Completion of Genetic Testing and Incidence of Pathogenic Germline Mutation among Patients with Early-Onset Colorectal Cancer: A Single Institution Analysis
Abstract
Simple Summary
Abstract
1. Introduction
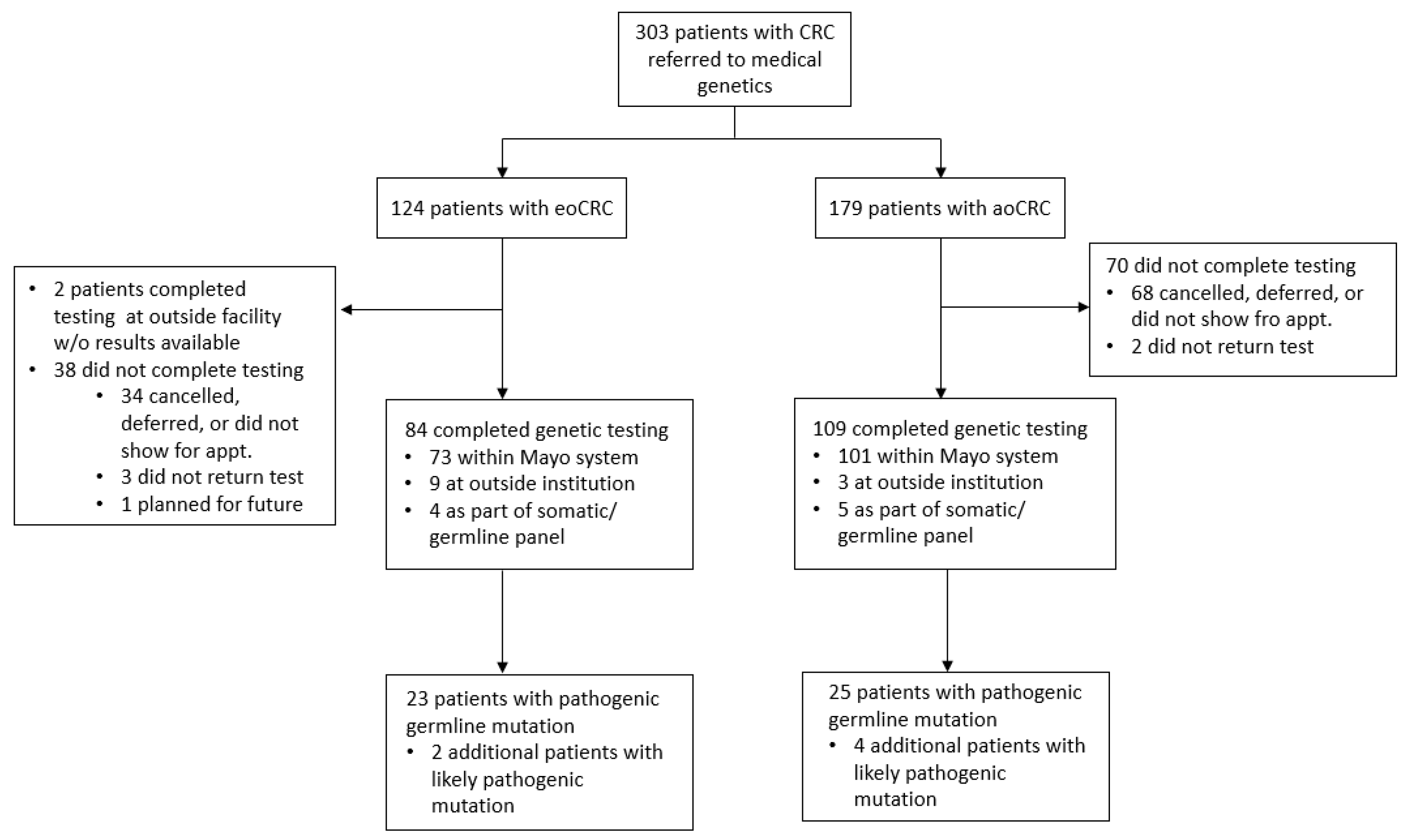
2. Materials and Methods
3. Results
3.1. Patient Demographics and Tumor Characteristics
3.2. Tumor Somatic Mutations
3.3. Patients with eoCRC Who Completed Germline Testing
3.4. Pathogenic Germline Variants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Araghi, M.; Soerjomataram, I.; Bardot, A.; Ferlay, J.; Cabasag, C.J.; Morrison, D.S.; De, P.; Tervonen, H.; Walsh, P.M.; Bucher, O.; et al. Changes in Colorectal Cancer Incidence in Seven High-Income Countries: A Population-Based Study. Lancet Gastroenterol. Hepatol. 2019, 4, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Torre, L.A.; Soerjomataram, I.; Hayes, R.B.; Bray, F.; Weber, T.K.; Jemal, A. Global Patterns and Trends in Colorectal Cancer Incidence in Young Adults. Gut 2019, 68, 2179–2185. [Google Scholar] [CrossRef] [PubMed]
- Zaborowski, A.M.; Abdile, A.; Adamina, M.; Aigner, F.; D’Allens, L.; Allmer, C.; Álvarez, A.; Anula, R.; Andric, M.; Atallah, S.; et al. Characteristics of Early-Onset vs Late-Onset Colorectal Cancer: A Review. JAMA Surg. 2021, 156, 865–874. [Google Scholar] [CrossRef]
- Sinicrope, F.A. Increasing Incidence of Early-Onset Colorectal Cancer. N. Engl. J. Med. 2022, 386, 1547–1558. [Google Scholar] [CrossRef]
- Patel, S.G.; Karlitz, J.J.; Yen, T.; Lieu, C.H.; Boland, C.R. The Rising Tide of Early-Onset Colorectal Cancer: A Comprehensive Review of Epidemiology, Clinical Features, Biology, Risk Factors, Prevention, and Early Detection. Lancet Gastroenterol. Hepatol. 2022, 7, 262–274. [Google Scholar] [CrossRef]
- Cercek, A.; Chatila, W.K.; Yaeger, R.; Walch, H.; Fernandes, G.D.S.; Krishnan, A.; Palmaira, L.; Maio, A.; Kemel, Y.; Srinivasan, P.; et al. A Comprehensive Comparison of Early-Onset and Average-Onset Colorectal Cancers. J. Natl. Cancer Inst. 2021, 113, 1683–1692. [Google Scholar] [CrossRef]
- Chang, D.T.; Pai, R.K.; Rybicki, L.A.; Dimaio, M.A.; Limaye, M.; Jayachandran, P.; Koong, A.C.; Kunz, P.A.; Fisher, G.A.; Ford, J.M.; et al. Clinicopathologic and Molecular Features of Sporadic Early-Onset Colorectal Adenocarcinoma: An Adenocarcinoma with Frequent Signet Ring Cell Differentiation, Rectal and Sigmoid Involvement, and Adverse Morphologic Features. Mod. Pathol. 2012, 25, 1128–1139. [Google Scholar] [CrossRef]
- Liu, P.H.; Wu, K.; Ng, K.; Zauber, A.G.; Nguyen, L.H.; Song, M.; He, X.; Fuchs, C.S.; Ogino, S.; Willett, W.C.; et al. Association of Obesity with Risk of Early-Onset Colorectal Cancer Among Women. JAMA Oncol. 2019, 5, 37–44. [Google Scholar] [CrossRef]
- Akimoto, N.; Ugai, T.; Zhong, R.; Hamada, T.; Fujiyoshi, K.; Giannakis, M.; Wu, K.; Cao, Y.; Ng, K.; Ogino, S. Rising Incidence of Early-Onset Colorectal Cancer—A Call to Action. Nat. Rev. Clin. Oncol. 2021, 18, 230–243. [Google Scholar] [CrossRef]
- Davidson, K.W.; Barry, M.J.; Mangione, C.M.; Cabana, M.; Caughey, A.B.; Davis, E.M.; Donahue, K.E.; Doubeni, C.A.; Krist, A.H.; Kubik, M.; et al. Screening for Colorectal Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2021, 325, 1965–1977. [Google Scholar] [CrossRef] [PubMed]
- Jasperson, K.W.; Tuohy, T.M.; Neklason, D.W.; Burt, R.W. Hereditary and Familial Colon Cancer. Gastroenterology 2010, 138, 2044–2058. [Google Scholar] [CrossRef]
- Ma, H.; Brosens, L.A.A.; Offerhaus, G.J.A.; Giardiello, F.M.; de Leng, W.W.J.; Montgomery, E.A. Pathology and Genetics of Hereditary Colorectal Cancer. Pathology 2018, 50, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Yurgelun, M.B.; Kulke, M.H.; Fuchs, C.S.; Allen, B.A.; Uno, H.; Hornick, J.L.; Ukaegbu, C.I.; Brais, L.K.; McNamara, P.G.; Mayer, R.J.; et al. Cancer Susceptibility Gene Mutations in Individuals with Colorectal Cancer. J. Clin. Oncol. 2017, 35, 1086–1095. [Google Scholar] [CrossRef] [PubMed]
- Pearlman, R.; Frankel, W.L.; Swanson, B.; Zhao, W.; Yilmaz, A.; Miller, K.; Bacher, J.; Bigley, C.; Nelsen, L.; Goodfellow, P.J.; et al. Prevalence and Spectrum of Germline Cancer Susceptibility Gene Mutations Among Patients with Early-Onset Colorectal Cancer. JAMA Oncol. 2017, 3, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Stoffel, E.M.; Koeppe, E.; Everett, J.; Ulintz, P.; Kiel, M.; Osborne, J.; Williams, L.; Hanson, K.; Gruber, S.B.; Rozek, L.S. Germline Genetic Features of Young Individuals with Colorectal Cancer. Gastroenterology 2018, 154, 897–905.e1. [Google Scholar] [CrossRef]
- Mikaeel, R.R.; Young, J.P.; Li, Y.; Smith, E.; Horsnell, M.; Uylaki, W.; Tapia Rico, G.; Poplawski, N.K.; Hardingham, J.E.; Tomita, Y.; et al. Survey of Germline Variants in Cancer-Associated Genes in Young Adults with Colorectal Cancer. Genes Chromosomes Cancer 2022, 61, 105–113. [Google Scholar] [CrossRef]
- You, Y.N.; Moskowitz, J.; Chang, G.J.; Mork, M.; Rodriguez-Bigas, M.; Bednarski, B.K.; Messick, C.; Tillman, M.; Skibber, J.; Nguyen, S.; et al. Germline Cancer Risk Profiles of Young-Onset Colorectal Cancer Patients: Findings from a Prospective Universal Germline Testing and Tele-Genetics Program. Dis. Colon Rectum 2022, 66, 531–542. [Google Scholar] [CrossRef]
- Gupta, S.; Weiss, J.M.; Axell, L.; Burke, C.A.; Chen, L.M.; Chung, D.C.; Clayback, K.M.; Dallas, S.; Felder, S.; Giardiello, F.M.; et al. NCCN Genetic/Familial High-Risk Assessment: Colorectal Version 2.2022. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) 2023. [Google Scholar]
- Lubbe, S.J.; Di Bernardo, M.C.; Chandler, I.P.; Houlston, R.S. Clinical Implications of the Colorectal Cancer Risk Associated with MUTYH Mutation. J. Clin. Oncol. 2009, 27, 3975–3980. [Google Scholar] [CrossRef]
- Daly, M.B.; Pal, T.; Berry, M.P.; Buys, S.S.; Dickson, P.; Domchek, S.M.; Elkhanany, A.; Friedman, S.; Goggins, M.; Hutton, M.L.; et al. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 77–102. [Google Scholar] [CrossRef] [PubMed]
- Uson, P.L.S.; Riegert-Johnson, D.; Boardman, L.; Kisiel, J.; Mountjoy, L.; Patel, N.; Lizaola-Mayo, B.; Borad, M.J.; Ahn, D.; Sonbol, M.B.; et al. Germline Cancer Susceptibility Gene Testing in Unselected Patients with Colorectal Adenocarcinoma: A Multicenter Prospective Study. Clin. Gastroenterol. Hepatol. 2022, 20, e508–e528. [Google Scholar] [CrossRef] [PubMed]
- Pearlman, R.; Frankel, W.L.; Swanson, B.J.; Jones, D.; Zhao, W.; Yilmaz, A.; Miller, K.; Bacher, J.; Bigley, C.; Nelsen, L.; et al. Prospective Statewide Study of Universal Screening for Hereditary Colorectal Cancer: The Ohio Colorectal Cancer Prevention Initiative. JCO Precis. Oncol. 2021, 5, 779–791. [Google Scholar] [CrossRef] [PubMed]
| All (n = 303) | eoCRC (n = 124) | aoCRC (n = 179) | |
|---|---|---|---|
| Age, median (IQR) | 51 (44, 65) | 42 (38, 46) | 62 (52, 70.5) |
| Sex | |||
| Male | 160 (53) | 73 (59) | 87 (49) |
| Female | 143 (47) | 51 (41) | 92 (51) |
| Race | |||
| White, non-Hispanic | 274 (90) | 107 (86) | 167 (93) |
| Black | 5 (2) | 3 (2) | 1 (1) |
| Hispanic | 7 (2) | 2 (2) | 5 (3) |
| Asian | 6 (2) | 3 (2) | 3 (2) |
| Am. Indian/Alaska Native | 2 (1) | 2 (2) | 0 (0) |
| Other/Unknown | 10 (3) | 7 (6) | 3 (2) |
| BMI near diagnosis | |||
| <25 | 76 (25) | 33 (27) | 43 (24) |
| ≥25, <30 | 74 (24) | 31 (25) | 43 (24) |
| ≥30 | 120 (40) | 44 (35) | 76 (42) |
| Unknown | 33 (11) | 16 (13) | 17 (9) |
| Tumor location | |||
| Right | 118 (39) | 32 (26) | 86 (48) |
| Left | 174 (57) | 86 (69) | 88 (49) |
| Rectum/rectosigmoid | 70 (23) | 36 (29) | 34 (19) |
| Unknown | 11 (4) | 6 (5) | 5 (3) |
| Stage at diagnosis | |||
| 0 | 9 (3) | 6 (5) | 3 (2) |
| 1 | 63 (21) | 13 (10) | 50 (28) |
| 2 | 66 (22) | 25 (20) | 41 (23) |
| 3 | 95 (31) | 44 (35) | 51 (28) |
| 4 | 53 (17) | 28 (23) | 25 (14) |
| Unknown | 17 (6) | 8 (6) | 9 (5) |
| Personal history of malignancy * | |||
| All | 102 (34) | 28 (23) | 74 (41) |
| Synchronous (colon) | 6 (2) | 1 (1) | 5 (3) |
| Metachronous (colon) | 12 (4) | 6 (5) | 6 (3) |
| Breast | 20 (7) | 6 (5) | 14 (8) |
| Ovary | 3 (1) | 0 (0) | 3 (2) |
| Uterus | 6 (2) | 0 (0) | 6 (3) |
| No/unknown | 201 (66) | 96 (77) | 105 (59) |
| Family history of malignancy ** | |||
| All | 274 (90) | 112 (90) | 162 (91) |
| Colon | 140 (46) | 59 (48) | 81 (45) |
| Breast | 135 (45) | 49 (40) | 86 (48) |
| Ovary | 22 (7) | 7 (6) | 15 (8) |
| Uterus | 15 (5) | 9 (7) | 6 (3) |
| Pancreas | 28 (9) | 12 (10) | 16 (9) |
| No/unknown | 29 (10) | 12 (10) | 17 (9) |
| eoCRC (n = 124) | aoCRC (n = 179) | |
|---|---|---|
| BRAF | ||
| WT | 70 (56) | 89 (50) |
| V600E | 3 (2) | 24 (13) |
| G469R | 0 (0) | 1 (1) |
| Unknown | 51 (41) | 65 (36) |
| KRAS | ||
| WT | 45 (36) | 68 (38) |
| Mutant | 27 (22) | 34 (19) |
| Unknown | 52 (42) | 77 (43) |
| NRAS | ||
| WT | 66 (53) | 97 (54) |
| Mutant | 2 (2) | 3 (2) |
| Unknown | 56 (45) | 79 (44) |
| Mismatch repair | ||
| pMMR | 91 (73) | 109 (61) |
| dMMR | 10 (8) | 50 (28) |
| MLH1/PMS2 loss, BRAF V600E or MLH1 promoter hypermethylation | 1 (1) | 23 (13) |
| MLH1/PMS2 loss, BRAF WT and MLH1 hypermethylation negative | 4 (3) | 5 (3) |
| MLH1/PMS2 loss, without BRAF or MLH1 hypermethylation test result | 2 (2) | 12 (7) |
| Other dMMR | 3 (2) | 9 (5) |
| Unknown dMMR | 0 (0) | 1 (1) |
| Unknown | 23 (19) | 20 (11) |
| Syndrome or Cancer(s) Associated with Gene | eoCRC (n = 84) | aoCRC (n = 109) | |
|---|---|---|---|
| Any | 23 (27.4) | 25 (23) | |
| MLH1 | Lynch syndrome | 3 (3.6) | 2 (1.8) |
| MSH2 | 2 (2.4) | 3 (2.8) | |
| PMS2 | 1 (1.2) | 3 (2.8) | |
| MSH6 | 1 (1.2) | 2 (1.8) | |
| MUTYH (biallelic) | Colorectal | 2 (2.4) | 2 (1.8) |
| MUTYH (monoallelic) * | 2 (2.4) | 2 (1.8) | |
| PTEN | Colorectal, kidney, thyroid, melanoma, breast, uterus | 0 (0) | 2 (1.8) |
| APC | Familial adenomatous polyposis | 1 (1.2) | 0 (0) |
| BMPR1A | Juvenile polyposis syndrome | 1 (1.2) | 0 (0) |
| NTHL1 (monoallelic) ** | Colorectal, breast | 0 (0) | 1 (0.9) |
| TP53 | Li–Fraumeni syndrome | 1 (1.2) | 0 (0) |
| CHEK2 | Breast, colorectal | 3 (3.6) | 5 (4.6) |
| ATM | Breast, pancreas, prostate, ovary | 3 (3.6) | 2 (1.8) |
| BRCA1 | Breast, uterus, ovary, prostate, pancreas | 1 (1.2) | 0 (0) |
| PALB2 | Breast, ovary, pancreas | 1 (1.2) | 0 (0) |
| SDHA | GIST, paraganglioma, pheochromocytoma, renal cell carcinoma | 1 (1.2) | 1 (0.9) |
| MITF | Melanoma, kidney | 0 (0) | 1 (0.9) |
| HOXB13 *** | Prostate | 0 (0) | 1 (0.9) |
| Likely Pathogenic | |||
| Any | 2 (2.4) | 4 (3.7) | |
| MSH2 | Lynch syndrome | 0 (0) | 2 (1.8) |
| PMS2 | 0 (0) | 1 (0.9) | |
| FH | Kidney | 1 (1.2) | 0 (0) |
| CDH1 | Stomach, breast | 1 (1.2) | 0 (0) |
| NBN (monoallelic) | Nijmegen breakage syndrome | 0 (0) | 1 (0.9) |
| dMMR Status | n | with PGV | PGVs Observed | |
|---|---|---|---|---|
| eoCRC | MLH1/PMS2 loss, BRAF V600E or MLH1 promoter hypermethylation | 1 | 1 | CHEK2 |
| MLH1/PMS2 loss, BRAF WT and MLH1 hypermethylation negative | 4 | 1 | MUTYH (biallelic) | |
| MLH1/PMS2 loss, without BRAF or MLH1 hypermethylation test result | 2 | 0 | N/A | |
| Other dMMR | 2 | 1 | MSH2 | |
| aoCRC | MLH1/PMS2 loss, BRAF V600E or MLH1 promoter hypermethylation | 15 | 1 | HOXB13 * |
| MLH1/PMS2 loss, BRAF WT and MLH1 hypermethylation negative | 3 | 1 | MLH1 | |
| MLH1/PMS2 loss, without BRAF or MLH1 hypermethylation test result | 8 | 2 | MSH6, ATM | |
| Other dMMR | 7 | 5 | PMS2 (2), MSH2 (2), MSH6 | |
| Unknown dMMR | 1 | 0 | N/A |
| eoCRC | aoCRC | |
|---|---|---|
| dMMR tumor | 3/9 (33) | 8/34 (24) |
| Personal history of additional LS malignancy | 4/9 (44) | 8/21 (38) |
| Family history of LS malignancy | 19/61 (31) | 19/85 (22) |
| One of the above | 20/64 (31) | 25/96 (26) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Storandt, M.H.; Rogen, K.R.; Iyyangar, A.; Schnell, R.R.; Mitchell, J.L.; Hubbard, J.M.; Sinicrope, F.A.; Jatoi, A.; Mahipal, A.; Shi, Q.; et al. Completion of Genetic Testing and Incidence of Pathogenic Germline Mutation among Patients with Early-Onset Colorectal Cancer: A Single Institution Analysis. Cancers 2023, 15, 3570. https://doi.org/10.3390/cancers15143570
Storandt MH, Rogen KR, Iyyangar A, Schnell RR, Mitchell JL, Hubbard JM, Sinicrope FA, Jatoi A, Mahipal A, Shi Q, et al. Completion of Genetic Testing and Incidence of Pathogenic Germline Mutation among Patients with Early-Onset Colorectal Cancer: A Single Institution Analysis. Cancers. 2023; 15(14):3570. https://doi.org/10.3390/cancers15143570
Chicago/Turabian StyleStorandt, Michael H., Kara R. Rogen, Anushka Iyyangar, Rylie R. Schnell, Jessica L. Mitchell, Joleen M. Hubbard, Frank A. Sinicrope, Aminah Jatoi, Amit Mahipal, Qian Shi, and et al. 2023. "Completion of Genetic Testing and Incidence of Pathogenic Germline Mutation among Patients with Early-Onset Colorectal Cancer: A Single Institution Analysis" Cancers 15, no. 14: 3570. https://doi.org/10.3390/cancers15143570
APA StyleStorandt, M. H., Rogen, K. R., Iyyangar, A., Schnell, R. R., Mitchell, J. L., Hubbard, J. M., Sinicrope, F. A., Jatoi, A., Mahipal, A., Shi, Q., & Jin, Z. (2023). Completion of Genetic Testing and Incidence of Pathogenic Germline Mutation among Patients with Early-Onset Colorectal Cancer: A Single Institution Analysis. Cancers, 15(14), 3570. https://doi.org/10.3390/cancers15143570








