Landscape of Genetic Mutations in Appendiceal Cancers
Abstract
Simple Summary
Abstract
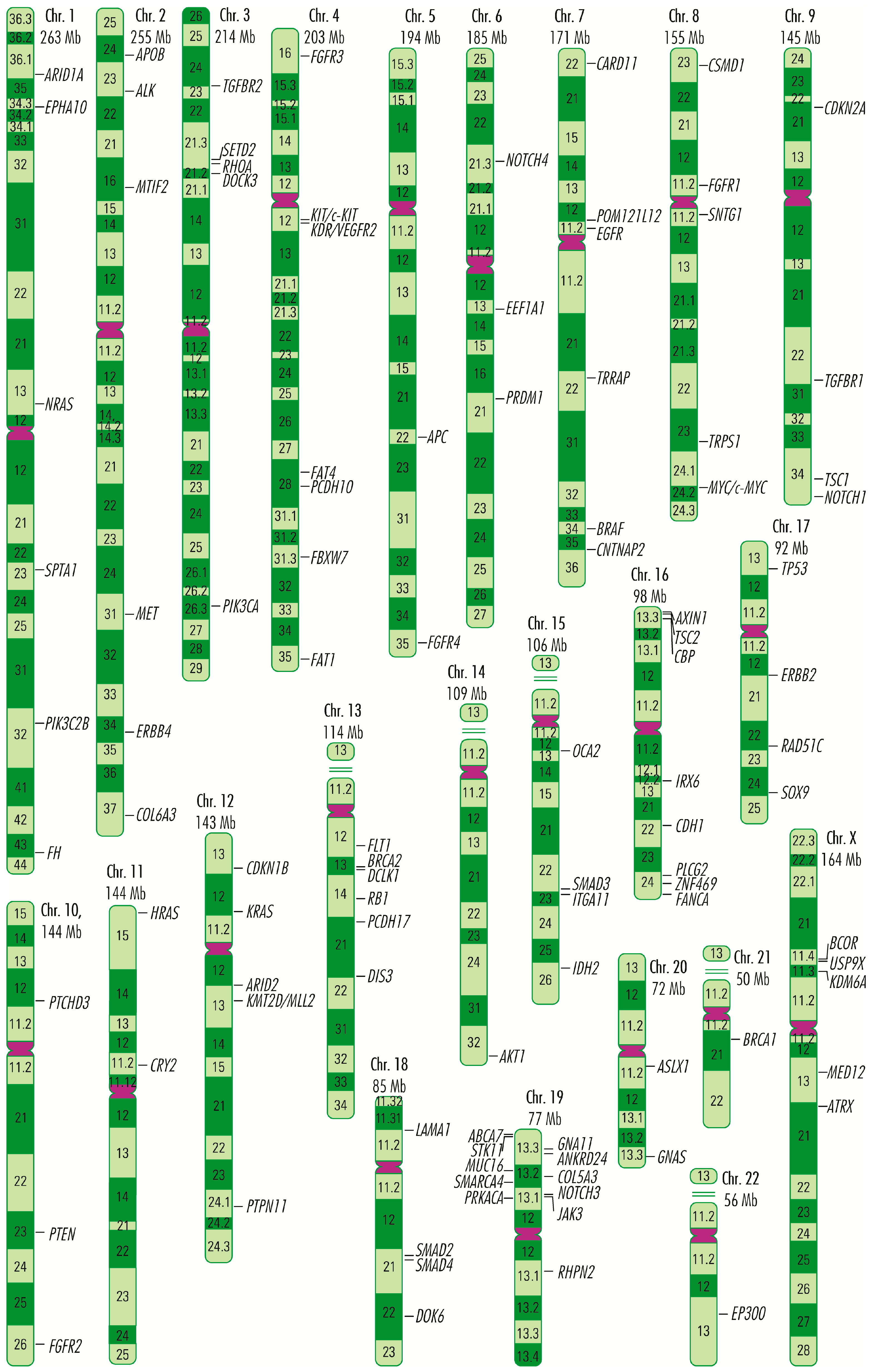
1. Introduction
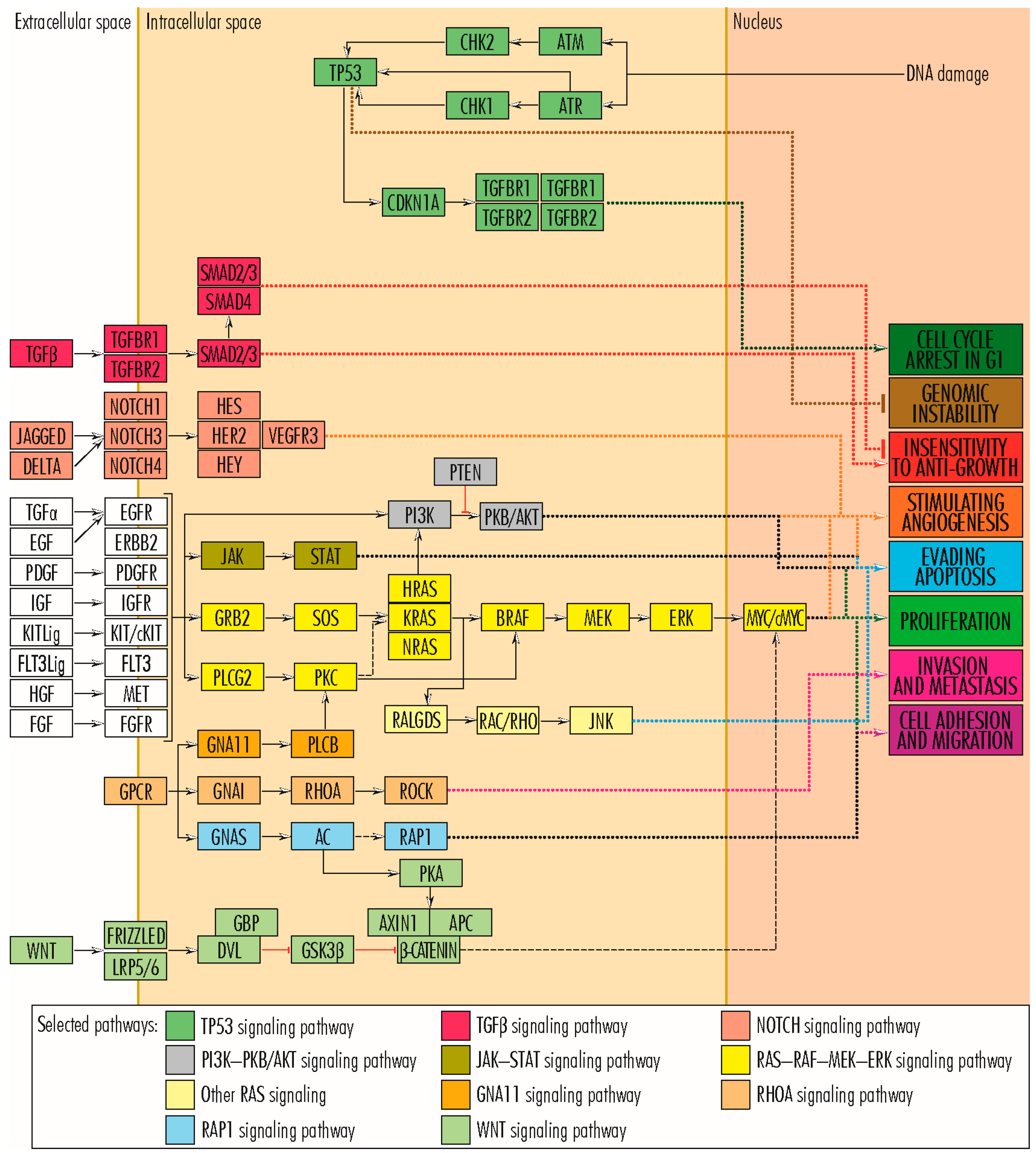
2. Mutations in Genes Whose Proteins Are Involved in Cell Proliferation
2.1. RAS–RAF–MEK–ERK Signaling Pathway
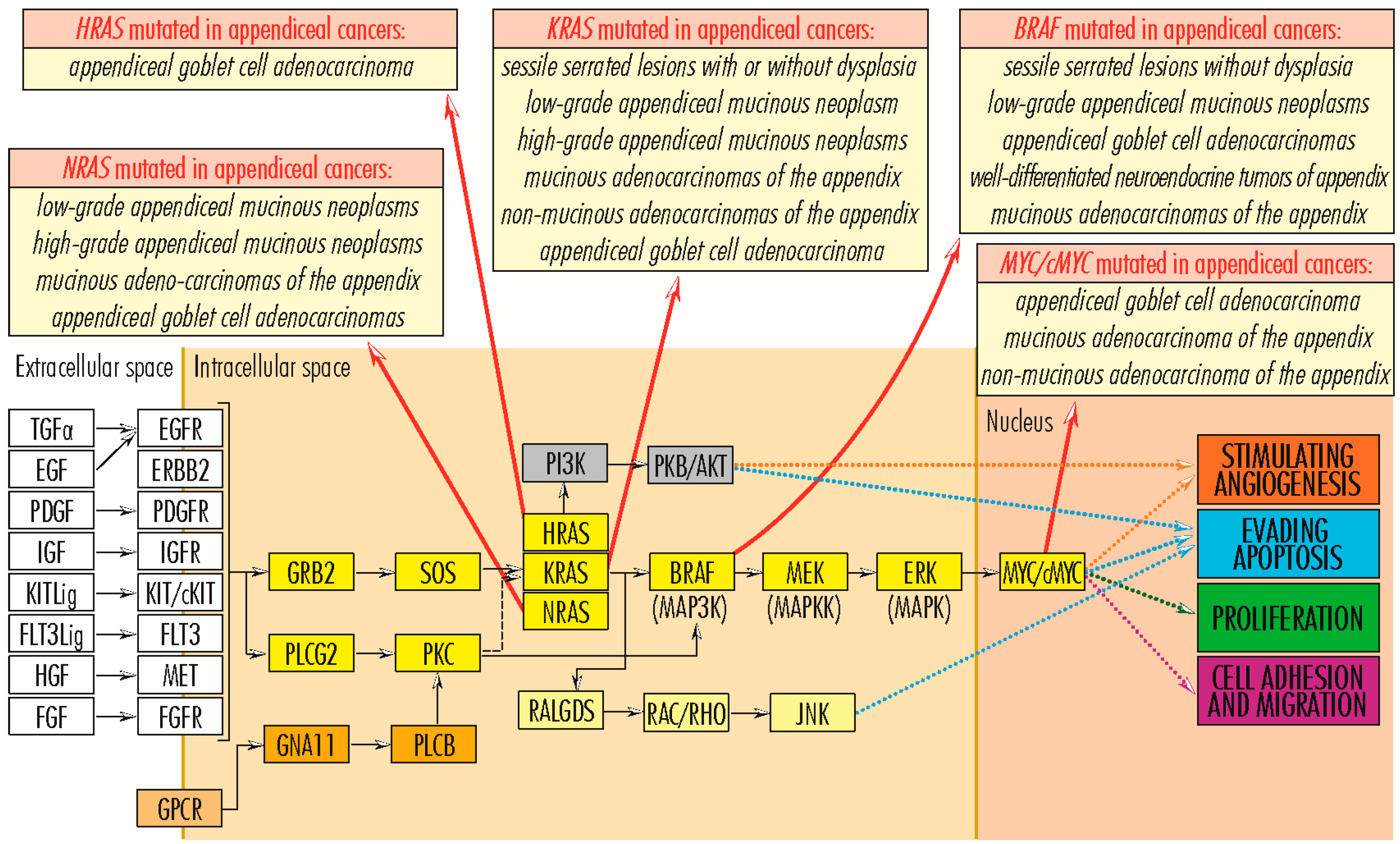
2.1.1. RAS Gene Family
2.1.2. RAF Gene Family
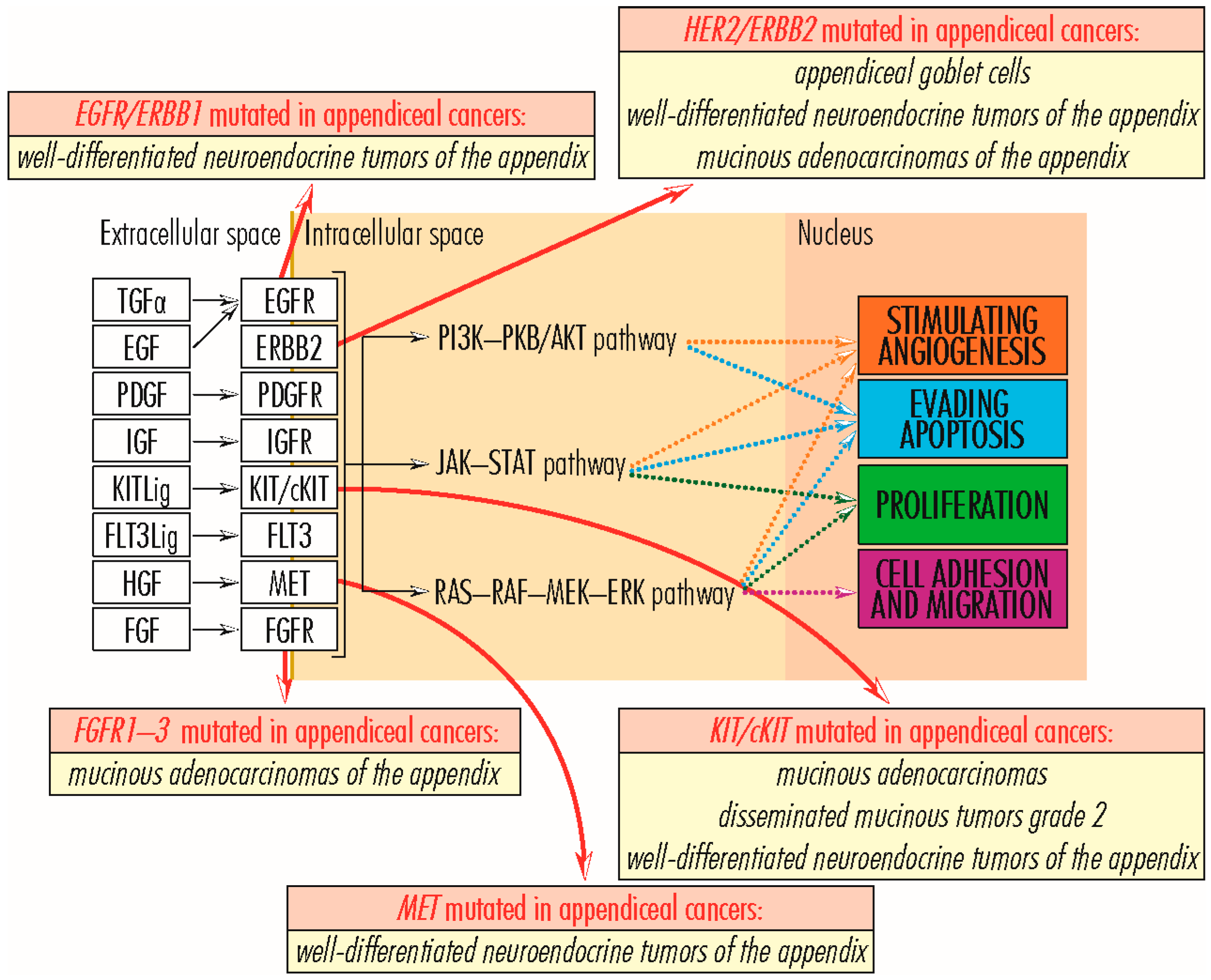
2.1.3. EGFR/ERBB Gene Family
2.1.4. MET Gene
2.1.5. FGFR Gene Family
2.1.6. KIT/cKIT Gene
2.1.7. MYC/c-MYC Gene
2.1.8. PLCG2 Gene
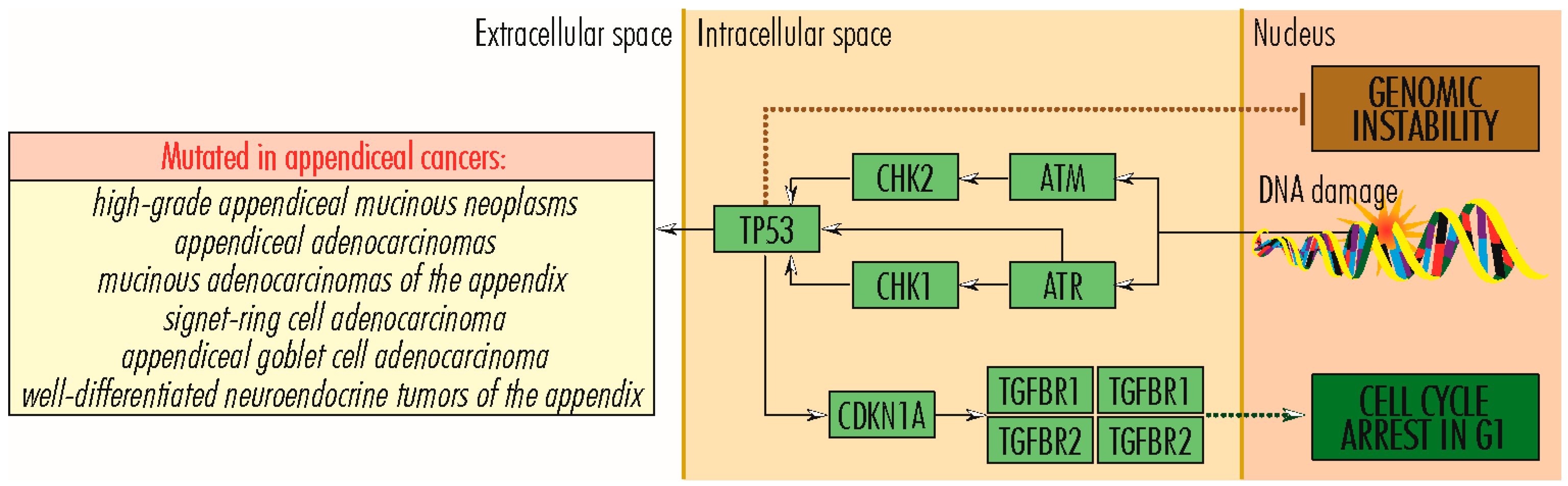
2.2. TP53 Signaling Pathway
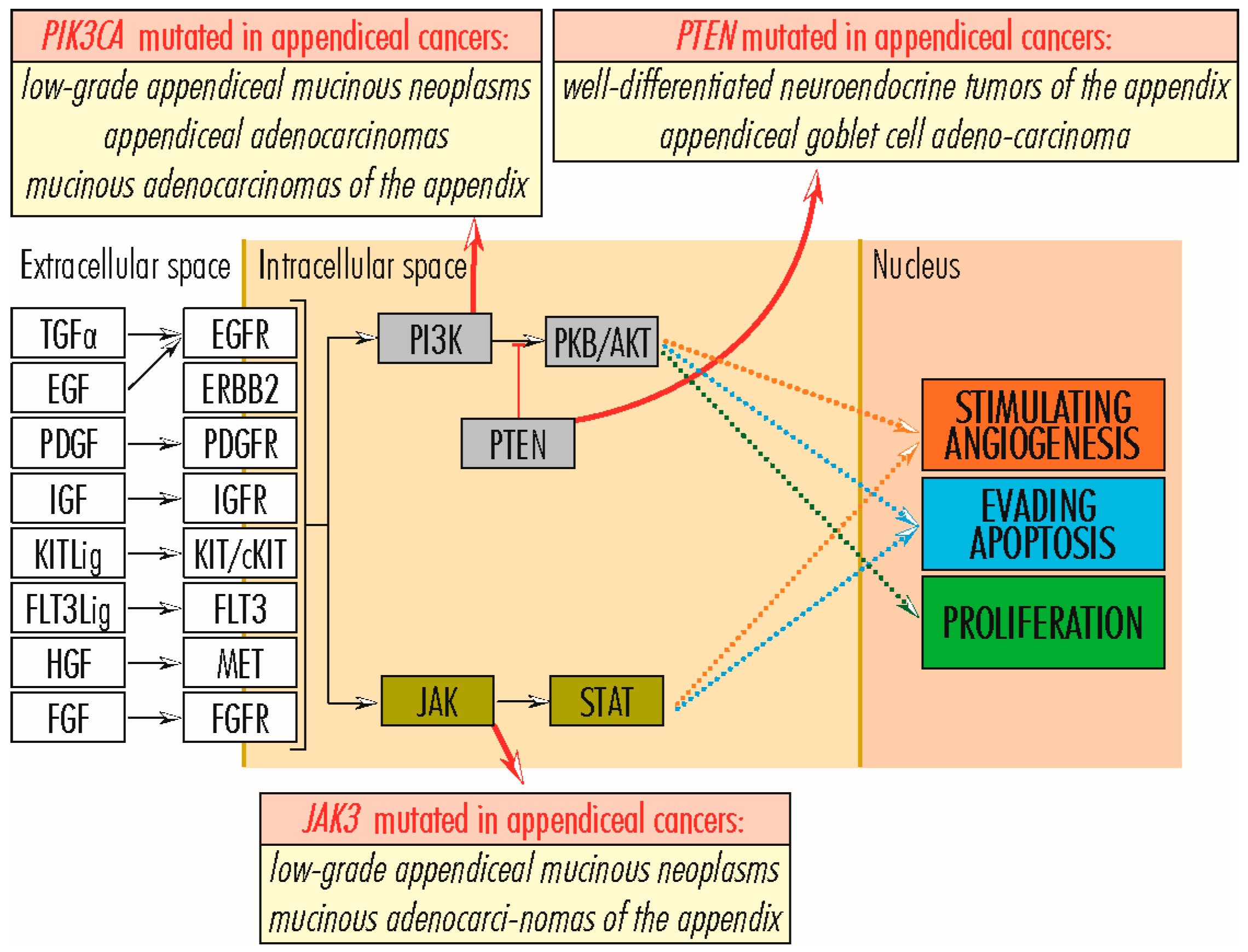
2.3. PI3K–PKB/AKT Signaling Pathway
2.3.1. AKT1 Gene
2.3.2. ITGA11 Gene
2.3.3. PIK3C2B and PIK3CA Genes
2.3.4. PTEN Gene
2.4. AKT-mTOR Signaling Pathway
2.5. RB1 Signaling Pathway
2.6. JAK3 Signaling Pathway
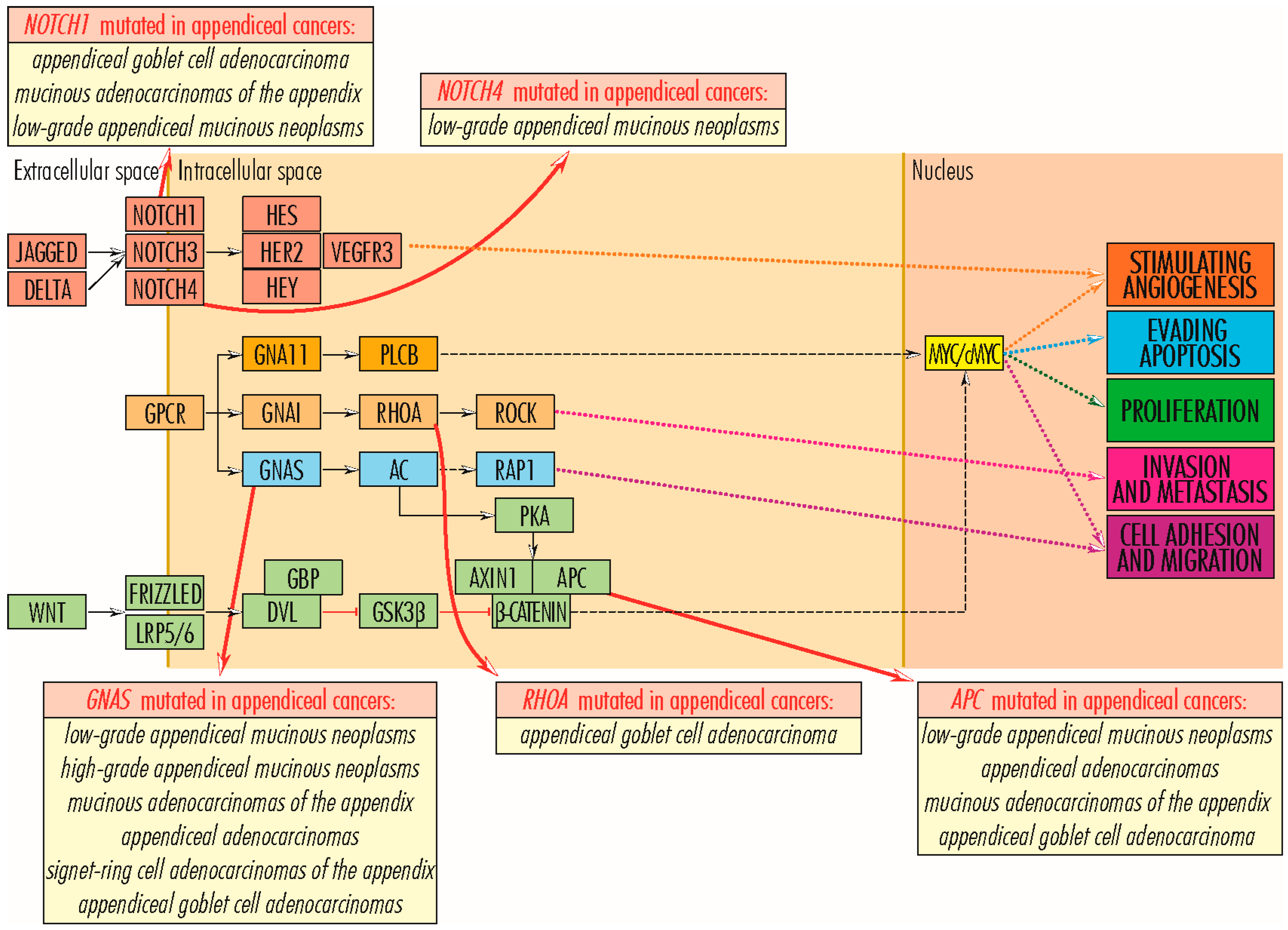
2.7. Wnt/β-Catenin Signaling Pathway
2.7.1. APC Gene
2.7.2. AXIN1 Gene
2.7.3. TCF7L2 Gene
3. Mutations in Genes Whose Proteins Are Involved in Cell Survival and Tumor Invasiveness/Metastasis
RHOA Gene
4. Mutations in Genes Regulating the Angiogenesis
4.1. GNA11 and GNAS Genes
4.2. EP300 and CPB/CREBBP Genes
4.3. KDR/VEGFR2 Gene
4.4. NOTCH Gene Family
4.5. FLT1/VEGFR1 Gene
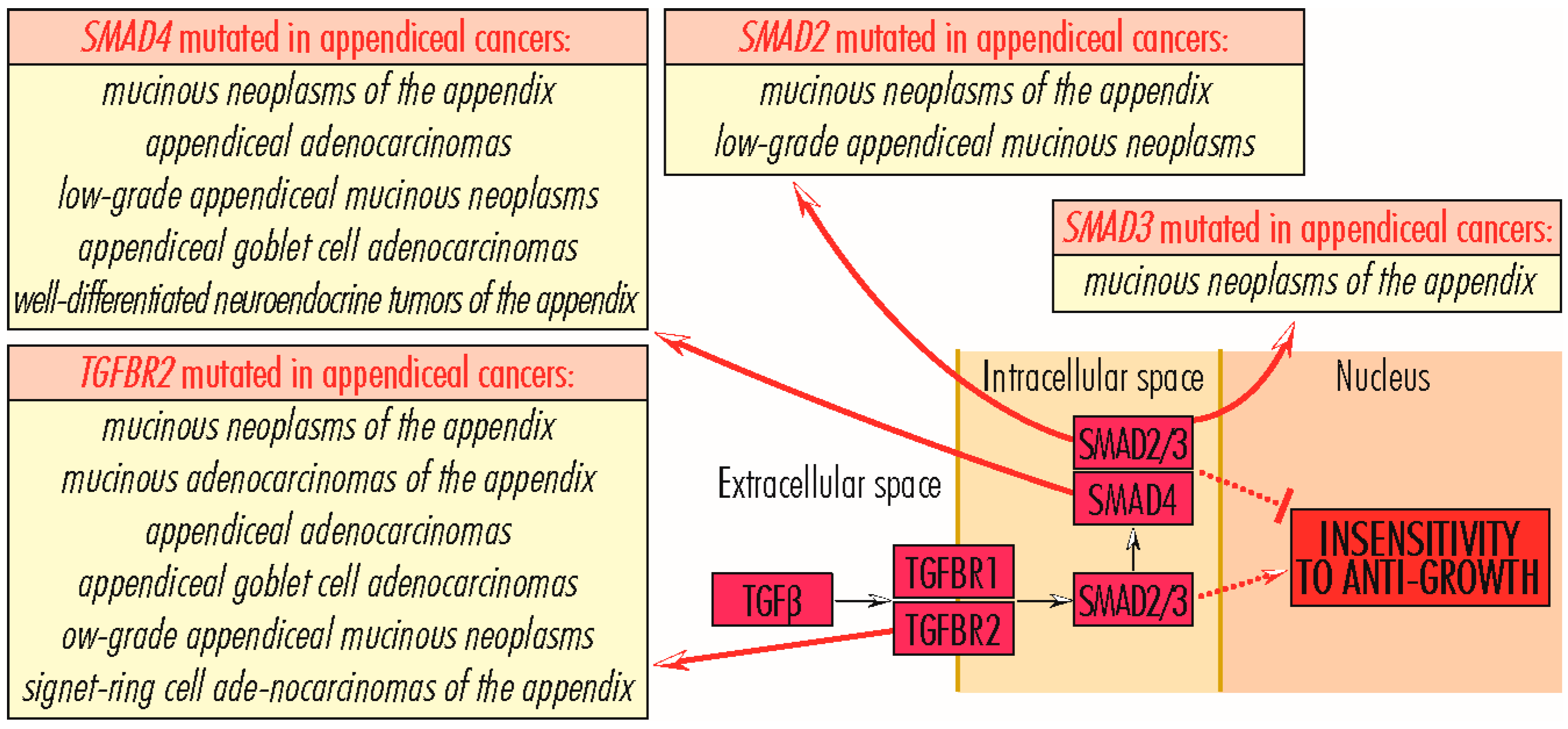
5. Mutations in Genes Whose Proteins Are Involved in Acquiring Insensitivity to Anti-Growth Signals
5.1. TGFBR Gene Family
5.2. SMAD Gene Family
6. Mutations in Genes Whose Proteins Are Involved in the Organization of the Extracellular Matrix
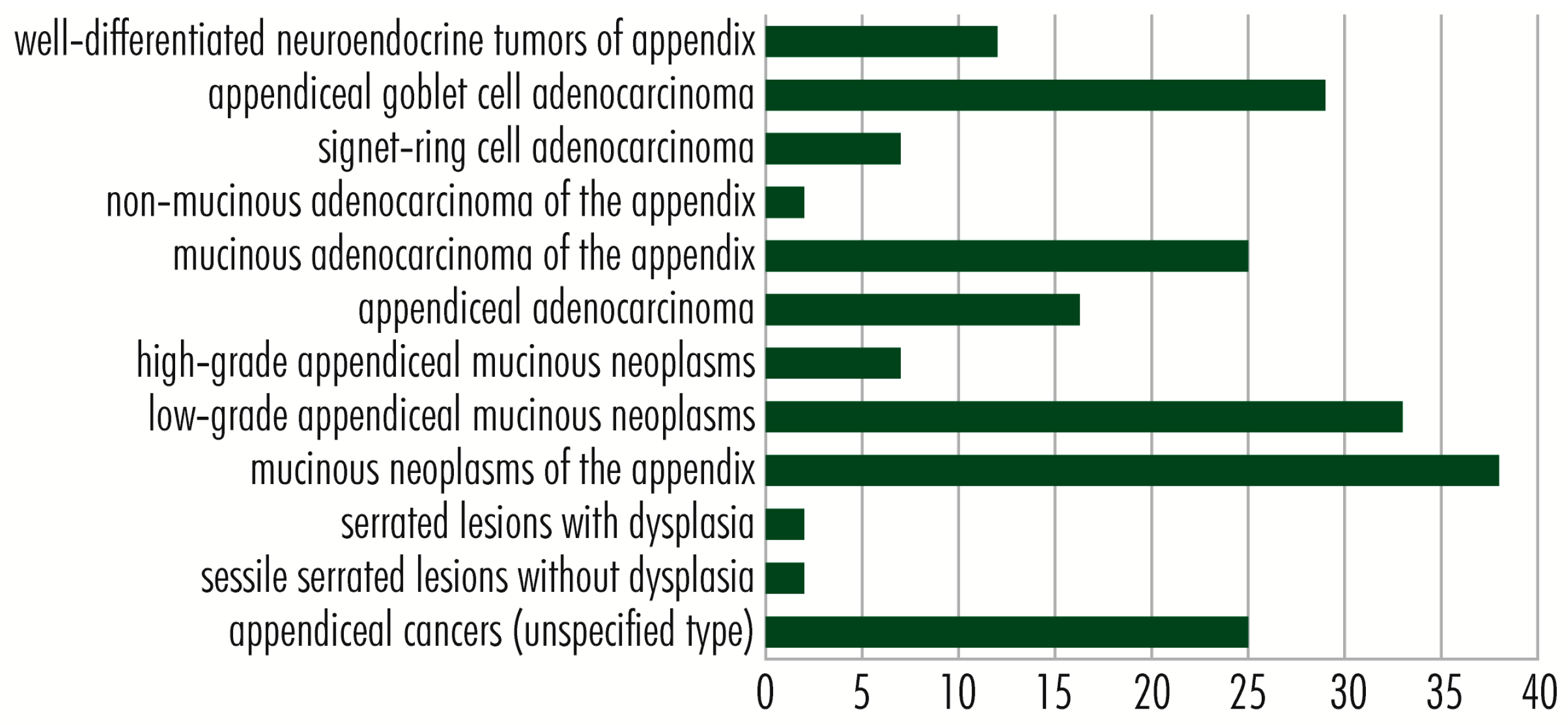
7. Other Genes Showing Mutations in Appendix Cancer
7.1. Genes Involved in DNA Metabolism/Expression
7.2. Genes Involved in Cell Junctions/Adhesion
7.3. Genes Encoding for Enzymes
7.4. Genes with Various Cellular Activities
8. Microsatellite Instability in Appendix Tumors
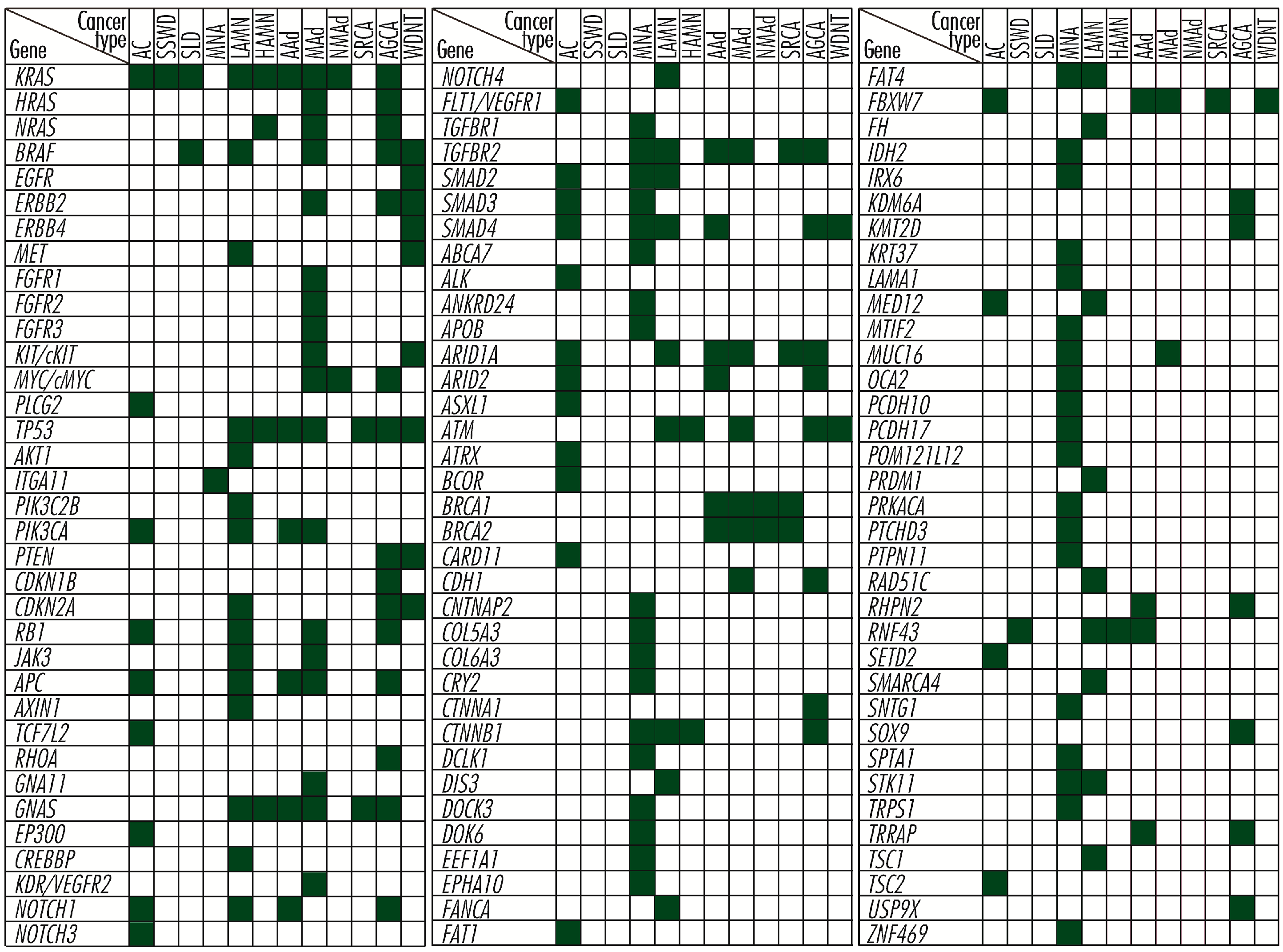
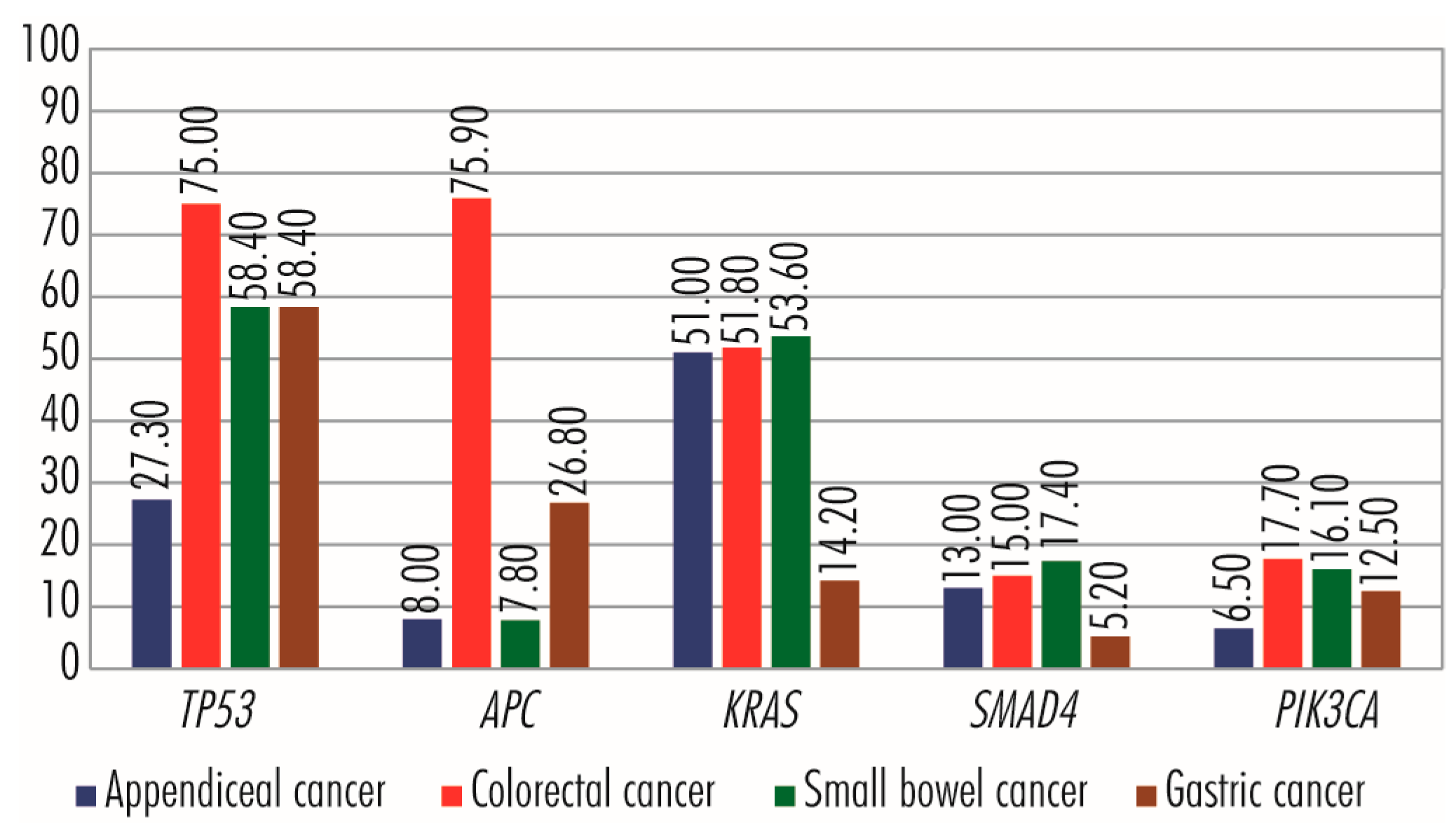
9. Comparison of Genetic Aberrations in Appendiceal, Colorectal, Small Bowel, and Gastric Cancers
10. Discussion
11. Future Perspectives
12. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Deshmukh, S.; Verde, F.; Johnson, P.T.; Fishman, E.K.; Macura, K.J. Anatomical variants and pathologies of the vermix. Emerg. Radiol. 2014, 21, 543–552. [Google Scholar] [CrossRef] [PubMed]
- Schumpelick, V.; Dreuw, B.; Ophoff, K.; Prescher, A. Appendix and cecum. Embryology, anatomy, and surgical applications. Surg. Clin. N. Am. 2000, 80, 295–318. [Google Scholar] [CrossRef] [PubMed]
- De Souza, S.C.; da Costa, S.R.M.R.; de Souza, I.G.S. Vermiform appendix: Positions and length—A study of 377 cases and literature review. J. Coloproctol. 2015, 35, 212–216. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. Appendix: Carcinoma. In AJCC Cancer Staging Manual, 8th ed.; Amin, M.B., Ed.; Springer: New York, NY, USA, 2017. [Google Scholar]
- Nagtegaal, I.D.; Klimstra, D.S.; Washington, M.K. Tumors of the appendix. In WHO Classification of Tumors: Digestive System Tumors, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2019. [Google Scholar]
- Ramnani, D.M.; Wistuba, I.I.; Behrens, C.; Gazdar, A.F.; Sobin, L.H.; Albores-Saavedra, J. K-ras and p53 mutations in the pathogenesis of classical and goblet cell carcinoids of the appendix. Cancer 1999, 86, 14–21. [Google Scholar] [CrossRef]
- Alakus, H.; Babicky, M.L.; Ghosh, P.; Yost, S.; Jepsen, K.; Dai, Y.; Arias, A.; Samuels, M.L.; Mose, E.S.; Schwab, R.B.; et al. Genome-wide mutational landscape of mucinous carcinomatosis peritonei of appendiceal origin. Genome Med. 2014, 6, 43. [Google Scholar] [CrossRef]
- Borazanci, E.; Millis, S.; Kimbrough, J.; Doll, N.; Hoff, D.; Ramanathan, R. Potential actionable targets in appendiceal cancer detected by immunohistochemistry, fluorescent in situ hybridization, and mutational analysis. J. Gastrointest. Oncol. 2017, 8, 164–172. [Google Scholar] [CrossRef]
- Ang, C.S.; Shen, J.P.; Hardy-Abeloos, C.J.; Huang, J.K.; Ross, J.S.; Miller, V.A.; Jacobs, M.T.; Chen, I.L.; Xu, D.; Ali, S.M.; et al. Genomic landscape of appendiceal neoplasms. JCO Precis. Oncol. 2018, 2, PO.17.00302. [Google Scholar] [CrossRef]
- Jesinghaus, M.; Konukiewitz, B.; Foersch, S.; Stenzinger, A.; Steiger, K.; Muckenhuber, A.; Groß, C.; Mollenhauer, M.; Roth, W.; Detlefsen, S.; et al. Appendiceal goblet cell carcinoids and adenocarcinomas ex-goblet cell carcinoid are genetically distinct from primary colorectal-type adenocarcinoma of the appendix. Mod. Pathol. 2018, 31, 829–839. [Google Scholar] [CrossRef]
- Johncilla, M.; Stachler, M.; Misdraji, J.; Lisovsky, M.; Yozu, M.; Lindeman, N.; Lauwers, G.Y.; Odze, R.D.; Srivastava, A. Mutational landscape of goblet cell carcinoids and adenocarcinoma ex goblet cell carcinoids of the appendix is distinct from typical carcinoids and colorectal adenocarcinomas. Mod. Pathol. 2018, 31, 989–996. [Google Scholar] [CrossRef]
- Liu, W.; Liu, L.; Wang, R.; Gong, G.; Ding, X.; Yang, B.; Bao, Y.; Wang, Z.; Zhang, B.; Zhao, D.; et al. Bevacizumab combined with oxaliplatin/capecitabine in patient with refractory and recurrent mucinous adenocarcinoma of the appendix: A case report. Front. Oncol. 2019, 9, 55. [Google Scholar] [CrossRef]
- Holowatyj, A.N.; Eng, C.; Wen, W.; Idrees, K.; Guo, X. Spectrum of somatic cancer gene variations among adults with appendiceal cancer by age at disease onset. JAMA Netw. Open 2021, 4, e216703. [Google Scholar] [CrossRef]
- KEGG Pathways in Cancer-Reference Pathway. Available online: https://www.genome.jp/kegg-bin/show_pathway?map05200 (accessed on 3 October 2022).
- Kang, D.W.; Kim, B.H.; Kim, J.M.; Kim, J.; Chang, H.J.; Chang, M.S.; Sohn, J.H.; Cho, M.Y.; Jin, S.Y.; Chang, H.K.; et al. Gastrointestinal pathology study group of the Korean society of pathologists. Standardization of the pathologic diagnosis of appendiceal mucinous neoplasms. J. Pathol. Transl. Med. 2021, 55, 247–264. [Google Scholar] [CrossRef]
- Copur, M.; Cushman-Vokoun, A.; Padussis, J.; Whitney, W.; Schroeder, C.; Herold, D.; Lintel, N.; Horn, A. Mucinous adenocarcinoma of the appendix with histologic response to neoadjuvant chemotherapy: Review of histologic and clinical spectrum of epithelial neoplastic mucinous lesions of the appendix. Oncology 2021, 35, 335–340. [Google Scholar] [CrossRef]
- Patel, S.; Alam, A.; Pant, R.; Chattopadhyay, S. Wnt signaling and Its significance within the tumor microenvironment: Novel therapeutic insights. Front. Immunol. 2019, 10, 2872. [Google Scholar] [CrossRef]
- Gremer, L.; Merbitz-Zahradnik, T.; Dvorsky, R.; Cirstea, I.C.; Kratz, C.P.; Zenker, M.; Wittinghofer, A.; Ahmadian, M.R. Germline KRAS mutations cause aberrant biochemical and physical properties leading to developmental disorders. Hum. Mutat. 2011, 32, 33–43. [Google Scholar] [CrossRef]
- Dobre, M.; Dinu, D.E.; Panaitescu, E.; Bîrlă, R.D.; Iosif, C.I.; Boeriu, M.; Constantinoiu, S.; Ivan, R.N.; Ardeleanu, C.M.; Costache, M. KRAS gene mutations—Prognostic factor in colorectal cancer? Rom. J. Morphol. Embryol. 2015, 56, 671–678. [Google Scholar]
- KRAS Gene—KRAS Proto-Oncogene, GTPase. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=KRAS&keywords=KRAS (accessed on 14 September 2022).
- Yantiss, R.K.; Panczykowski, A.; Misdraji, J.; Hahn, H.P.; Odze, R.D.; Rennert, H.; Chen, Y.T. A comprehensive study of nondysplastic and dysplastic serrated polyps of the vermiform appendix. Am. J. Surg. Pathol. 2008, 32, 175. [Google Scholar] [CrossRef]
- Pai, R.K.; Hartman, D.J.; Gonzalo, D.H.; Lai, K.K.; Downs-Kelly, E.; Goldblum, J.R.; Liu, X.; Patil, D.T.; Bennett, A.E.; Plesec, T.P.; et al. Serrated lesions of the appendix frequently harbor KRAS mutations and not BRAF mutations indicating a distinctly different serrated neoplastic pathway in the appendix. Hum. Pathol. 2014, 45, 227–235. [Google Scholar] [CrossRef]
- Munari, G.; Businello, G.; Mattiolo, P.; Pennelli, G.; Sbaraglia, M.; Borga, C.; Pucciarelli, S.; Spolverato, G.; Mescoli, C.; Galuppini, F.; et al. Molecular profiling of appendiceal serrated lesions, polyps and mucinous neoplasms: A single-centre experience. J. Cancer Res. Clin. Oncol. 2021, 147, 1897–1904. [Google Scholar] [CrossRef]
- Zauber, P.; Berman, E.; Marotta, S.; Sabbath-Solitare, M.; Bishop, T. Ki-ras gene mutations are invariably present in low-grade mucinous tumors of the vermiform appendix. Scand. J. Gastroenterol. 2011, 46, 869–874. [Google Scholar] [CrossRef]
- Shetty, S.; Thomas, P.; Ramanan, B.; Sharma, P.; Govindarajan, V.; Loggie, B. Kras mutations and p53 overexpression in pseudomyxoma peritonei: Association with phenotype and prognosis. J. Surg. Res. 2013, 180, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Mody, K.; de Abreu, F.B.; Pipas, J.M.; Peterson, J.D.; Gallagher, T.L.; Suriawinata, A.A.; Ripple, G.H.; Hourdequin, K.C.; Smith, K.D.; et al. Molecular profiling of appendiceal epithelial tumors using massively parallel sequencing to identify somatic mutations. Clin. Chem. 2014, 60, 1004–1011. [Google Scholar] [CrossRef] [PubMed]
- Singhi, A.D.; Davison, J.M.; Choudry, H.A.; Pingpank, J.F.; Ahrendt, S.A.; Holtzman, M.P.; Zureikat, A.H.; Zeh, H.J.; Ramalingam, L.; Mantha, G.; et al. GNAS is frequently mutated in both low-grade and high-grade disseminated appendiceal mucinous neoplasms but does not affect survival. Hum. Pathol. 2014, 45, 1737–1743. [Google Scholar] [CrossRef] [PubMed]
- Davison, J.M.; Choudry, H.A.; Pingpank, J.F.; Ahrendt, S.A.; Holtzman, M.P.; Zureikat, A.H.; Zeh, H.J.; Ramalingam, L.; Zhu, B.; Nikiforova, M.; et al. Clinicopathologic and molecular analysis of disseminated appendiceal mucinous neoplasms: Identification of factors predicting survival and proposed criteria for a three-tiered assessment of tumor grade. Mod. Pathol. 2014, 27, 1521–1539. [Google Scholar] [CrossRef]
- Noguchi, R.; Yano, H.; Gohda, Y.; Suda, R.; Igari, T.; Ohta, Y.; Yamashita, N.; Yamaguchi, K.; Terakado, Y.; Ikenoue, T.; et al. Molecular profiles of high-grade and low-grade pseudomyxoma peritonei. Cancer Med. 2015, 4, 1809–1816. [Google Scholar] [CrossRef]
- Pengelly, R.J.; Rowaiye, B.; Pickard, K.; Moran, B.; Dayal, S.; Tapper, W.; Mirnezami, A.; Cecil, T.; Mohamed, F.; Carr, N.; et al. Analysis of mutation and loss of heterozygosity by whole-exome sequencing yields insights into Pseudomyxoma peritonei. J. Mol. Diagn. 2018, 20, 635–642. [Google Scholar] [CrossRef]
- Schrader, L.; Choo, E. 330 low-grade mucinous neoplasm of the appendix in the setting of a CDKN2A gene mutation: A potential association risk factor. Am. J. Clin. Pathol. 2018, 149, 142. [Google Scholar] [CrossRef]
- King, M.C.; Munoz-Zuluaga, C.; Ledakis, P.; Studeman, K.; Sittig, M.; Gushchin, V.; Sardi, A. Germline and somatic genetic alterations in two first-degree relatives with appendiceal low-grade mucinous carcinoma peritonei. Clin. Case. Rep. 2020, 8, 3168–3177. [Google Scholar] [CrossRef]
- Mikaeel, R.R.; Young, J.P.; Tapia Rico, G.; Hewett, P.J.; Hardingham, J.E.; Uylaki, W.; Horsnell, M.; Price, T.J. Immunohistochemistry features and molecular pathology of appendiceal neoplasms. Crit. Rev. Clin. Lab. Sci. 2021, 58, 369–384. [Google Scholar] [CrossRef]
- Flatmark, K.; Torgunrud, A.; Fleten, K.G.; Davidson, B.; Juul, H.V.; Mensali, N.; Lund-Andersen, C.; Inderberg, E.M. Peptide vaccine targeting mutated GNAS: A potential novel treatment for pseudomyxoma peritonei. J. Immunother. Cancer 2021, 9, e003109. [Google Scholar] [CrossRef]
- Kabbani, W.; Houlihan, P.S.; Luthra, R.; Hamilton, S.R.; Rashid, A. Mucinous and nonmucinous appendiceal adenocarcinomas: Different clinicopathological features but similar genetic alterations. Mod. Pathol. 2002, 15, 599–605. [Google Scholar] [CrossRef] [PubMed]
- Arai, H.; Baca, Y.; Battaglin, F.; Kawanishi, N.; Wang, J.; Soni, S.; Zhang, W.; Millstein, J.; Johnston, C.; Goldberg, R.M.; et al. Molecular characterization of appendiceal goblet cell carcinoid. Mol. Cancer Ther. 2020, 19, 2634–2640. [Google Scholar] [CrossRef]
- Ottaiano, A.; Santorsola, M.; Perri, F.; Pace, U.; Marra, B.; Correra, M.; Sabbatino, F.; Cascella, M.; Petrillo, N.; Ianniello, M.; et al. Clinical and molecular characteristics of rare malignant tumors of colon and rectum. Biology 2022, 11, 267. [Google Scholar] [CrossRef] [PubMed]
- HRAS Gene—HRas Proto-Oncogene, GTPase. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=HRAS (accessed on 3 October 2022).
- NRAS Gene—NRAS Proto-Oncogene, GTPase. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=NRAS&keywords=NRAS (accessed on 3 October 2022).
- Raghav, K.P.; Shetty, A.V.; Kazmi, S.M.; Zhang, N.; Morris, J.; Taggart, M.; Fournier, K.; Royal, R.; Mansfield, P.; Eng, C.; et al. Impact of molecular alterations and targeted therapy in appendiceal adenocarcinomas. Oncologist 2013, 18, 1270–1277. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Huang, X.; Liu, H.; Wei, C.; Ru, H.; Qin, H.; Lai, H.; Meng, Y.; Wu, G.; Xie, W.; et al. Immune landscape and prognostic immune-related genes in KRAS-mutant colorectal cancer patients. J. Transl. Med. 2021, 19, 27. [Google Scholar] [CrossRef]
- Matas, J.; Kohrn, B.; Fredrickson, J.; Carter, K.; Yu, M.; Wang, T.; Gui, X.; Soussi, T.; Moreno, V.; Grady, W.M.; et al. Colorectal cancer is associated with the presence of cancer driver mutations in normal colon. Cancer Res. 2022, 82, 1492–1502. [Google Scholar] [CrossRef]
- Matallanas, D.; Birtwistle, M.; Romano, D.; Zebisch, A.; Rauch, J.; von Kriegsheim, A.; Kolch, W. Raf family kinases: Old dogs have learned new tricks. Genes Cancer 2011, 2, 232–260. [Google Scholar] [CrossRef]
- BRAF Gene—B-Raf Proto-Oncogene, Serine/Threonine Kinase. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=BRAF&keywords=BRAF (accessed on 3 October 2022).
- Ueda, K.; Yamada, T.; Ohta, R.; Matsuda, A.; Sonoda, H.; Kuriyama, S.; Takahashi, G.; Iwai, T.; Takeda, K.; Miyasaka, T.; et al. BRAF V600E mutations in right-side colon cancer: Heterogeneity detected by liquid biopsy. Eur. J. Surg. Oncol. 2022, 48, 1375–1383. [Google Scholar] [CrossRef]
- Asako, K.; Hayama, T.; Hashiguchi, Y.; Miyata, T.; Fukushima, Y.; Shimada, R.; Kaneko, K.; Nozawa, K.; Matsuda, K.; Fukagawa, T. Prognostic value of KRAS exon-specific mutations in patients with colorectal cancer. Anticancer Res. 2023, 43, 1563–1568. [Google Scholar] [CrossRef]
- EGFR Gene—Epidermal Growth Factor Receptor. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=EGFR (accessed on 4 October 2022).
- Roskoski, R., Jr. The ErbB/HER family of protein-tyrosine kinases and cancer. Pharmacol. Res. 2014, 79, 34–74. [Google Scholar] [CrossRef]
- ERBB2 Gene—Erb-B2 Receptor Tyrosine Kinase 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ERBB2 (accessed on 4 October 2022).
- Arteaga, C.L.; Engelman, J.A. ERBB receptors: From oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell 2014, 25, 282–303. [Google Scholar] [CrossRef]
- ERBB4 Gene—Erb-B2 Receptor Tyrosine Kinase. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ERBB4 (accessed on 4 October 2022).
- Petrini, I. Biology of MET: A double life between normal tissue repair and tumor progression. Ann. Transl. Med. 2015, 3, 82. [Google Scholar] [CrossRef]
- MET Gene—MET Proto-Oncogene, Receptor Tyrosine Kinase. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MET (accessed on 5 October 2022).
- Li, J.; Li, Z.; Ding, Y.; Xu, Y.; Zhu, X.; Cao, N.; Huang, C.; Qin, M.; Liu, F.; Zhao, A. TP53 mutation and MET amplification in circulating tumor DNA analysis predict disease progression in patients with advanced gastric cancer. PeerJ 2021, 9, e11146. [Google Scholar] [CrossRef]
- FGFR1 Gene—Fibroblast Growth Factor Receptor 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FGFR1 (accessed on 5 October 2022).
- FGFR4 Gene—Fibroblast Growth Factor Receptor 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FGFR4 (accessed on 5 October 2022).
- Krook, M.A.; Reeser, J.W.; Ernst, G.; Barker, H.; Wilberding, M.; Li, G.; Chen, H.Z.; Roychowdhury, S. Fibroblast growth factor receptors in cancer: Genetic alterations, diagnostics, therapeutic targets and mechanisms of resistance. Br. J. Cancer 2021, 124, 880–892. [Google Scholar] [CrossRef]
- FGFR2 Gene—Fibroblast Growth Factor Receptor 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FGFR2 (accessed on 5 October 2022).
- FGFR3 Gene—Fibroblast Growth Factor Receptor 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FGFR3 (accessed on 5 October 2022).
- Jogo, T.; Nakamura, Y.; Shitara, K.; Bando, H.; Yasui, H.; Esaki, T.; Terazawa, T.; Satoh, T.; Shinozaki, E.; Nishina, T.; et al. Circulating tumor DNA analysis detects FGFR2 amplification and concurrent genomic alterations associated with FGFR inhibitor efficacy in advanced gastric cancer. Clin. Cancer Res. 2021, 27, 5619–5627. [Google Scholar] [CrossRef]
- Miller, D.M.; Thomas, S.D.; Islam, A.; Muench, D.; Sedoris, K. c-Myc and cancer metabolism. Clin. Cancer Res. 2012, 18, 5546–5553. [Google Scholar] [CrossRef]
- MYC Gene—MYC Proto-Oncogene, BHLH Transcription Factor. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MYC (accessed on 6 October 2022).
- Riviere, P.; Fanta, P.T.; Ikeda, S.; Baumgartner, J.; Heestand, G.M.; Kurzrock, R. The mutational landscape of gastrointestinal malignancies as reflected by circulating tumor DNA. Mol. Cancer Ther. 2018, 17, 297–305. [Google Scholar] [CrossRef]
- Lin, X.; Lin, X.; Guo, L.; Wang, Y.; Zhang, G. Distinct clinicopathological characteristics, genomic alteration and prognosis in breast cancer with concurrent TP53 mutation and MYC amplification. Thorac. Cancer. 2022, 13, 3441–3450. [Google Scholar] [CrossRef]
- Jackson, J.T.; Mulazzani, E.; Nutt, S.L.; Masters, S.L. The role of PLCγ2 in immunological disorders, cancer, and neurodegeneration. J. Biol. Chem. 2021, 297, 100905. [Google Scholar] [CrossRef]
- PLCG2 Gene—Phospholipase C Gamma 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PLCG2 (accessed on 6 October 2022).
- Peng, Q.; Luo, D.; Yang, Y.; Zhu, Y.; Luo, Q.; Chen, H.; Chen, D.; Zhou, Z.; Lu, X. Clinical and immunological features of an APLAID patient caused by a novel mutation in PLCG2. Front. Immunol. 2023, 14, 1014150. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, R.; Yang, W.; Li, C.; Huang, J.; Wen, Z.; Du, G.; Jiang, L. PLCG2 as a potential indicator of tumor microenvironment remodeling in soft tissue sarcoma. Medicine 2021, 100, e25008. [Google Scholar] [CrossRef] [PubMed]
- Hernandez Borrero, L.J.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188556. [Google Scholar] [CrossRef] [PubMed]
- TP53 Gene—Tumor Protein P53. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TP53&keywords=TP (accessed on 30 September 2022).
- Oka, K.; Ueda, M.; Ikenaga, M.; Tanida, T.; Taguchi, D.; Fukushima, S.; Yoshida, S.; Yukimoto, R.; Iede, K.; Tsuda, Y.; et al. A Case of Early Appendiceal Adenocarcinoma Coexisting with High-Grade Appendiceal Mucinous Neoplasm. Cancer Chemother. 2022, 49, 1714–1716. [Google Scholar]
- Yanai, Y.; Saito, T.; Hayashi, T.; Akazawa, Y.; Yatagai, N.; Tsuyama, S.; Tomita, S.; Hirai, S.; Ogura, K.; Matsumoto, T.; et al. Molecular and clinicopathological features of appendiceal mucinous neoplasms. Virchows Arch. 2021, 478, 413–426. [Google Scholar] [CrossRef]
- Yan, F.; Shi, F.; Li, X.; Chang, H.; Jin, M.; Li, Y. Prognostic significance of CEA, Ki67 and p53 in pseudomyxoma peritonei of appendiceal origin. J. Int. Med. Res. 2021, 49, 3000605211022297. [Google Scholar] [CrossRef]
- AKT1 Gene—AKT Serine/Threonine Kinase. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=AKT1 (accessed on 7 October 2022).
- Chen, Y.; Huang, L.; Dong, Y.; Tao, C.; Zhang, R.; Shao, H.; Shen, H. Effect of AKT1 (p. E17K) hotspot mutation on malignant tumorigenesis and prognosis. Front. Cell Dev. Biol. 2020, 8, 573599. [Google Scholar] [CrossRef]
- Mundi, P.S.; Sachdev, J.; McCourt, C.; Kalinsky, K. AKT in cancer: New molecular insights and advances in drug development. Br. J. Clin. Pharmacol. 2016, 82, 943–956. [Google Scholar] [CrossRef]
- Kalinsky, K.; Hong, F.; McCourt, C.K.; Sachdev, J.C.; Mitchell, E.P.; Zwiebel, J.A.; Doyle, L.A.; McShane, L.M.; Li, S.; Gray, R.J.; et al. Effect of capivasertib in patients with an AKT1 E17K-mutated tumor: NCI-MATCH subprotocol EAY131-Y nonrandomized trial. JAMA Oncol. 2021, 7, 271–278. [Google Scholar] [CrossRef]
- ITGA11 Gene—Integrin Subunit Alpha 11. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ITGA11 (accessed on 7 October 2022).
- Primac, I.; Maquoi, E.; Blacher, S.; Heljasvaara, R.; Van Deun, J.; Smeland, H.Y.; Canale, A.; Louis, T.; Stuhr, L.; Sounni, N.E.; et al. Stromal integrin α11 regulates PDGFR-β signaling and promotes breast cancer progression. J. Clin. Investig. 2019, 129, 4609–4628. [Google Scholar] [CrossRef]
- Iwai, M.; Tulafu, M.; Togo, S.; Kawaji, H.; Kadoya, K.; Namba, Y.; Jin, J.; Watanabe, J.; Okabe, T.; Hidayat, M.; et al. Cancer-associated fibroblast migration in non-small cell lung cancers is modulated by increased integrin α11 expression. Mol. Oncol. 2021, 15, 1507–1527. [Google Scholar] [CrossRef]
- PIK3CA Gene—Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PIK3CA (accessed on 7 October 2022).
- Zhang, Y.; Ng, P.K.-S.; Kucherlapati, M.; Chen, F.; Liu, Y.; Tsang, Y.H.; De Velasco, G.; Jeong, K.J.; Akbani, R.; Hadjipanayis, A.; et al. A pan-cancer proteogenomic atlas of PI3K/AKT/mTOR pathway alterations. Cancer Cell. 2017, 31, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Thorpe, L.M.; Yuzugullu, H.; Zhao, J.J. PI3K in cancer: Divergent roles of isoforms, modes of activation and therapeutic targeting. Nat. Rev. Cancer. 2015, 15, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Giudice, F.S.; Squarize, C.H. The determinants of head and neck cancer: Unmasking the PI3K pathway mutations. J. Carcinog. Mutagen. 2013, 3. [Google Scholar] [CrossRef]
- Tokunaga, R.; Xiu, J.; Johnston, C.; Goldberg, R.M.; Philip, P.A.; Seeber, A.; Naseem, M.; Lo, J.H.; Arai, H.; Battaglin, F.; et al. Molecular profiling of appendiceal adenocarcinoma and comparison with right-sided and left-sided colorectal cancer. Clin. Cancer Res. 2019, 25, 3096–3103. [Google Scholar] [CrossRef] [PubMed]
- Samuels, Y.; Waldman, T. Oncogenic mutations of PIK3CA in human cancers. Curr. Top. Microbiol. Immunol. 2010, 347, 21–41. [Google Scholar] [CrossRef]
- PIK3C2B Gene—Phosphatidylinositol-4-Phosphate 3-Kinase Catalytic Subunit Type 2 Beta. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PIK3C2B (accessed on 7 October 2022).
- PTEN Gene—Phosphatase and Tensin Homolog. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PTEN (accessed on 7 October 2022).
- Belletti, B.; Baldassarre, G. Roles of CDKN1B in cancer? Aging 2015, 7, 529–530. [Google Scholar] [CrossRef]
- CDKN1B Gene—Cyclin Dependent Kinase Inhibitor 1B. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CDKN1B (accessed on 7 October 2022).
- Chan, S.H.; Chiang, J.; Ngeow, J. CDKN2A germline alterations and the relevance of genotype-phenotype associations in cancer predisposition. Hered. Cancer Clin. Pract. 2021, 19, 21. [Google Scholar] [CrossRef]
- CDKN2A Gene—Cyclin Dependent Kinase Inhibitor 2A. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CDKN2A (accessed on 7 October 2022).
- Yun, J.; Li, Y.; Xu, C.T.; Pan, B.R. Epidemiology and Rb1 gene of retinoblastoma. Int. J. Ophthalmol. 2011, 4, 103–109. [Google Scholar] [CrossRef]
- Lan, X.; Xu, W.; Tang, X.; Ye, H.; Song, X.; Lin, L.; Ren, X.; Yu, G.; Zhang, H.; Wu, S. Spectrum of RB1 germline mutations and clinical features in unrelated chinese patients with retinoblastoma. Front. Genet. 2020, 11, 142. [Google Scholar] [CrossRef]
- Knudsen, E.S.; Nambiar, R.; Rosario, S.R.; Smiraglia, D.J.; Goodrich, D.W.; Witkiewicz, A.K. Pan-cancer molecular analysis of the RB tumor suppressor pathway. Commun. Biol. 2020, 3, 158. [Google Scholar] [CrossRef]
- RB1 Gene—RB Transcriptional Corepressor 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=RB1 (accessed on 14 October 2022).
- JAK3 Gene—Janus Kinase 3. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=JAK3 (accessed on 14 October 2022).
- Nairismägi, M.; Gerritsen, M.E.; Li, Z.M.; Wijaya, G.C.; Chia, B.K.H.; Laurensia, Y.; Lim, J.Q.; Yeoh, K.W.; Yao, X.S.; Pang, W.L.; et al. Oncogenic activation of JAK3-STAT signaling confers clinical sensitivity to PRN371, a novel selective and potent JAK3 inhibitor, in natural killer/T-cell lymphoma. Leukemia 2018, 32, 1147–1156. [Google Scholar] [CrossRef]
- Vadivel, C.K.; Gluud, M.; Torres-Rusillo, S.; Boding, L.; Willerslev-Olsen, A.; Buus, T.B.; Nielsen, T.K.; Persson, J.L.; Bonefeld, C.M.; Geisler, C.; et al. JAK3 is expressed in the nucleus of malignant T cells in cutaneous T cell lymphoma (CTCL). Cancers 2021, 13, 280. [Google Scholar] [CrossRef]
- APC Gene—APC Regulator of WNT Signaling Pathway. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=APC (accessed on 14 October 2022).
- Palacio-Rúa, K.A.; Isaza-Jiménez, L.F.; Ahumada-Rodríguez, E.; Muñetón-Peña, C.M. Genetic analysis in APC, KRAS, and TP53 in patients with stomach and colon cancer. Rev. Gastroenterol. Mex. 2014, 79, 79–89. [Google Scholar] [CrossRef]
- Zhang, L.; Shay, J.W. Multiple roles of APC and its therapeutic implications in colorectal cancer. J. Natl. Cancer Inst. 2017, 109, djw332. [Google Scholar] [CrossRef]
- AXIN1 Gene—Axin 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=AXIN1 (accessed on 14 October 2022).
- Schalken, J.A. Editorial comment on: Mutations in the AXIN1 gene in advanced prostate cancer. Eur. Urol. 2009, 56, 494. [Google Scholar] [CrossRef]
- Picco, G.; Petti, C.; Centonze, A.; Torchiaro, E.; Crisafulli, G.; Novara, L.; Acquaviva, A.; Bardelli, A.; Medico, E. Loss of AXIN1 drives acquired resistance to WNT pathway blockade in colorectal cancer cells carrying RSPO3 fusions. EMBO Mol. Med. 2017, 9, 293–303. [Google Scholar] [CrossRef]
- TCF7L2 Gene—Transcription Factor 7 like 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TCF7L2 (accessed on 14 October 2022).
- Wenzel, J.; Rose, K.; Haghighi, E.B.; Lamprecht, C.; Rauen, G.; Freihen, V.; Kesselring, R.; Boerries, M.; Hecht, A. Loss of the nuclear Wnt pathway effector TCF7L2 promotes migration and invasion of human colorectal cancer cells. Oncogene 2020, 39, 3893–3909. [Google Scholar] [CrossRef]
- Mitroi, A.F.; Leopa, N.; Dumitru, E.; Brînzan, C.; Tocia, C.; Dumitru, A.; Popescu, R.C. Association of TCF7L2, CASC8 and GREM1 polymorphisms in patients with colorectal cancer and type II diabetes mellitus. Genes 2022, 13, 1297. [Google Scholar] [CrossRef]
- RHOA Gene—Ras Homolog Family Member A. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=RHOA (accessed on 14 October 2022).
- Schaefer, A.; Der, C.J. RHOA takes the RHOad less traveled to cancer. Trends Cancer 2022, 8, 655–669. [Google Scholar] [CrossRef]
- Jeong, D.; Park, S.; Kim, H.; Kim, C.J.; Ahn, T.S.; Bae, S.B.; Kim, H.J.; Kim, T.H.; Im, J.; Lee, M.S.; et al. RhoA is associated with invasion and poor prognosis in colorectal cancer. Int. J. Oncol. 2016, 48, 714–722. [Google Scholar] [CrossRef]
- Kalpana, G.; Figy, C.; Yeung, M.; Yeung, K.C. Reduced RhoA expression enhances breast cancer metastasis with a concomitant increase in CCR5 and CXCR4 chemokines signaling. Sci. Rep. 2019, 9, 16351. [Google Scholar] [CrossRef] [PubMed]
- GNA11 Gene—G Protein Subunit Alpha 11. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GNA11 (accessed on 14 October 2022).
- Jansen, P.; Müller, H.; Lodde, G.C.; Zaremba, A.; Möller, I.; Sucker, A.; Paschen, A.; Esser, S.; Schaller, J.; Gunzer, M.; et al. GNA14, GNA11, and GNAQ mutations are frequent in benign but not malignant cutaneous vascular tumors. Front. Genet. 2021, 12, 663272. [Google Scholar] [CrossRef] [PubMed]
- GNAS Gene—GNAS Complex Locus. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=GNAS (accessed on 14 October 2022).
- Turan, S.; Bastepe, M. The GNAS complex locus and human diseases associated with loss-of-function mutations or epimutations within this imprinted gene. Horm. Res. Paediatr. 2013, 80, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Zhu, L.; Cui, Z.; Tang, J.; Xie, M.; Ren, G. Elevated expression of GNAS promotes breast cancer cell proliferation and migration via the PI3K/AKT/Snail1/E-cadherin axis. Clin. Transl. Oncol. 2019, 21, 1207–1219. [Google Scholar] [CrossRef] [PubMed]
- Iyer, N.G.; Ozdag, H.; Caldas, C. p300/CBP and cancer. Oncogene 2004, 23, 4225–4231. [Google Scholar] [CrossRef]
- EP300 Gene—E1A Binding Protein P300. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=EP300 (accessed on 14 October 2022).
- CREBBP Gene—CREB Binding Protein. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CREBBP (accessed on 14 October 2022).
- Tang, Z.; Yu, W.; Zhang, C.; Zhao, S.; Yu, Z.; Xiao, X.; Tang, R.; Xuan, Y.; Yang, W.; Hao, J.; et al. CREB-binding protein regulates lung cancer growth by targeting MAPK and CPSF4 signaling pathway. Mol. Oncol. 2016, 10, 317–329. [Google Scholar] [CrossRef]
- Durbin, A.D.; Wang, T.; Wimalasena, V.K.; Zimmerman, M.W.; Li, D.; Dharia, N.V.; Mariani, L.; Shendy, N.A.M.; Nance, S.; Patel, A.G.; et al. EP300 selectively controls the enhancer landscape of MYCN-amplified Nneuroblastoma. Cancer Discov. 2022, 12, 730–751. [Google Scholar] [CrossRef]
- Bi, Y.; Kong, P.; Zhang, L.; Cui, H.; Xu, X.; Chang, F.; Yan, T.; Li, J.; Cheng, C.; Song, B.; et al. EP300 as an oncogene correlates with poor prognosis in esophageal squamous carcinoma. J. Cancer 2019, 10, 5413–5426. [Google Scholar] [CrossRef]
- Rezaei, F.M.; Hashemzadeh, S.; Gavgani, R.R.; Feizi, M.H.; Pouladi, N.; Kafil, H.S.; Rostamizadeh, L.; Oskooei, V.K.; Taheri, M.; Sakhinia, E. Dysregulated KDR and FLT1 gene expression in colorectal cancer patients. Rep. Biochem. Mol. Biol. 2019, 8, 244–252. [Google Scholar]
- KDR Gene—Kinase Insert Domain Receptor. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=KDR (accessed on 14 October 2022).
- Antonescu, C.R.; Yoshida, A.; Guo, T.; Chang, N.E.; Zhang, L.; Agaram, N.P.; Qin, L.X.; Brennan, M.F.; Singer, S.; Maki, R.G. KDR activating mutations in human angiosarcomas are sensitive to specific kinase inhibitors. Cancer Res. 2009, 69, 7175–7179. [Google Scholar] [CrossRef]
- Wang, F.; Liu, G. Influence of KDR genetic variation on the effectiveness and safety of bevacizumab in the first-line treatment for patients with advanced colorectal cancer. Int. J. Gen. Med. 2022, 15, 5651–5659. [Google Scholar] [CrossRef]
- Nilsson, M.B.; Giri, U.; Gudikote, J.; Tang, X.; Lu, W.; Tran, H.; Fan, Y.; Koo, A.; Diao, L.; Tong, P.; et al. KDR amplification is associated with VEGF-induced activation of the mTOR and invasion pathways but does not predict clinical benefit to the VEGFR TKI vandetanib. Clin. Cancer Res. 2016, 22, 1940–1950. [Google Scholar] [CrossRef]
- Geng, N.; Su, J.; Liu, Z.; Ding, C.; Xie, S.; Hu, W. The influence of KDR genetic variation on the efficacy and safety of patients with advanced NSCLC receiving first-line bevacizumab plus chemotherapy regimen. Technol. Cancer Res. Treat. 2021, 20, 15330338211019433. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, P.; Liang, X.; Xu, J.; Liu, X.; Wu, Y.; Zhang, J.; Wang, W.; Zhang, F.; Guo, R. Association of KDR mutation with better clinical outcomes in pan-cancer for immune checkpoint inhibitors. Am. J. Cancer Res. 2022, 12, 1766–1783. [Google Scholar] [CrossRef]
- NOTCH1 Gene—Notch Receptor 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=NOTCH1 (accessed on 14 October 2022).
- Anusewicz, D.; Orzechowska, M.; Bednarek, A.K. Notch signaling pathway in cancer-review with bioinformatic analysis. Cancers 2021, 13, 768. [Google Scholar] [CrossRef]
- NOTCH3 Gene—Notch Receptor 3. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=NOTCH3 (accessed on 14 October 2022).
- Xiu, M.; Wang, Y.; Li, B.; Wang, X.; Xiao, F.; Chen, S.; Zhang, L.; Zhou, B.; Hua, F. The role of Notch3 signaling in cancer stemness and chemoresistance: Molecular mechanisms and targeting strategies. Front. Mol. Biosci. 2021, 8, 694141. [Google Scholar] [CrossRef]
- NOTCH4 Gene—Notch Receptor 4. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=NOTCH4 (accessed on 14 October 2022).
- Zhou, B.; Lin, W.; Long, Y.; Yang, Y.; Zhang, H.; Wu, K.; Chu, Q. Notch signaling pathway: Architecture, disease, and therapeutics. Signal Transduct. Target. Ther. 2022, 7, 95. [Google Scholar] [CrossRef]
- D’Assoro, A.B.; Leon-Ferre, R.; Braune, E.B.; Lendahl, U. Roles of Notch signaling in the tumor microenvironment. Int. J. Mol. Sci. 2022, 23, 6241. [Google Scholar] [CrossRef]
- FLT1 Gene—Fms Related Receptor Tyrosine Kinase 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FLT1 (accessed on 14 October 2022).
- Zhang, S.D.; McCrudden, C.M.; Meng, C.; Lin, Y.; Kwok, H.F. The significance of combining VEGFA, FLT1, and KDR expressions in colon cancer patient prognosis and predicting response to bevacizumab. OncoTargets Ther. 2015, 8, 835–843. [Google Scholar] [CrossRef]
- Vander Ark, A.; Cao, J.; Li, X. TGF-β receptors: In and beyond TGF-β signaling. Cell. Signal. 2018, 52, 112–120. [Google Scholar] [CrossRef]
- Bharathy, S.; Xie, W.; Yingling, J.M.; Reiss, M. Cancer-associated transforming growth factor beta type II receptor gene mutant causes activation of bone morphogenic protein-Smads and invasive phenotype. Cancer Res. 2008, 68, 1656–1666. [Google Scholar] [CrossRef] [PubMed]
- TGFBR2 Gene—Transforming Growth Factor Beta Receptor 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TGFBR2 (accessed on 14 October 2022).
- Li, T.; Wang, H.; Xu, J.; Li, C.; Zhang, Y.; Wang, G.; Liu, Y.; Cai, S.; Fang, W.; Li, J.; et al. TGFBR2 mutation predicts resistance to immune checkpoint inhibitors in patients with non-small cell lung cancer. Ther. Adv. Med. Oncol. 2021, 13, 17588359211038477. [Google Scholar] [CrossRef] [PubMed]
- TGFBR1 Gene—Transforming Growth Factor Beta Receptor 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TGFBR1 (accessed on 14 October 2022).
- Wang, Y.Q.; Qi, X.W.; Wang, F.; Jiang, J.; Guo, Q.N. Association between TGFBR1 polymorphisms and cancer risk: A meta-analysis of 35 case-control studies. PLoS ONE 2012, 7, e42899. [Google Scholar] [CrossRef] [PubMed]
- Pasche, B.; Pennison, M.J.; Jimenez, H.; Wang, M. TGFBR1 and cancer susceptibility. Trans. Am. Clin. Climatol. Assoc. 2014, 125, 300–312. [Google Scholar] [PubMed]
- Zhou, R.; Huang, Y.; Cheng, B.; Wang, Y.; Xiong, B. TGFBR1*6A is a potential modifier of migration and invasion in colorectal cancer cells. Oncol. Lett. 2018, 15, 3971–3976. [Google Scholar] [CrossRef] [PubMed]
- Attisano, L.; Lee-Hoeflich, S.T. The Smads. Genome Biol. 2001, 2, REVIEWS3010. [Google Scholar] [CrossRef]
- SMAD2 Gene—SMAD Family Member 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SMAD2 (accessed on 14 October 2022).
- Samanta, D.; Datta, P.K. Alterations in the Smad pathway in human cancers. Front. Biosci. 2012, 17, 1281–1293. [Google Scholar] [CrossRef]
- Fleming, N.I.; Jorissen, R.N.; Mouradov, D.; Christie, M.; Sakthianandeswaren, A.; Palmieri, M.; Day, F.; Li, S.; Tsui, C.; Lipton, L.; et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013, 73, 725–735. [Google Scholar] [CrossRef]
- SMAD3 Gene—SMAD Family Member 3. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SMAD3 (accessed on 14 October 2022).
- SMAD4 Gene—SMAD Family Member 4. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SMAD4 (accessed on 14 October 2022).
- Bertrand-Chapel, A.; Caligaris, C.; Fenouil, T.; Savary, C.; Aires, S.; Martel, S.; Huchedé, P.; Chassot, C.; Chauvet, V.; Cardot-Ruffino, V.; et al. SMAD2/3 mediate oncogenic effects of TGF-β in the absence of SMAD4. Commun. Biol. 2022, 5, 1068. [Google Scholar] [CrossRef]
- Manou, D.; Caon, I.; Bouris, P.; Triantaphyllidou, I.E.; Giaroni, C.; Passi, A.; Karamanos, N.K.; Vigetti, D.; Theocharis, A.D. The complex interplay between extracellular matrix and cells in tissues. Methods Mol Biol. 2019, 1952, 1–20. [Google Scholar] [CrossRef]
- Karamanos, N.K.; Theocharis, A.D.; Piperigkou, Z.; Manou, D.; Passi, A.; Skandalis, S.S.; Vynios, D.H.; Orian-Rousseau, V.; Ricard-Blum, S.; Schmelzer, C.E.H.; et al. A guide to the composition and functions of the extracellular matrix. FEBS J. 2021, 288, 6850–6912. [Google Scholar] [CrossRef]
- Theocharis, A.D.; Gialeli, C.; Bouris, P.; Giannopoulou, E.; Skandalis, S.S.; Aletras, A.J.; Iozzo, R.V.; Karamanos, N.K. Cell-matrix interactions: Focus on proteoglycan-proteinase interplay and pharmacological targeting in cancer. FEBS J. 2014, 281, 5023–5042. [Google Scholar] [CrossRef]
- Kai, F.; Drain, A.P.; Weaver, V.M. The extracellular matrix modulates the metastatic journey. Dev. Cell. 2019, 49, 332–346. [Google Scholar] [CrossRef]
- Acerbi, I.; Cassereau, L.; Dean, I.; Shi, Q.; Au, A.; Park, C.; Chen, Y.Y.; Liphardt, J.; Hwang, E.S.; Weaver, V.M. Human breast cancer invasion and aggression correlates with ECM stiffening and immune cell infiltration. Integr. Biol 2015, 7, 1120–1134. [Google Scholar] [CrossRef]
- Miroshnikova, Y.A.; Rozenberg, G.I.; Cassereau, L.; Pickup, M.; Mouw, J.K.; Ou, G.; Templeman, K.L.; Hannachi, E.I.; Gooch, K.J.; Sarang-Sieminski, A.L.; et al. α5β1-Integrin promotes tension-dependent mammary epithelial cell invasion by engaging the fibronectin synergy site. Mol. Biol. Cell 2017, 28, 2958–2977. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Chen, S.; Yuan, W.; Fan, Q.; Tian, J.; Wang, X.; Chen, L.; Zhang, X.; Wei, W.; Liu, R.; et al. Oriented collagen fibers direct tumor cell intravasation. Proc. Natl. Acad. Sci. USA 2016, 113, 11208–11213. [Google Scholar] [CrossRef]
- Goddard, E.T.; Bozic, I.; Riddell, S.R.; Ghajar, C.M. Dormant tumour cells, their niches and the influence of immunity. Nat. Cell Biol. 2018, 20, 1240–1249. [Google Scholar] [CrossRef]
- COL5A3 Gene—Collagen Type V Alpha 3 Chain. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=COL5A3 (accessed on 17 October 2022).
- COL6A3 Gene—Collagen Type VI Alpha 3 Chain. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=COL6A3 (accessed on 17 October 2022).
- ZNF469 Gene—Zinc Finger Protein 469. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ZNF4692022 (accessed on 30 August 2022).
- ATRX Gene—ATRX Chromatin Remodeler. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ATRX (accessed on 17 October 2022).
- BRCA1 Gene—BRCA1 DNA Repair Associated. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=BRCA1 (accessed on 17 October 2022).
- BRCA2 Gene—BRCA2 DNA Repair Associated. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=BRCA2 (accessed on 17 October 2022).
- FANCA Gene—FA Complementation Group A. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FANCA (accessed on 17 October 2022).
- KDM6A Gene—Lysine Demethylase 6A. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=KDM6A (accessed on 17 October 2022).
- KMT2D Gene—Lysine Methyltransferase 2D. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=KMT2D (accessed on 17 October 2022).
- RAD51C Gene—RAD51 Paralog C. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=RAD51C (accessed on 18 October 2022).
- SETD2 Gene—SET Domain Containing 2, Histone Lysine Methyltransferase. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SETD2 (accessed on 18 October 2022).
- TRPS1 Gene—Transcriptional Repressor GATA Binding 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TRPS1 (accessed on 30 August 2022).
- CDH1 Gene—Cadherin 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CDH1 (accessed on 17 October 2022).
- CSMD1 Gene—CUB and Sushi Multiple Domains 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CSMD1 (accessed on 17 October 2022).
- CTNNA1 Gene—Catenin Alpha 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CTNNA1 (accessed on 17 October 2022).
- CTNNB1 Gene—Catenin Beta 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CTNNB1 (accessed on 17 October 2022).
- CNTNAP2 Gene—Contactin Associated Protein 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CNTNAP2 (accessed on 17 October 2022).
- LAMA1 Gene—Laminin Subunit Alpha 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=LAMA1 (accessed on 18 October 2022).
- SPTA1 Gene—Spectrin Alpha, Erythrocytic 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SPTA1 (accessed on 18 October 2022).
- ALK Gene—ALK Receptor Tyrosine Kinase. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ALK (accessed on 17 October 2022).
- ARID1A Gene—AT-Rich Interaction Domain 1A. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ARID1A (accessed on 17 October 2022).
- ARID2 Gene—AT-Rich Interaction Domain 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ARID2 (accessed on 17 October 2022).
- ATM Gene—ATM Serine/Threonine Kinase. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ATM (accessed on 17 October 2022).
- CARD11 Gene—Caspase Recruitment Domain Family Member 11. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CARD11 (accessed on 17 October 2022).
- DCLK1 Gene—Doublecortin Like Kinase 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=DCLK1 (accessed on 17 October 2022).
- DIS3 Gene—DIS3 Homolog, Exosome Endoribonuclease and 3′-5′ Exoribonuclease. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=DIS3 (accessed on 17 October 2022).
- FH Gene—Fumarate Hydratase. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FH (accessed on 17 October 2022).
- IDH2 Gene—Isocitrate Dehydrogenase (NADP+) 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=IDH2 (accessed on 17 October 2022).
- PRKACA Gene—Protein Kinase CAMP-Activated Catalytic Subunit Alpha. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PRKACA (accessed on 18 October 2022).
- PTPN11 Gene—Protein Tyrosine Phosphatase Non-Receptor Type 11. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PTPN11 (accessed on 18 October 2022).
- RHPN2 Gene—Rhophilin Rho GTPase Binding Protein 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=RHPN2 (accessed on 18 October 2022).
- SMARCA4 Gene—SWI/SNF Related, Matrix Associated, Actin Dependent Regulator of Chromatin, Subfamily A, Member 4. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SMARCA4 (accessed on 18 October 2022).
- TRRAP Gene—Transformation/Transcription Domain Associated Protein. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TRRAP (accessed on 30 August 2022).
- USP9X Gene—Ubiquitin Specific Peptidase 9 X-Linked. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=USP9X (accessed on 30 August 2022).
- ABCA7 Gene—ATP Binding Cassette Subfamily A Member 7. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ABCA7 (accessed on 17 October 2022).
- ANKRD24 Gene—Ankyrin Repeat Domain 24. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ANKRD24 (accessed on 17 October 2022).
- APOB Gene—Apolipoprotein B. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=APOB (accessed on 17 October 2022).
- ASXL1 Gene—ASXL Transcriptional Regulator 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=ASXL1 (accessed on 17 October 2022).
- BCOR Gene—BCL6 Corepressor. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=BCOR (accessed on 17 October 2022).
- CRY2 Gene—Cryptochrome Circadian Regulator 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=CRY2 (accessed on 17 October 2022).
- DOCK3—Dedicator of Cytokinesis 3. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=DOCK3 (accessed on 17 October 2022).
- DOK6 Gene—Docking Protein 6. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=DOK6 (accessed on 17 October 2022).
- EEF1A1 Gene—Eukaryotic Translation Elongation Factor 1 Alpha 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=EEF1A1 (accessed on 17 October 2022).
- EPHA10 Gene—EPH Receptor A10. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=EPHA10 (accessed on 17 October 2022).
- FAT1 Gene—FAT Atypical Cadherin 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAT1 (accessed on 17 October 2022).
- FAT4 Gene—FAT Atypical Cadherin 4. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FAT4 (accessed on 17 October 2022).
- FBXW7 Gene—F-Box and WD Repeat Domain Containing 7. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FBXW7 (accessed on 17 October 2022).
- IRX6 Gene—Iroquois Homeobox 6. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=IRX6 (accessed on 17 October 2022).
- KRT37 Gene—Keratin 37. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=KRT37 (accessed on 18 October 2022).
- MED12 Gene—Mediator Complex Subunit 12. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MED12 (accessed on 18 October 2022).
- MTIF2 Gene—Mitochondrial Translational Initiation Factor 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MTIF2 (accessed on 18 October 2022).
- MUC16 Gene—Mucin 16, Cell Surface Associated. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=MUC16 (accessed on 18 October 2022).
- Shibahara, H.; Higashi, M.; Yokoyama, S.; Rousseau, K.; Kitazono, I.; Osako, M.; Shirahama, H.; Tashiro, Y.; Kurumiya, Y.; Narita, M.; et al. A comprehensive expression analysis of mucins in appendiceal carcinoma in a multicenter study: MUC3 is a novel prognostic factor. PLoS ONE 2014, 9, e115613. [Google Scholar] [CrossRef]
- OCA2 Gene—OCA2 Melanosomal Transmembrane Protein. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=OCA2 (accessed on 18 October 2022).
- PCDH10 Gene—Protocadherin 10. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PCDH10 (accessed on 18 October 2022).
- PCDH17 Gene—Protocadherin 17. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PCDH17 (accessed on 18 October 2022).
- POM121L12 Gene—POM121 Transmembrane Nucleoporin Like 12. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=POM121L12 (accessed on 18 October 2022).
- PRDM1 Gene—PR/SET Domain 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PRDM1 (accessed on 18 October 2022).
- PTCHD3 Gene—Patched Domain Containing 3 (Gene/Pseudogene). Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=PTCHD3 (accessed on 18 October 2022).
- RNF43 Gene—Ring Finger Protein 43. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=RNF43 (accessed on 18 October 2022).
- SNTG1 Gene—Syntrophin Gamma 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=SNTG1 (accessed on 18 October 2022).
- SOX9 Gene—SRY-Box Transcription Factor 9. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=2022 (accessed on 18 October 2022).
- STK11 Gene—Serine/Threonine Kinase 11. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=STK11 (accessed on 30 August 2022).
- TSC1 Gene—TSC Complex Subunit 1. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TSC12022 (accessed on 30 August 2022).
- TSC2 Gene—TSC Complex Subunit 2. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TSC2 (accessed on 30 August 2022).
- Iacopetta, B.; Grieu, F.; Amanuel, B. Microsatellite instability in colorectal cancer. Asia Pac. J. Clin. Oncol. 2010, 6, 260–269. [Google Scholar] [CrossRef]
- Amato, M.; Franco, R.; Facchini, G.; Addeo, R.; Ciardiello, F.; Berretta, M.; Vita, G.; Sgambato, A.; Pignata, S.; Caraglia, M.; et al. Microsatellite Instability: From the Implementation of the Detection to a Prognostic and Predictive Role in Cancers. Int. J. Mol. Sci. 2022, 23, 8726. [Google Scholar] [CrossRef]
- Bonneville, R.; Krook, M.A.; Chen, H.Z.; Smith, A.; Samorodnitsky, E.; Wing, M.R.; Reeser, J.W.; Roychowdhury, S. Detection of Microsatellite Instability Biomarkers via Next-Generation Sequencing. Methods Mol. Biol. 2020, 2055, 119–132. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Luo, H.; Huang, L.; Luo, H.; Zhu, X. Microsatellite instability: A review of what the oncologist should know. Cancer Cell Int. 2020, 20, 16. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-Valentin, M.; Sampson, J.R.; Seppälä, T.T.; Ten Broeke, S.W.; Plazzer, J.P.; Nakken, S.; Engel, C.; Aretz, S.; Jenkins, M.A.; Sunde, L.; et al. Cancer risks by gene, age, and gender in 6350 carriers of pathogenic mismatch repair variants: Findings from the Prospective Lynch Syndrome Database. Genet. Med. 2020, 22, 1569. [Google Scholar] [CrossRef] [PubMed]
- Bucksch, K.; Zachariae, S.; Aretz, S.; Büttner, R.; Holinski-Feder, E.; Holzapfel, S.; Hüneburg, R.; Kloor, M.; von Knebel Doeberitz, M.; Morak, M.; et al. Cancer risks in Lynch syndrome, Lynch-like syndrome, and familial colorectal cancer type X: A prospective cohort study. BMC Cancer. 2020, 20, 460. [Google Scholar] [CrossRef]
- Morales-Miranda, A.; Rosado, I.D.; Núñez, C.C.; Montero, F.C. Appendiceal carcinoma associated with microsatellite instability. Mol. Clin. Oncol. 2018, 8, 694–698. [Google Scholar] [CrossRef]
- Taggart, M.W.; Galbincea, J.; Mansfield, P.F.; Fournier, K.F.; Royal, R.E.; Overman, M.J.; Rashid, A.; Abraham, S.C. High-level microsatellite instability in appendiceal carcinomas. Am. J. Surg. Pathol. 2013, 37, 1192–1200. [Google Scholar] [CrossRef]
- Bonneville, R.; Krook, M.A.; Kautto, E.A.; Miya, J.; Wing, M.R.; Chen, H.Z.; Reeser, J.W.; Yu, L.; Roychowdhury, S. Landscape of microsatellite instability across 39 cancer types. JCO Precis. Oncol. 2017, 2017. [Google Scholar] [CrossRef]
- Misdraji, J.; Burgart, L.J.; Lauwers, G.Y. Defective mismatch repair in the pathogenesis of low-grade appendiceal mucinous neoplasms and adenocarcinomas. Mod. Pathol. 2004, 17, 1447–1454. [Google Scholar] [CrossRef]
- Komm, M.; Kronawitter-Fesl, M.; Kremer, M.; Lutz, L.; Holinski-Feder, E.; Kopp, R. Primary mucinous adenocarcinoma of the vermiform appendix with high grade microsatellite instability. J. Cancer 2011, 2, 302–306. [Google Scholar] [CrossRef]
- Emile, S.H.; Horesh, N.; Garoufalia, Z.; Gefen, R.; Zhou, P.; Wexner, S.D. The Prognostic Impact of Microsatellite Instability on the Outcome of Appendiceal Adenocarcinoma: A National Cancer Database Analysis. J. Gastrointest. Surg. 2023, 27, 354–362. [Google Scholar] [CrossRef]
- Pandya, K.; Overman, M.J.; Gulhati, P. Molecular Landscape of Small Bowel Adenocarcinoma. Cancers 2022, 14, 1287. [Google Scholar] [CrossRef]
- Davison, J.M.; Hartman, D.A.; Singhi, A.D.; Choudry, H.A.; Ahrendt, S.A.; Zureikat, A.H.; Ramalingam, L.; Nikiforova, M.; Pai, R.K. Loss of SMAD4 protein expression is associated with high tumor grade and poor prognosis in disseminated appendiceal mucinous neoplasms. Am. J. Surg. Pathol. 2014, 38, 583–592. [Google Scholar] [CrossRef]
- Singhi, A.D.; Foxwell, T.J.; Nason, K.; Cressman, K.L.; McGrath, K.M.; Sun, W.; Bahary, N.; Zeh, H.J.; Levy, R.M.; Luketich, J.D.; et al. Smad4 loss in esophageal adenocarcinoma is associated with an increased propensity for disease recurrence and poor survival. Am. J. Surg. Pathol. 2015, 39, 487–495. [Google Scholar] [CrossRef]
- Maru, D.; Wu, T.T.; Canada, A.; Houlihan, P.S.; Hamilton, S.R.; Rashid, A. Loss of chromosome 18q and DPC4 (Smad4) mutations in appendiceal adenocarcinomas. Oncogene 2004, 23, 859–864. [Google Scholar] [CrossRef]
- Roncati, L.; Gasparri, P.; Gallo, G.; Bernardelli, G.; Zanelli, G.; Manenti, A. Appendix Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1226, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.; Wu, S.; Hamid, M.; Mahipal, A.; Cjakrabarti, S.; Bajor, D.; Selfridge, J.E.; Asa, S.L. Management of Appendix Neuroendocrine Neoplasms: Insights on the Current Guidelines. Cancers 2022, 15, 295. [Google Scholar] [CrossRef]
- Alhadid, D.; AlShammari, A.; Almana, H.; Aburahmah, M. Missed Gastric Cancer Metastasis to the Appendix: Case Report and Literature Review. Am. J. Case. Rep. 2020, 21, e920010. [Google Scholar] [CrossRef]
- Kondo, T. Current status and future outlook for patient-derived cancer models from a rare cancer research perspective. Cancer Sci. 2021, 112, 953–961. [Google Scholar] [CrossRef]
| First Mutated Gene | Second Mutated Gene | Relation |
|---|---|---|
| Early-onset appendiceal cancers | ||
| P53 | GNAS | Mutual exclusivity |
| SOX9 | KRAS/TP53 | Mutual exclusivity |
| SMAD4 | PIK3CA | Co-occurrence |
| Late-onset appendiceal cancers | ||
| GNAS | KRAS | Co-occurrence |
| PI3KCA | APC/KRAS | Co-occurrence |
| TP53 | KRAS | Co-occurrence |
| GNAS | TP53 | Mutual exclusivity |
| SMAD4 | TP53/ATM | Co-occurrence |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Constantin, M.; Mătanie, C.; Petrescu, L.; Bolocan, A.; Andronic, O.; Bleotu, C.; Mitache, M.M.; Tudorache, S.; Vrancianu, C.O. Landscape of Genetic Mutations in Appendiceal Cancers. Cancers 2023, 15, 3591. https://doi.org/10.3390/cancers15143591
Constantin M, Mătanie C, Petrescu L, Bolocan A, Andronic O, Bleotu C, Mitache MM, Tudorache S, Vrancianu CO. Landscape of Genetic Mutations in Appendiceal Cancers. Cancers. 2023; 15(14):3591. https://doi.org/10.3390/cancers15143591
Chicago/Turabian StyleConstantin, Marian, Cristina Mătanie, Livia Petrescu, Alexandra Bolocan, Octavian Andronic, Coralia Bleotu, Mihaela Magdalena Mitache, Sorin Tudorache, and Corneliu Ovidiu Vrancianu. 2023. "Landscape of Genetic Mutations in Appendiceal Cancers" Cancers 15, no. 14: 3591. https://doi.org/10.3390/cancers15143591
APA StyleConstantin, M., Mătanie, C., Petrescu, L., Bolocan, A., Andronic, O., Bleotu, C., Mitache, M. M., Tudorache, S., & Vrancianu, C. O. (2023). Landscape of Genetic Mutations in Appendiceal Cancers. Cancers, 15(14), 3591. https://doi.org/10.3390/cancers15143591









