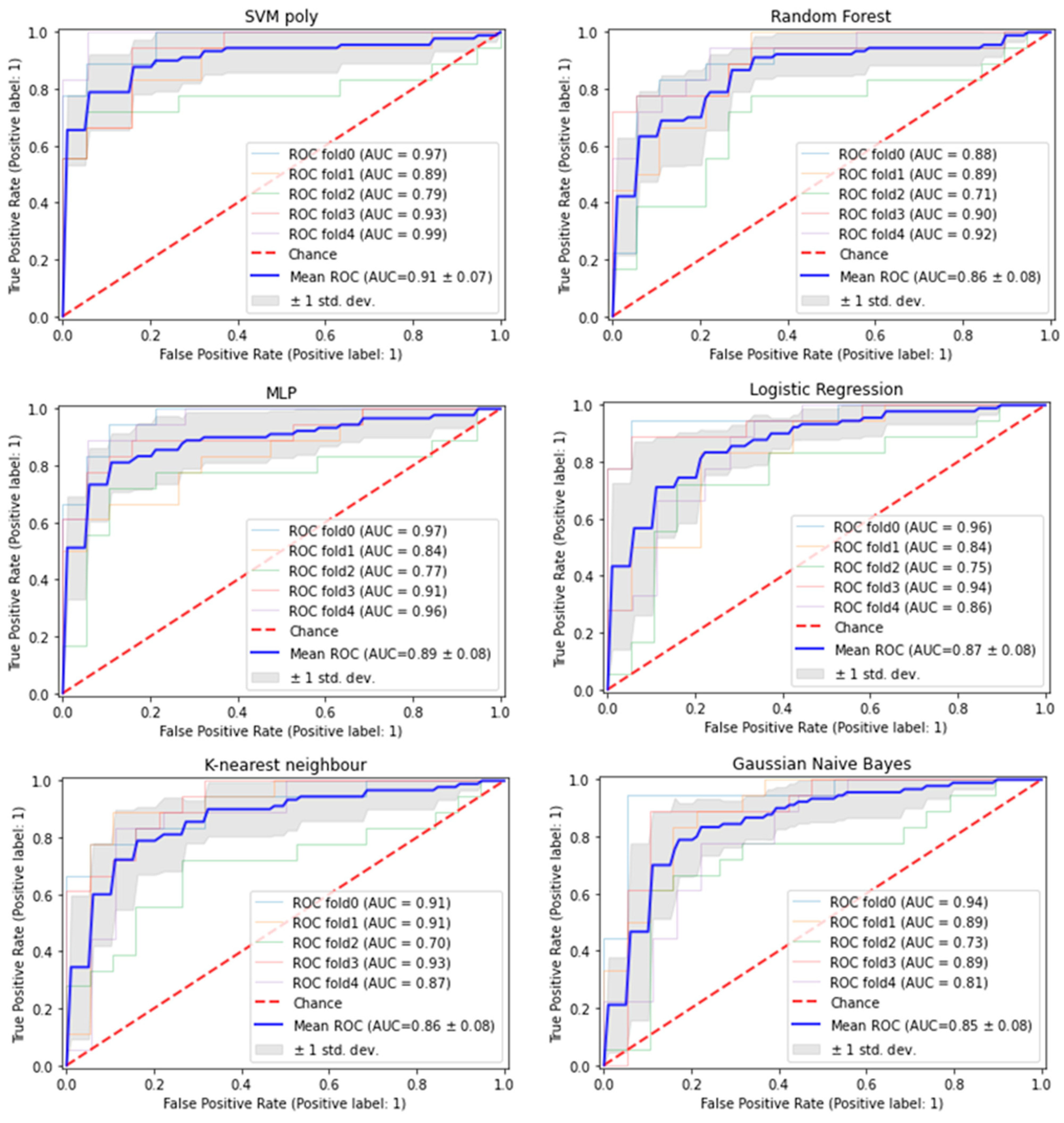
2.4.2. Evaluation across 6 Different Models
2.5. Analysing Differential Gene Expression in Exosomal RNA Expression Data
2.6. Validation of Differential Expression and Predictive Performance of Potential Predictors in Tissues Samples
2.7. Pathway Enrichment Analysis
2.8. Text Mining Analysis
3. Results
3.1. Nine Exosomal RNA Signatures Selected by Machine Learning Approach Have Good Predictive Performance in Predicting HCC
3.2. The Nine ML Selected Exosomal RNA Signatures Performs Better than Top Nine Differentially Expressed RNAs
3.3. Majority of the Exosomal RNA Signatures Are also Differentially Expressed in Tumour Tissues as Compared to Adjacent Non-Tumourous Tissues
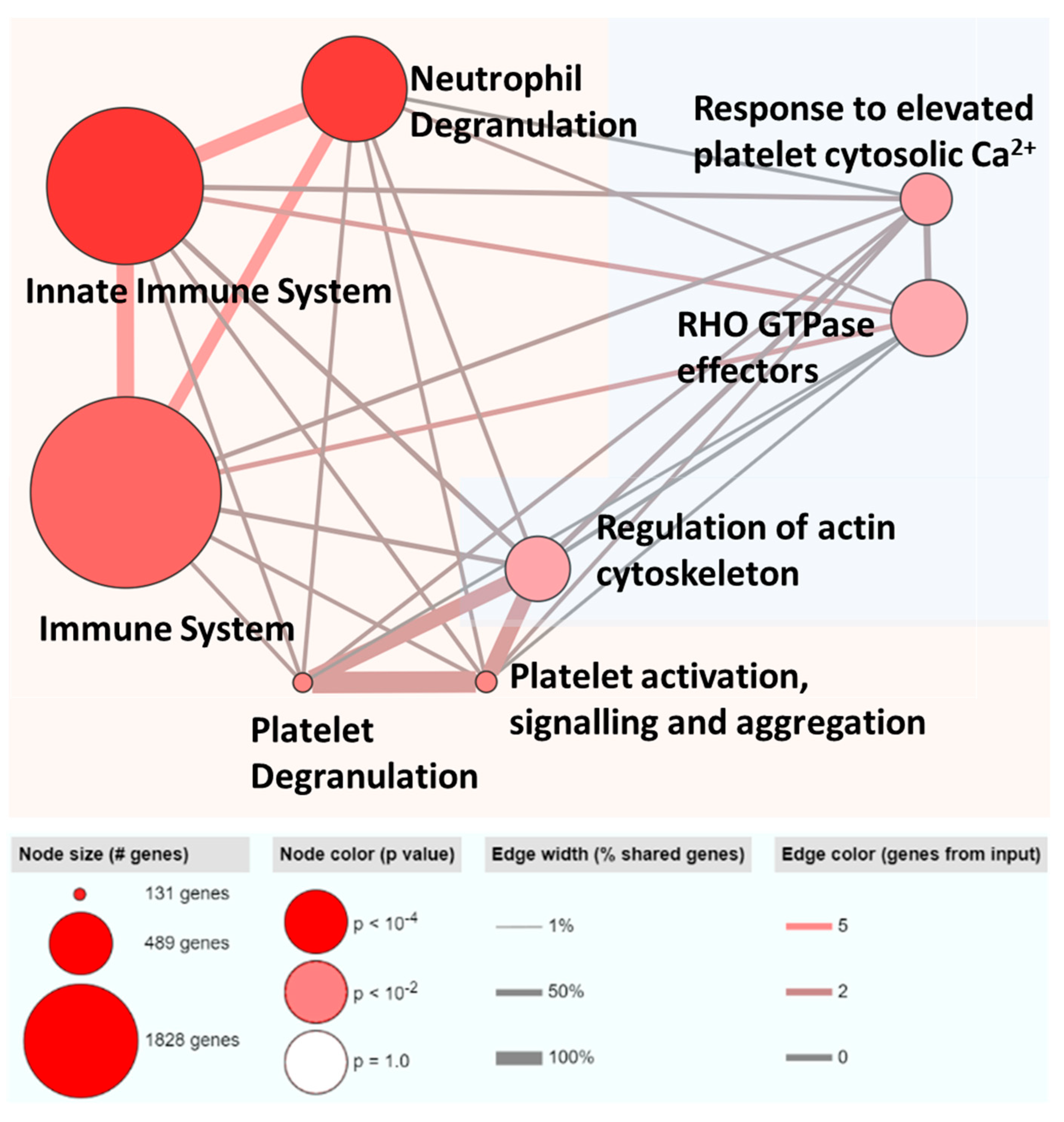
3.4. ML-Selected Exosomal RNA Signatures Are Mainly Implicated in Immune Pathways and Majority Are Previously Reported to Be Associated with HCC
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Llovet, J.M.; Kelley, R.K.; Villanueva, A.; Singal, A.G.; Pikarsky, E.; Roayaie, S.; Lencioni, R.; Koike, K.; Zucman-Rossi, J.; Finn, R.S. Hepatocellular carcinoma. Nat. Rev. Dis. Primers 2021, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Rumgay, H.; Arnold, M.; Ferlay, J.; Lesi, O.; Cabasag, C.J.; Vignat, J.; Laversanne, M.; McGlynn, K.A.; Soerjomataram, I. Global burden of primary liver cancer in 2020 and predictions to 2040. J. Hepatol. 2022, 77, 1598–1606. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, N.D.; Pillai, A.; Singal, A.G. Update on the Diagnosis and Treatment of Hepatocellular Carcinoma. Gastroenterol. Hepatol. 2020, 16, 506–516. [Google Scholar]
- Wang, W.; Wei, C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 2020, 7, 308–319. [Google Scholar] [CrossRef]
- Hanif, H.; Ali, M.J.; Susheela, A.T.; Khan, I.W.; Luna-Cuadros, M.A.; Khan, M.M.; Lau, D.T. Update on the applications and limitations of alpha-fetoprotein for hepatocellular carcinoma. World J. Gastroenterol. 2022, 28, 216–229. [Google Scholar] [CrossRef]
- Lee, C.W.; Tsai, H.I.; Lee, W.C.; Huang, S.W.; Lin, C.Y.; Hsieh, Y.C.; Kuo, T.; Chen, C.W.; Yu, M.C. Normal Alpha-Fetoprotein Hepatocellular Carcinoma: Are They Really Normal? J. Clin. Med. 2019, 8, 1736. [Google Scholar] [CrossRef]
- Adigun, O.O.; Yarrarapu, S.N.S.; Khetarpal, S. Alpha Fetoprotein; StatPearls: Treasure Island, FL, USA, 2022. [Google Scholar]
- Atiq, O.; Tiro, J.; Yopp, A.C.; Muffler, A.; Marrero, J.A.; Parikh, N.D.; Murphy, C.; McCallister, K.; Singal, A.G. An assessment of benefits and harms of hepatocellular carcinoma surveillance in patients with cirrhosis. Hepatology 2017, 65, 1196–1205. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, G.; Zhang, P.; Zhang, J.; Li, X.; Gan, D.; Cao, X.; Han, M.; Du, H.; Ye, Y. The threshold of alpha-fetoprotein (AFP) for the diagnosis of hepatocellular carcinoma: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0228857. [Google Scholar] [CrossRef]
- Chanteloup, G.; Cordonnier, M.; Isambert, N.; Bertaut, A.; Marcion, G.; Garrido, C.; Gobbo, J. Membrane-bound exosomal HSP70 as a biomarker for detection and monitoring of malignant solid tumours: A pilot study. Pilot Feasibility Stud. 2020, 6, 35. [Google Scholar] [CrossRef]
- Makler, A.; Asghar, W. Exosomal biomarkers for cancer diagnosis and patient monitoring. Expert. Rev. Mol. Diagn. 2020, 20, 387–400. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef]
- Wen, S.W.; Lima, L.G.; Lobb, R.J.; Norris, E.L.; Hastie, M.L.; Krumeich, S.; Moller, A. Breast Cancer-Derived Exosomes Reflect the Cell-of-Origin Phenotype. Proteomics 2019, 19, e1800180. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell. Biosci. 2019, 9, 19. [Google Scholar] [CrossRef]
- Cui, X.; Fu, Q.; Wang, X.; Xia, P.; Cui, X.; Bai, X.; Lu, Z. Molecular mechanisms and clinical applications of exosomes in prostate cancer. Biomark. Res. 2022, 10, 56. [Google Scholar] [CrossRef]
- Desdin-Mico, G.; Mittelbrunn, M. Role of exosomes in the protection of cellular homeostasis. Cell. Adh Migr. 2017, 11, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Verrilli, M.A.; Picou, F.; Court, F.A. Schwann cell-derived exosomes enhance axonal regeneration in the peripheral nervous system. Glia 2013, 61, 1795–1806. [Google Scholar] [CrossRef]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef]
- Abd Elmageed, Z.Y.; Yang, Y.; Thomas, R.; Ranjan, M.; Mondal, D.; Moroz, K.; Fang, Z.; Rezk, B.M.; Moparty, K.; Sikka, S.C.; et al. Neoplastic reprogramming of patient-derived adipose stem cells by prostate cancer cell-associated exosomes. Stem Cells 2014, 32, 983–997. [Google Scholar] [CrossRef] [PubMed]
- Ge, Q.; Zhou, Y.; Lu, J.; Bai, Y.; Xie, X.; Lu, Z. miRNA in plasma exosome is stable under different storage conditions. Molecules 2014, 19, 1568–1575. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, C.; Zhang, P.; Guo, G.; Jiang, T.; Zhao, X.; Jiang, J.; Huang, X.; Tong, H.; Tian, Y. Serum exosomal microRNAs combined with alpha-fetoprotein as diagnostic markers of hepatocellular carcinoma. Cancer Med. 2018, 7, 1670–1679. [Google Scholar] [CrossRef]
- Sohn, W.; Kim, J.; Kang, S.H.; Yang, S.R.; Cho, J.Y.; Cho, H.C.; Shim, S.G.; Paik, Y.H. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp. Mol. Med. 2015, 47, e184. [Google Scholar] [CrossRef] [PubMed]
- Bzdok, D.; Altman, N.; Krzywinski, M. Statistics versus machine learning. Nat. Methods 2018, 15, 233–234. [Google Scholar] [CrossRef] [PubMed]
- Emmert-Streib, F.; Yang, Z.; Feng, H.; Tripathi, S.; Dehmer, M. An Introductory Review of Deep Learning for Prediction Models With Big Data. Front. Artif. Intell. 2020, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Vadapalli, S.; Abdelhalim, H.; Zeeshan, S.; Ahmed, Z. Artificial intelligence and machine learning approaches using gene expression and variant data for personalized medicine. Brief. Bioinform. 2022, 23, bbac191. [Google Scholar] [CrossRef] [PubMed]
- Andrades, R.; Recamonde-Mendoza, M. Machine learning methods for prediction of cancer driver genes: A survey paper. Brief. Bioinform. 2022, 23, bbac062. [Google Scholar] [CrossRef]
- Tan, P.-N.; Steinbach, M.; Kumar, V. Introduction to Data Mining; Pearson Education India: Boston, MA, USA, 2016. [Google Scholar]
- Cervantes, J.; Garcia-Lamont, F.; Rodríguez-Mazahua, L.; Lopez, A. A comprehensive survey on support vector machine classification: Applications, challenges and trends. Neurocomputing 2020, 408, 189–215. [Google Scholar] [CrossRef]
- Cervantes, J.; García Lamont, F.; López-Chau, A.; Rodríguez Mazahua, L.; Sergio Ruíz, J. Data selection based on decision tree for SVM classification on large data sets. Appl. Soft Comput. 2015, 37, 787–798. [Google Scholar] [CrossRef]
- Raheja, J.L.; Mishra, A.; Chaudhary, A. Indian sign language recognition using SVM. Pattern Recognit. Image Anal. 2016, 26, 434–441. [Google Scholar] [CrossRef]
- Bing-Yu, S.; De-Shuang, H.; Hai-Tao, F. Lidar signal denoising using least-squares support vector machine. IEEE Signal Process. Lett. 2005, 12, 101–104. [Google Scholar] [CrossRef]
- Liang, X.; Zhu, L.; Huang, D.-S. Multi-task ranking SVM for image cosegmentation. Neurocomputing 2017, 247, 126–136. [Google Scholar] [CrossRef]
- Chen, P.; Wang, B.; Wong, H.S.; Huang, D.S. Prediction of protein B-factors using multi-class bounded SVM. Protein Pept. Lett. 2007, 14, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Bhowmik, T.K.; Ghanty, P.; Roy, A.; Parui, S.K. SVM-based hierarchical architectures for handwritten Bangla character recognition. Int. Journal. Doc. Anal. Recognit. (IJDAR) 2009, 12, 97–108. [Google Scholar] [CrossRef]
- Zhu, K.; Tao, Q.; Yan, J.; Lang, Z.; Li, X.; Li, Y.; Fan, C.; Yu, Z. Machine learning identifies exosome features related to hepatocellular carcinoma. Front. Cell. Dev. Biol. 2022, 10, 1020415. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, S.; Xi, X.; Zhang, M.; Liu, X.; Tang, W.; Cai, P.; Xing, S.; Bao, P.; Jin, Y.; et al. Integrative analysis of long extracellular RNAs reveals a detection panel of noncoding RNAs for liver cancer. Theranostics 2021, 11, 181–193. [Google Scholar] [CrossRef]
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018, 46, D106–D112. [Google Scholar] [CrossRef]
- Lai, H.; Li, Y.; Zhang, H.; Hu, J.; Liao, J.; Su, Y.; Li, Q.; Chen, B.; Li, C.; Wang, Z.; et al. exoRBase 2.0: An atlas of mRNA, lncRNA and circRNA in extracellular vesicles from human biofluids. Nucleic Acids Res. 2022, 50, D118–D128. [Google Scholar] [CrossRef]
- Abraham, A.; Pedregosa, F.; Eickenberg, M.; Gervais, P.; Mueller, A.; Kossaifi, J.; Gramfort, A.; Thirion, B.; Varoquaux, G. Machine learning for neuroimaging with scikit-learn. Front. Neuroinform. 2014, 8, 14. [Google Scholar] [CrossRef]
- Shahriyari, L. Effect of normalization methods on the performance of supervised learning algorithms applied to HTSeq-FPKM-UQ data sets: 7SK RNA expression as a predictor of survival in patients with colon adenocarcinoma. Brief. Bioinform. 2019, 20, 985–994. [Google Scholar] [CrossRef]
- Kamburov, A.; Wierling, C.; Lehrach, H.; Herwig, R. ConsensusPathDB-a database for integrating human functional interaction networks. Nucleic Acids Res. 2009, 37, D623–D628. [Google Scholar] [CrossRef]
- Kamburov, A.; Pentchev, K.; Galicka, H.; Wierling, C.; Lehrach, H.; Herwig, R. ConsensusPathDB: Toward a more complete picture of cell biology. Nucleic Acids Res. 2011, 39, D712–D717. [Google Scholar] [CrossRef]
- Croft, D.; O’Kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B.; et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2011, 39, D691–D697. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, D109–D114. [Google Scholar] [CrossRef]
- Cock, P.J.; Antao, T.; Chang, J.T.; Chapman, B.A.; Cox, C.J.; Dalke, A.; Friedberg, I.; Hamelryck, T.; Kauff, F.; Wilczynski, B.; et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 2009, 25, 1422–1423. [Google Scholar] [CrossRef]
- Hardy-Sosa, A.; Leon-Arcia, K.; Llibre-Guerra, J.J.; Berlanga-Acosta, J.; Baez, S.C.; Guillen-Nieto, G.; Valdes-Sosa, P.A. Diagnostic Accuracy of Blood-Based Biomarker Panels: A Systematic Review. Front. Aging Neurosci. 2022, 14, 683689. [Google Scholar] [CrossRef]
- Zhu, C.S.; Pinsky, P.F.; Cramer, D.W.; Ransohoff, D.F.; Hartge, P.; Pfeiffer, R.M.; Urban, N.; Mor, G.; Bast, R.C., Jr.; Moore, L.E.; et al. A framework for evaluating biomarkers for early detection: Validation of biomarker panels for ovarian cancer. Cancer Prev. Res. 2011, 4, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Chen, Y.; Wu, W.; Chen, L.; Yang, X.; Zhang, S. Identification and validation of four hub genes involved in the plaque deterioration of atherosclerosis. Aging 2019, 11, 6469–6489. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Jain, V.V.; Finn, P.W.; Perkins, D.L. Hubs in biological interaction networks exhibit low changes in expression in experimental asthma. Mol. Syst. Biol. 2007, 3, 98. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Zhao, H.; Jiang, Y.; Yin, C.; Zhang, J. HCC-Derived Exosomes: Critical Player and Target for Cancer Immune Escape. Cells 2019, 8, 558. [Google Scholar] [CrossRef] [PubMed]
- Foerster, F.; Hess, M.; Gerhold-Ay, A.; Marquardt, J.U.; Becker, D.; Galle, P.R.; Schuppan, D.; Binder, H.; Bockamp, E. The immune contexture of hepatocellular carcinoma predicts clinical outcome. Sci. Rep. 2018, 8, 5351. [Google Scholar] [CrossRef]
- Azadeh, M.; Salehzadeh, A.; Ghaedi, K.; Talesh Sasani, S. NEAT1 can be a diagnostic biomarker in the breast cancer and gastric cancer patients by targeting XIST, hsa-miR-612, and MTRNR2L8: Integrated RNA targetome interaction and experimental expression analysis. Genes. Environ. 2022, 44, 16. [Google Scholar] [CrossRef] [PubMed]
- Willis-Owen, S.A.G.; Domingo-Sabugo, C.; Starren, E.; Liang, L.; Freidin, M.B.; Arseneault, M.; Zhang, Y.; Lu, S.K.; Popat, S.; Lim, E.; et al. Y disruption, autosomal hypomethylation and poor male lung cancer survival. Sci. Rep. 2021, 11, 12453. [Google Scholar] [CrossRef] [PubMed]
| Model | SVM | MLP | Random Forest | Logistic Regression | K-Nearest Neighbour | Gaussian Naïve Bayes |
|---|---|---|---|---|---|---|
| Accuracy | 0.761 | 0.761 | 0.848 | 0.783 | 0.848 | 0.761 |
| Precision | 0.790 | 0.739 | 0.941 | 0.833 | 0.941 | 0.867 |
| Sensitivity | 0.682 | 0.773 | 0.727 | 0.682 | 0.727 | 0.591 |
| Specificity | 0.833 | 0.750 | 0.958 | 0.875 | 0.958 | 0.917 |
| FPR | 0.167 | 0.250 | 0.042 | 0.125 | 0.0417 | 0.083 |
| F1-Score | 0.732 | 0.756 | 0.821 | 0.750 | 0.821 | 0.703 |
| AUC | 0.840 | 0.850 | 0.870 | 0.810 | 0.880 | 0.790 |
| Exosome RNA | Gene Ensemble ID | Name | Mean Importance | Importance Rank |
|---|---|---|---|---|
| MTRNR2L8 | ENSG00000255823.4 | MT-RNR2 Like 8 | 0.162 | 1 |
| FTL | ENSG00000087086.14 | Ferritin Light Chain | 0.090 | 2 |
| PPBP | ENSG00000163736.3 | Pro-Platelet Basic Protein | 0.027 | 4 |
| TMSB4X | ENSG00000205542.10 | Thymosin Beta 4 X-Linked | 0.018 | 5 |
| S100A11 | ENSG00000163191.5 | S100 Calcium Binding Protein A11 | 0.018 | 6 |
| S100A9 | ENSG00000163220.10 | S100 Calcium Binding Protein A9 | 0.009 | 7 |
| ACTB | ENSG00000075624.14 | Actin Beta | 0.009 | 8 |
| exoRBase circID | circBase ID | Genomic Position | Strand | Parent Gene Symbol | Parent Gene Type | Mean Importance | Importance Rank |
|---|---|---|---|---|---|---|---|
| exo_circ_22106 | hsa_circ_000072 | chr16:85633914-85634132 (exon) | + | GSE1 | protein coding | 0.036 | 3 |
| exo_circ_79050 | hsa_circ_0009024 | chrY:19587210-19587507 (exon) | + | TXLNGY | pseudogene | 3.70 × 10−17 | 9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yap, J.Y.Y.; Goh, L.S.H.; Lim, A.J.W.; Chong, S.S.; Lim, L.J.; Lee, C.G. Machine Learning Identifies a Signature of Nine Exosomal RNAs That Predicts Hepatocellular Carcinoma. Cancers 2023, 15, 3749. https://doi.org/10.3390/cancers15143749
Yap JYY, Goh LSH, Lim AJW, Chong SS, Lim LJ, Lee CG. Machine Learning Identifies a Signature of Nine Exosomal RNAs That Predicts Hepatocellular Carcinoma. Cancers. 2023; 15(14):3749. https://doi.org/10.3390/cancers15143749
Chicago/Turabian StyleYap, Josephine Yu Yan, Laura Shih Hui Goh, Ashley Jun Wei Lim, Samuel S. Chong, Lee Jin Lim, and Caroline G. Lee. 2023. "Machine Learning Identifies a Signature of Nine Exosomal RNAs That Predicts Hepatocellular Carcinoma" Cancers 15, no. 14: 3749. https://doi.org/10.3390/cancers15143749
APA StyleYap, J. Y. Y., Goh, L. S. H., Lim, A. J. W., Chong, S. S., Lim, L. J., & Lee, C. G. (2023). Machine Learning Identifies a Signature of Nine Exosomal RNAs That Predicts Hepatocellular Carcinoma. Cancers, 15(14), 3749. https://doi.org/10.3390/cancers15143749







