Molecular Subtypes and Tumor Microenvironment Characteristics of Small-Cell Lung Cancer Associated with Platinum-Resistance
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Enrollment and Sample Collection for the NCC Cohort
2.2. Gene Expression Analysis of SCLC to Identify Molecular Subtypes
2.3. Gene Expression Comparison between TP53/RB1-Mutated NSCLC and SCLC
2.4. Single-Cell Transcriptome Analysis to Dissect the SCLC TME
2.5. Survival Analysis to Identify Molecular Features Related to SCLC Outcomes
2.6. IHC
2.7. Multiplex Immunofluorescence (mIF) Staining and Analysis of the Immune Cell Population
2.8. Drug Response Screening of PDCs
2.9. Drug Susceptibility Investigation for SCLC Cell Lines and PDCs
3. Results
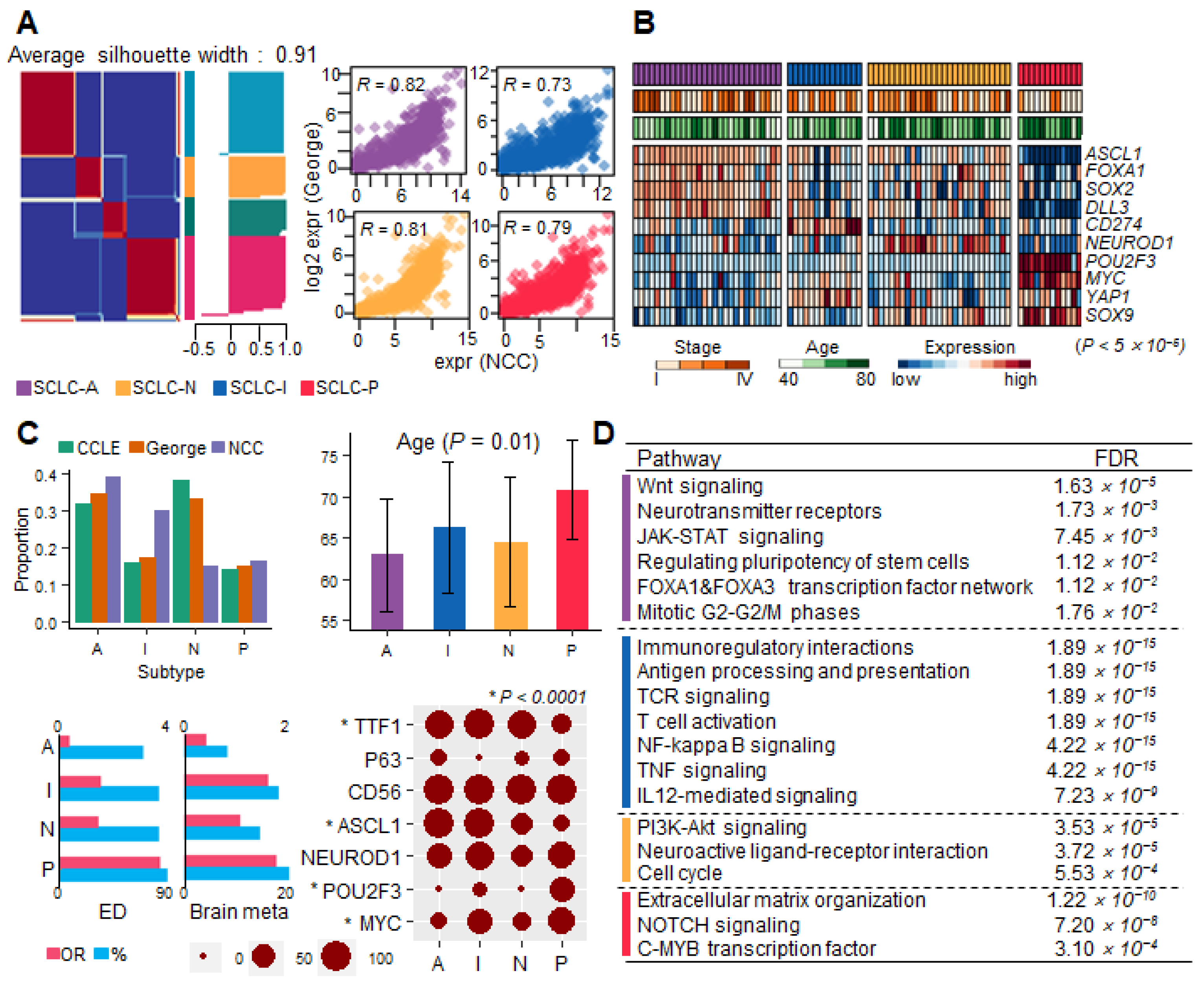
3.1. Molecular Subtypes of SCLC and Their Clinical Relevance
3.2. Early Developmental Status of SCLC Identified from TP53/RB1-Mutated NSCLC Resembles That of SCLC-A
3.3. Single-Cell Clusters to Dissect Endothelial-to-Mesenchymal Transition (EndMT) Phenotypes in SCLC-I Type
3.4. SCLC TME Cell Cluster Signatures to Predict the Worst Outcome
3.5. SCLC Molecular Features and Drug Candidates for Resistance to Platinum-Based Chemotherapy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rudin, C.M.; Brambilla, E.; Faivre-Finn, C.; Sage, J. Small-Cell Lung Cancer. Nat. Rev. Dis. Primers 2021, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Horn, L.; Mansfield, A.S.; Szczȩsna, A.; Havel, L.; Krzakowski, M.; Hochmair, M.J.; Huemer, F.; Losonczy, G.; Johnson, M.L.; Nishio, M.; et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379, 2220–2229. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, L.; Dvorkin, M.; Chen, Y.Y.H.Y.; Reinmuth, N.; Hotta, K.; Trukhin, D.; Statsenko, G.; Hochmair, M.J.; Özgüroğlu, M.; Ji, J.H.; et al. Durvalumab plus Platinum–Etoposide versus Platinum–Etoposide in First-Line Treatment of Extensive-Stage Small-Cell Lung Cancer (CASPIAN): A Randomised, Controlled, Open-Label, Phase 3 Trial. Lancet 2019, 394, 1929–1939. [Google Scholar] [CrossRef] [PubMed]
- Gay, C.M.; Stewart, C.A.; Park, E.M.; Diao, L.; Groves, S.M.; Heeke, S.; Nabet, B.Y.; Fujimoto, J.; Solis, L.M.; Lu, W.; et al. Patterns of Transcription Factor Programs and Immune Pathway Activation Define Four Major Subtypes of SCLC with Distinct Therapeutic Vulnerabilities. Cancer Cell 2021, 39, 346–360.e7. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Lee, J.; Kim, S.; Kim, S.; Youk, J.; Park, S.; An, Y.; Keam, B.; Kim, D.W.; Heo, D.S.; et al. ClonalHistory & Genetic Predictors of Transformation into Small-Cell Carcinomas from Lung Adenocarcinomas. J. Clin. Oncol. 2017, 35, 3065–3074. [Google Scholar]
- Jin, Y.; Chen, Y.; Tang, H.; Hu, X.; Hubert, S.M.; Li, Q.; Su, D.; Xu, H.; Fan, Y.; Yu, X.; et al. Activation of PI3K/AKT Pathway Is a Potential Mechanism of Treatment Resistance in Small Cell Lung Cancer. Clin. Cancer Res. 2022, 28, 526–539. [Google Scholar] [CrossRef]
- Chan, J.M.; Quintanal-Villalonga, Á.; Gao, V.R.; Xie, Y.; Allaj, V.; Chaudhary, O.; Masilionis, I.; Egger, J.; Chow, A.; Walle, T.; et al. Signatures of Plasticity, Metastasis, and Immunosuppression in an Atlas of Human Small Cell Lung Cancer. Cancer Cell 2021, 39, 1479–1496.e18. [Google Scholar] [CrossRef]
- Tlemsani, C.; Pongor, L.; Elloumi, F.; Girard, L.; Huffman, K.E.; Roper, N.; Varma, S.; Luna, A.; Rajapakse, V.N.; Sebastian, R.; et al. SCLC-CellMiner: A Resource for Small Cell Lung Cancer Cell Line Genomics and Pharmacology Based on Genomic Signatures. Cell Rep. 2020, 33, 108296. [Google Scholar] [CrossRef]
- Poirier, J.T.; George, J.; Owonikoko, T.K.; Berns, A.; Brambilla, E.; Byers, L.A.; Carbone, D.; Chen, H.J.; Christensen, C.L.; Dive, C.; et al. New Approaches to SCLC Therapy: From the Laboratory to the Clinic. J. Thorac. Oncol. 2020, 15, 520–540. [Google Scholar] [CrossRef]
- Aissa, A.F.; Islam, A.B.M.M.K.; Ariss, M.M.; Go, C.C.; Rader, A.E.; Conrardy, R.D.; Gajda, A.M.; Rubio-Perez, C.; Valyi-Nagy, K.; Pasquinelli, M.; et al. Single-Cell Transcriptional Changes Associated with Drug Tolerance and Response to Combination Therapies in Cancer. Nat. Commun. 2021, 12, 1628. [Google Scholar] [CrossRef]
- Pavan, A.; Attili, I.; Pasello, G.; Guarneri, V.; Conte, P.F.; Bonanno, L. Immunotherapy in Small-Cell Lung Cancer: From Molecular Promises to Clinical Challenges. J. Immunother. Cancer 2019, 7, 205. [Google Scholar] [CrossRef]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretia, L.; Kong, G.; Leenders, F.; Lu, X.; Fernández-Cuesta, L.; Bosco, G.; et al. Comprehensive Genomic Profiles of Small Cell Lung Cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Dawson, E.; Duong, A.; Haw, R.; Stein, L. ReactomeFIViz: A Cytoscape App for Pathway and Network-Based Data Analysis. F1000Research 2014, 3, 146. [Google Scholar] [PubMed]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene Set Variation Analysis for Microarray and RNA-Seq Data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R.; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-Generation Characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef] [PubMed]
- Waggott, D.; Chu, K.; Yin, S.; Wouters, B.G.; Liu, F.F.; Boutros, P.C. NanoStringNorm: An Extensible R Package for the Pre-Processing of Nanostring MRNA and MiRNA Data. Bioinformatics 2012, 28, 1546–1548. [Google Scholar] [CrossRef]
- Borromeo, M.D.; Savage, T.K.; Kollipara, R.K.; He, M.; Augustyn, A.; Osborne, J.K.; Girard, L.; Minna, J.D.; Gazdar, A.F.; Cobb, M.H.; et al. ASCL1 and NEUROD1 Reveal Heterogeneity in Pulmonary Neuroendocrine Tumors and Regulate Distinct Genetic Programs. Cell Rep. 2016, 16, 1259–1272. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Huang, Y.H.; Klingbeil, O.; He, X.Y.; Wu, X.S.; Arun, G.; Lu, B.; Somerville, T.D.D.; Milazzo, J.P.; Wilkinson, J.E.; Demerdash, O.E.; et al. POU2F3 Is a Master Regulator of a Tuft Cell-like Variant of Small Cell Lung Cancer. Genes. Dev. 2018, 32, 915–928. [Google Scholar] [CrossRef]
- Zhang, W.; Girard, L.; Zhang, Y.A.; Haruki, T.; Papari-Zareei, M.; Stastny, V.; Ghayee, H.K.; Pacak, K.; Oliver, T.G.; Minna, J.D.; et al. Small Cell Lung Cancer Tumors and Preclinical Models Display Heterogeneity of Neuroendocrine Phenotypes. Transl. Lung Cancer Res. 2018, 7, 32–49. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Campbell, J.D.; Brooks, A.N.; Berger, A.H.; Lee, W.; Chmielecki, J.; Beer, D.G.; Cope, L.; Creighton, C.J.; Danilova, L.; et al. Comprehensive Molecular Profiling of Lung Adenocarcinoma: The Cancer Genome Atlas Research Network. Nature 2014, 511, 543–550. [Google Scholar]
- Hammerman, P.S.; Voet, D.; Lawrence, M.S.; Voet, D.; Jing, R.; Cibulskis, K.; Sivachenko, A.; Stojanov, P.; McKenna, A.; Lander, E.S.; et al. Comprehensive Genomic Characterization of Squamous Cell Lung Cancers. Nature 2012, 489, 519–525. [Google Scholar]
- Chen, C.; Grennan, K.; Badner, J.; Zhang, D.; Gershon, E.; Jin, L.; Liu, C. Removing Batch Effects in Analysis of Expression Microarray Data: An Evaluation of Six Batch Adjustment Methods. PLoS ONE 2011, 6, e17238. [Google Scholar] [CrossRef] [PubMed]
- Butler, A.; Hoffman, P.; Smibert, P.; Papalexi, E.; Satija, R. Integrating Single-Cell Transcriptomic Data across Different Conditions, Technologies, and Species. Nat. Biotechnol. 2018, 36, 411–420. [Google Scholar] [CrossRef]
- Ireland, A.S.; Micinski, A.M.; Kastner, D.W.; Guo, B.; Wait, S.J.; Spainhower, K.B.; Conley, C.C.; Chen, O.S.; Guthrie, M.R.; Soltero, D.; et al. MYC Drives Temporal Evolution of Small Cell Lung Cancer Subtypes by Reprogramming Neuroendocrine Fate. Cancer Cell 2020, 38, 60–78.e12. [Google Scholar] [CrossRef]
- Qiu, X.; Hill, A.; Packer, J.; Lin, D.; Ma, Y.A.; Trapnell, C. Single-Cell MRNA Quantification and Differential Analysis with Census. Nat. Methods 2017, 14, 309–315. [Google Scholar] [CrossRef]
- Tsunoda, T.; Koga, H.; Yokomizo, A.; Tatsugami, K.; Eto, M.; Inokuchi, J.; Hirata, A.; Masuda, K.; Okumura, K.; Naito, S. Inositol 1,4,5-Trisphosphate (IP3) Receptor Type1 (IP3R1) Modulates the Acquisition of Cisplatin Resistance in Bladder Cancer Cell Lines. Oncogene 2005, 24, 1396–1402. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Pietanza, M.C.; Waqar, S.N.; Krug, L.M.; Dowlati, A.; Hann, C.L.; Chiappori, A.; Owonikoko, T.K.; Woo, K.M.; Cardnell, R.J.; Fujimoto, J.; et al. Randomized, Double-Blind, Phase II Study of Temozolomide in Combination with Either Veliparib or Placebo in Patients with Relapsed-Sensitive or Refractory Small-Cell Lung Cancer. J. Clin. Oncol. 2018, 36, 2386–2394. [Google Scholar] [CrossRef]
- Bid, H.K.; Phelps, D.A.; Xaio, L.; Guttridge, D.C.; Lin, J.; London, C.; Baker, L.H.; Mo, X.; Houghton, P.J. The Bromodomain BET Inhibitor JQ1 Suppresses Tumor Angiogenesis in Models of Childhood Sarcoma. Mol. Cancer Ther. 2016, 15, 1018–1028. [Google Scholar] [CrossRef] [PubMed]
- Quintanal-Villalonga, A.; Taniguchi, H.; Zhan, Y.A.; Hasan, M.M.; Chavan, S.S.; Meng, F.; Uddin, F.; Manoj, P.; Donoghue, M.T.A.; Won, H.H.; et al. Multiomic Analysis of Lung Tumors Defines Pathways Activated in Neuroendocrine Transformation. Cancer Discov. 2021, 11, 3028–3047. [Google Scholar] [CrossRef] [PubMed]
- Mollaoglu, G.; Guthrie, M.R.; Böhm, S.; Brägelmann, J.; Can, I.; Ballieu, P.M.; Marx, A.; George, J.; Heinen, C.; Chalishazar, M.D.; et al. MYC Drives Progression of Small Cell Lung Cancer to a Variant Neuroendocrine Subtype with Vulnerability to Aurora Kinase Inhibition. Cancer Cell 2017, 31, 270–285. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, H.K.; Lee, K.; Hong, Y.; Cho, J.H.; Choi, J.W.; Lee, J.I.; Suh, Y.L.; Ku, B.M.; Eum, H.H.; et al. Single-Cell RNA Sequencing Demonstrates the Molecular and Cellular Reprogramming of Metastatic Lung Adenocarcinoma. Nat. Commun. 2020, 11, 2285. [Google Scholar] [CrossRef]
- Welch-Reardon, K.M.; Wu, N.; Hughes, C.C.W. A Role for Partial Endothelial-Mesenchymal Transitions in Angiogenesis? Arter. Thromb. Vasc. Biol. 2015, 35, 303. [Google Scholar] [CrossRef] [PubMed]
- Schupp, J.C.; Adams, T.S.; Cosme, C.; Raredon, M.S.B.; Yuan, Y.; Omote, N.; Poli, S.; Chioccioli, M.; Rose, K.A.; Manning, E.P.; et al. Integrated Single-Cell Atlas of Endothelial Cells of the Human Lung. Circulation 2021, 144, 286–302. [Google Scholar] [CrossRef]
- Da Motta, L.L.; Ledaki, I.; Purshouse, K.; Haider, S.; De Bastiani, M.A.; Baban, D.; Morotti, M.; Steers, G.; Wigfield, S.; Bridges, E.; et al. The BET Inhibitor JQ1 Selectively Impairs Tumour Response to Hypoxia and Downregulates CA9 and Angiogenesis in Triple Negative Breast Cancer. Oncogene 2017, 36, 122–132. [Google Scholar] [CrossRef]
- Lenhart, R.; Kirov, S.; Desilva, H.; Cao, J.; Lei, M.; Johnston, K.; Peterson, R.; Schweizer, L.; Purandare, A.; Ross-Macdonald, P.; et al. Sensitivity of Small Cell Lung Cancer to BET Inhibition Is Mediated by Regulation of ASCL1 Gene Expression. Mol. Cancer Ther. 2015, 14, 2167–2174. [Google Scholar] [CrossRef]
- Chen, H.; Gesumaria, L.; Park, Y.-K.; Oliver, T.G.; Singer, D.S.; Ge, K.; Schrump, D.S. BET Inhibitors Target the SCLC-N Subtype of Small-Cell Lung Cancer by Blocking NEUROD1 Transactivation. Mol. Cancer Res. 2023, 21, 91–101. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Kim, S.; Park, S.-Y.; Lee, G.K.; Lim, K.Y.; Kim, J.Y.; Hwang, J.-A.; Yu, N.; Kang, E.H.; Hwang, M.; et al. Molecular Subtypes and Tumor Microenvironment Characteristics of Small-Cell Lung Cancer Associated with Platinum-Resistance. Cancers 2023, 15, 3568. https://doi.org/10.3390/cancers15143568
Kim J, Kim S, Park S-Y, Lee GK, Lim KY, Kim JY, Hwang J-A, Yu N, Kang EH, Hwang M, et al. Molecular Subtypes and Tumor Microenvironment Characteristics of Small-Cell Lung Cancer Associated with Platinum-Resistance. Cancers. 2023; 15(14):3568. https://doi.org/10.3390/cancers15143568
Chicago/Turabian StyleKim, Jihyun, Sunshin Kim, Seog-Yun Park, Geon Kook Lee, Kun Young Lim, Jin Young Kim, Jung-Ah Hwang, Namhee Yu, Eun Hye Kang, Mihwa Hwang, and et al. 2023. "Molecular Subtypes and Tumor Microenvironment Characteristics of Small-Cell Lung Cancer Associated with Platinum-Resistance" Cancers 15, no. 14: 3568. https://doi.org/10.3390/cancers15143568
APA StyleKim, J., Kim, S., Park, S.-Y., Lee, G. K., Lim, K. Y., Kim, J. Y., Hwang, J.-A., Yu, N., Kang, E. H., Hwang, M., Song, B. R., Park, C., & Han, J.-Y. (2023). Molecular Subtypes and Tumor Microenvironment Characteristics of Small-Cell Lung Cancer Associated with Platinum-Resistance. Cancers, 15(14), 3568. https://doi.org/10.3390/cancers15143568





