The Local Microbiome in Esophageal Cancer and Treatment Response: A Review of Emerging Data and Future Directions
Abstract
Simple Summary
Abstract
1. Introduction
2. Defining “Microbiome”
3. Detecting the Microbiome
4. Describing the Microbiome
5. Esophageal Microbiome
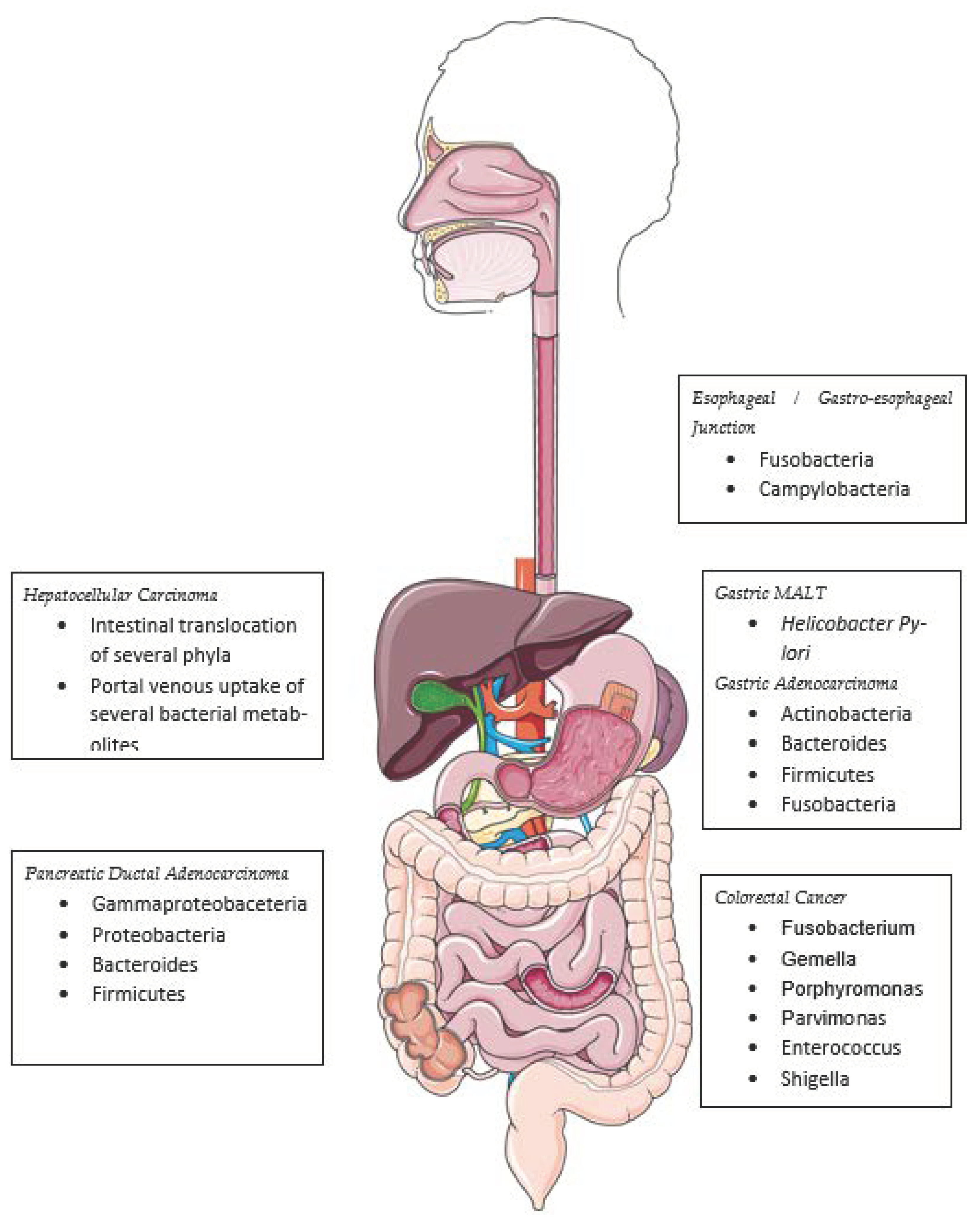
6. The Microbiome in the Carcinogenesis of Other Gastrointestinal Cancers
7. Mb Signaling, Immune Cell Interactions & Treatment Responses
7.1. Chemotherapy
7.2. Immunotherapy
7.3. Cellular Therapy
7.4. Fecal Microbiome Transplant
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hvid-Jensen, F.; Pedersen, L.; Drewes, A.M.; Sørensen, H.T.; Funch-Jensen, P. Incidence of adenocarcinoma among patients with Barrett’s esophagus. N. Engl. J. Med. 2011, 365, 1375–1383. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.M.; Shen, L.; Shah, M.A.; Enzinger, P.; Adenis, A.; Doi, T.; Kojima, T.; Metges, J.P.; Li, Z.; Kim, S.B.; et al. Pembrolizumab plus chemotherapy versus chemotherapy alone for first-line treatment of advanced oesophageal cancer (KEYNOTE-590): A randomised, placebocontrolled, phase 3 study. Lancet 2021, 398, 759–771. [Google Scholar] [CrossRef] [PubMed]
- Kelly, R.J.; Ajani, J.A.; Kuzdzal, J.; Zander, T.; Van Cutsem, E.; Piessen, G.; Mendez, G.; Feliciano, J.; Motoyama, S.; Lièvre, A.; et al. Adjuvant Nivolumab in Resected Esophageal or Gastroesophageal Junction Cancer. N. Engl. J. Med. 2021, 384, 1191–1203. [Google Scholar] [CrossRef]
- Yap, D.W.T.; Leone, A.G.; Wong, N.Z.H.; Zhao, J.J.; Tey, J.C.S.; Sundar, R.; Pietrantonio, F. Effectiveness of Immune Checkpoint Inhibitors in Patients with Advanced Esophageal Squamous Cell Carcinoma: A Meta-analysis Including Low PD-L1 Subgroups. JAMA Oncol. 2023, 9, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- Cho, I.; Blaser, M.J. The human microbiome: At the interface of health and disease. Nat. Rev. Genet. 2012, 13, 260–270. [Google Scholar] [CrossRef]
- Ahmad, A.F.; Dwivedi, G.; O’Gara, F.; Caparros-Martin, J.; Ward, N.C. The gut microbiome and cardiovascular disease: Current knowledge and clinical potential. Am. J. Physiol. Heart Circ. Physiol. 2019, 317, H923–H938. [Google Scholar] [CrossRef]
- Sittipo, P.; Lobionda, S.; Lee, Y.K.; Maynard, C.L. Intestinal microbiota and the immune system in metabolic diseases. J. Microbiol. 2018, 56, 154–162. [Google Scholar] [CrossRef]
- Holmes, A.; Finger, C.; Morales-Scheihing, D.; Lee, J.; McCullough, L.D. Gut dysbiosis and age-related neurological diseases; an innovative approach for therapeutic interventions. Transl. Res. 2020, 226, 39–56. [Google Scholar] [CrossRef]
- Peterson, D.A.; Frank, D.N.; Pace, N.R.; Gordon, J.I. Metagenomic approaches for defining the pathogenesis of inflammatory bowel diseases. Cell. Host Microbe 2008, 3, 417–427. [Google Scholar] [CrossRef] [PubMed]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Peters, B.A.; Wu, J.; Pei, Z.; Yang, L.; Purdue, M.P.; Freedman, N.D.; Jacobs, E.J.; Gapstur, S.M.; Hayes, R.B.; Ahn, J. Oral Microbiome Composition Reflects Prospective Risk for Esophageal Cancers. Cancer Res. 2017, 77, 6777–6787. [Google Scholar] [CrossRef]
- Bassis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 2015, 6, e00037. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.; Yin, J.; Zhao, J.; Ma, S.R.; Wang, H.R.; Wang, M.; Chen, W.; Wei, W.Q. Microbial Similarity and Preference for Specific Sites in Healthy Oral Cavity and Esophagus. Front. Microbiol. 2018, 9, 1603. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, N.P.; Riordan, S.M.; Gorman, C.J.; Nielsen, S.; Russell, T.L.; Correa-Ospina, C.; Fernando, B.S.M.; Waters, S.A.; Castaño-Rodríguez, N.; Man, S.M.; et al. Multi-omics of the esophageal microenvironment identifies signatures associated with progression of Barrett’s esophagus. Genome Med. 2021, 13, 133. [Google Scholar] [CrossRef] [PubMed]
- Pisani, A.; Rausch, P.; Bang, C.; Ellul, S.; Tabone, T.; Marantidis Cordina, C.; Zahra, G.; Franke, A.; Ellul, P. Dysbiosis in the Gut Microbiota in Patients with Inflammatory Bowel Disease during Remission. Microbiol. Spectr. 2022, 10, e0061622. [Google Scholar] [CrossRef]
- Liou, J.M.; Chen, C.C.; Chang, C.M.; Fang, Y.J.; Bair, M.J.; Chen, P.Y.; Chang, C.Y.; Hsu, Y.C.; Chen, M.J.; Chen, C.C.; et al. Taiwan Gastrointestinal Disease and Helicobacter Consortium. Long-term changes of gut microbiota, antibiotic resistance, and metabolic parameters after Helicobacter pylori eradication: A multicentre, open-label, randomised trial. Lancet Infect. Dis. 2019, 19, 1109–1120. [Google Scholar] [CrossRef]
- Spohn, S.N.; Young, V.B. Gastrointestinal Microbial Ecology with Perspectives on Health and Disease. In Physiology of the Gastrointestinal Tract, 6th ed.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 737–753. [Google Scholar]
- Wilmanski, T.; Rappaport, N.; Diener, C.; Gibbons, S.M.; Price, N.D. From taxonomy to metabolic output: What factors define gut microbiome health? Gut Microbes 2021, 13, 1. [Google Scholar] [CrossRef]
- Pei, Z.; Bini, E.J.; Yang, L.; Zhou, M.; Francois, F.; Blaser, M.J. Bacterial microbiome in the human distal esophagus. Proc. Natl. Acad. Sci. USA 2004, 101, 4250–4255. [Google Scholar] [CrossRef]
- Yang, L.; Lu, X.; Nossa, C.W.; Francois, F.; Peek, R.M.; Pei, Z. Inflammation and intestinal metaplasia of the distal esophagus are associated with alterations in the microbiome. Gastroenterology 2009, 137, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, A.H.; Kelly, L.A.; Kreft, R.E.; Barlek, M.; Omstead, A.N.; Matsui, D.; Boyd, N.H.; Gazarik, K.E.; Heit, M.I.; Nistico, L.; et al. Associations of microbiota and toll-like receptor signaling pathway in esophageal adenocarcinoma. BMC Cancer 2016, 16, 52. [Google Scholar] [CrossRef] [PubMed]
- Amir, I.; Konikoff, F.M.; Oppenheim, M.; Gophna, U.; Half, E.E. Gastric microbiota is altered in oesophagitis and Barrett’s oesophagus and further modified by proton pump inhibitors. Environ. Microbiol. 2014, 16, 2905–2914. [Google Scholar] [CrossRef] [PubMed]
- Elliott, D.R.F.; Walker, A.W.; O’Donovan, M.; Parkhill, J.; Fitzgerald, R.C. A non-endoscopic device to sample the oesophageal microbiota: A case-control study. Lancet Gastroenterol. Hepatol. 2017, 2, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, K.; Baba, Y.; Nakagawa, S.; Mima, K.; Miyake, K.; Nakamura, K.; Sawayama, H.; Kinoshita, K.; Ishimoto, T.; Iwatsuki, M.; et al. Human microbiome fusobacterium nucleatum in esophageal cancer tissue is associated with prognosis. Clin. Cancer Res. 2016, 22, 5574–5581. [Google Scholar] [CrossRef]
- Davakis, S.; Kapelouzou, A.; Liakakos, T.; Mpoura, M.; Stergiou, D.; Sakellariou, S.; Charalabopoulos, A. The Role of Toll-like Receptors in Esophageal Cancer. Anticancer. Res. 2022, 42, 2813–2818. [Google Scholar] [CrossRef]
- Burrell, R.A.; McClelland, S.E.; Endesfelder, D.; Groth, P.; Weller, M.-C.; Shaikh, N.; Domingo, E.; Kanu, N.; Dewhurst, S.M.; Gronroos, E.; et al. Replication stress links structural and numerical cancer chromosomal instability. Nature 2013, 494, 492–496. [Google Scholar] [CrossRef]
- Cover, T.L.; Blaser, M.J. Helicobacter pylori in health and disease. Gastroenterology 2009, 136, 1863–1873. [Google Scholar] [CrossRef]
- Hatakeyama, M. Structure and function of Helicobacter pylori CagA, the first-identified bacterial protein involved in human cancer. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017, 93, 196–219. [Google Scholar] [CrossRef]
- Wang, Z.; Gao, X.; Zeng, R.; Wu, Q.; Sun, H.; Wu, W.; Zhang, X.; Sun, G.; Yan, B.; Wu, L.; et al. Changes of the Gastric Mucosal Microbiome Associated with Histological Stages of Gastric Carcinogenesis. Front. Microbiol. 2020, 11, 997. [Google Scholar] [CrossRef]
- Nakatsu, G.; Li, X.; Zhou, H.; Sheng, J.; Wong, S.H.; Wu, W.K.; Ng, S.C.; Tsoi, H.; Dong, Y.; Zhang, N.; et al. Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat. Commun. 2015, 6, 8727. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2011, 6, 320–329. [Google Scholar] [CrossRef] [PubMed]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef]
- Xu, M.; Yamada, M.; Li, M.; Liu, H.; Chen, S.G.; Han, Y.W. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J. Biol. Chem. 2007, 282, 25000–25009. [Google Scholar] [CrossRef]
- Engevik, M.A.; Danhof, H.A.; Ruan, W.; Engevik, A.C.; Chang-Graham, A.L.; Engevik, K.A.; Shi, Z.; Zhao, Y.; Brand, C.K.; Krystofiak, E.S.; et al. Fusobacterium nucleatum secretes outer membrane vesicles and promotes intestinal inflammation. mBio 2021, 12, 12.e02706–20. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Khalyfa, A.A.; Punatar, S.; Yarbrough, A. Hepatocellular Carcinoma: Understanding the Inflammatory Implications of the Microbiome. Int. J. Mol. Sci. 2022, 23, 8164. [Google Scholar] [CrossRef]
- Schneider, K.M.; Mohs, A.; Gui, W.; Galvez, E.J.C.; Candels, L.S.; Hoenicke, L.; Muthukumarasamy, U.; Holland, C.H.; Elfers, C.; Kilic, K.; et al. Imbalanced gut microbiota fuels hepatocellular carcinoma development by shaping the hepatic inflammatory microenvironment. Nat. Commun. 2022, 13, 3964. [Google Scholar] [CrossRef]
- Pleguezuelos-Manzano, C.; Puschhof, J.; Rosendahl Huber, A.; van Hoeck, A.; Wood, H.M.; Nomburg, J.; Gurjao, C.; Manders, F.; Dalmasso, G.; Stege, P.B.; et al. Mutational signature in colorectal cancer caused by genotoxic pks(+) E. coli. Nature 2022, 580, 269–273. [Google Scholar] [CrossRef]
- Iftekhar, A.; Berger, H.; Bouznad, N.; Heuberger, J.; Boccellato, F.; Dobrindt, U.; Hermeking, H.; Sigal, M.; Meyer, T.F. Genomic aberrations after short-term exposure to colibactin-producing E. coli transform primary colon epithelial cells. Nat. Commun. 2021, 12, 1003. [Google Scholar] [CrossRef] [PubMed]
- Scanu, T.; Spaapen, R.M.; Bakker, J.M.; Pratap, C.B.; Wu, L.E.; Hofland, I.; Broeks, A.; Shukla, V.K.; Kumar, M.; Janssen, H.; et al. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell. Host Microbe 2015, 17, 763–774. [Google Scholar] [CrossRef]
- Cheng, W.T.; Kantilal, H.K.; Davamani, F. The mechanism of Bacteroides fragilis toxin contributes to colon cancer formation. Malays. J. Med. Sci. 2020, 27, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Gregory, A.C.; Zablocki, O.; Zayed, A.A.; Howell, A.; Bolduc, B.; Sullivan, M.B. The gut virome database reveals age-dependent patterns of virome diversity in the human gut. Cell. Host Microbe 2020, 28, 724–740.e8. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.Z.; Zhang, H.; Yang, J.; Zeng, J.; Wang, H. Preliminary assessment of viral metagenome from cancer tissue and blood from patients with lung adenocarcinoma. J. Med. Virol. 2021, 93, 5126–5133. [Google Scholar] [CrossRef] [PubMed]
- Kapitan, M.; Niemiec, M.J.; Steimle, A.; Frick, J.S.; Jacobsen, I.D. Fungi as part of the microbiota and interactions with intestinal bacteria. Curr. Top. Microbiol. Immunol. 2019, 422, 265–301. [Google Scholar]
- Leung, J.M.; Graham, A.L.; Knowles, S.C.L. Parasite-Microbiota interactions with the vertebrate gut: Synthesis through an ecological lens. Front. Microbiol. 2018, 9, 843. [Google Scholar] [CrossRef]
- Kalaora, S.; Nagler, A.; Wargo, J.A.; Samuels, Y. Mechanisms of immune activation and regulation: Lessons from melanoma. Nat. Rev. Cancer 2022, 22, 195–207. [Google Scholar] [CrossRef]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.A.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M.; et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef]
- Kespohl, M.; Vachharajani, N.; Luu, M.; Harb, H.; Pautz, S.; Wolff, S.; Sillner, N.; Walker, A.; Schmitt-Kopplin, P.; Boettger, T.; et al. The microbial metabolite butyrate induces expression of Th1-associated factors in CD4+ T Cells. Front. Immunol. 2017, 8, 1036. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; DeRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Mathewson, N.D.; Jenq, R.; Mathew, A.V.; Koenigsknecht, M.; Hanash, A.; Toubai, T.; Oravecz-Wilson, K.; Wu, S.-R.; Sun, Y.; Rossi, C.; et al. Gut microbiome-derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat. Immunol. 2016, 17, 505–513. [Google Scholar] [CrossRef]
- Bachem, A.; Makhlouf, C.; Binger, K.J.; de Souza, D.P.; Tull, D.; Hochheiser, K.; Whitney, P.G.; Fernandez-Ruiz, D.; Dähling, S.; Kastenmüller, W.; et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8(+) T cells. Immunity 2019, 51, 285–297.e5. [Google Scholar] [CrossRef] [PubMed]
- Ida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteriacontrol cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Viaud, S.; Daillère, R.; Boneca, I.G.; Lepage, P.; Langella, P.; Chamaillard, M.; Pittet, M.J.; Ghiringhelli, F.; Trinchieri, G.; Goldszmid, R.; et al. Gut microbiome and anticancer immune response: Really hot Sh*t! Cell. Death Differ. 2015, 22, 199–214. [Google Scholar] [CrossRef]
- Le Bastard, Q.; Ward, T.; Sidiropoulos, D.; Hillmann, B.M.; Chun, C.L.; Sadowsky, M.J.; Knights, D.; Montassier, E. Fecal microbiota transplantation reverses antibiotic and chemotherapy-induced gut dysbiosis in mice. Sci. Rep. 2018, 8, 6219. [Google Scholar] [CrossRef]
- Luchner, M.; Reinke, S.; Milicic, A. TLR agonists as vaccine adjuvants targeting cancer and infectious diseases. Pharmaceutics 2021, 13, 142. [Google Scholar] [CrossRef]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, N.; Zhou, L.; Wang, J.; Zhou, Y.; Zhang, T.; Fang, Y.; Deng, J.; Gao, Y.; Liang, X.; et al. IL-2 regulates tumor-reactive CD8+ T cell exhaustion by activating the aryl hydrocarbon receptor. Nat. Immunol. 2021, 22, 358–369. [Google Scholar] [CrossRef] [PubMed]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2021, 350, 1084–1089. [Google Scholar] [CrossRef]
- Vetizou, M.; Pitt, J.M.; Daillére, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef]
- Chaput, N.; Lepage, P.; Coutzac, C.; Soularue, E.; Le Roux, K.; Monot, C.; Boselli, L.; Routier, E.; Cassard, L.; Collins, M.; et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017, 28, 1368–1379. [Google Scholar] [CrossRef] [PubMed]
- Frankel, A.E.; Coughlin, L.A.; Kim, J.; Froehlich, T.W.; Xie, Y.; Frenkel, E.P.; Koh, A.Y. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia 2017, 19, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillére, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Tanoue, T.; Morita, S.; Plichta, D.R.; Skelly, A.N.; Suda, W.; Sugiura, Y.; Narushima, S.; Vlamakis, H.; Motoo, I.; Sugita, K.; et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 2019, 565, 600–605. [Google Scholar] [CrossRef]
- Baruch, E.N.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021, 371, 602–609. [Google Scholar] [CrossRef]
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Luu, M.; Riester, Z.; Baldrich, A.; Reichardt, N.; Yuille, S.; Busetti, A.; Klein, M.; Wempe, A.; Leister, H.; Raifer, H.; et al. Microbial short-chain fatty acids modulate CD8+ T cell responses and improve adoptive immunotherapy for cancer. Nat. Commun. 2021, 12, 4077. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.; Dai, A.; Ghilardi, G.; Amelsberg, K.V.; Devlin, S.M.; Pajarillo, R.; Slingerland, J.B.; Beghi, S.; Herrera, P.S.; Giardina, P.; et al. Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat. Med. 2022, 28, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Fillon, S.A.; Harris, J.K.; Wagner, B.D.; Kelly, C.J.; Stevens, M.J.; Moore, W.; Fang, R.; Schroeder, S.; Masterson, J.C.; Robertson, C.E.; et al. Novel Device to Sample the Esophageal Microbiome—The Esophageal String Test. PLoS ONE 2012, 7, e42938. [Google Scholar] [CrossRef]
- Zaidi, A.H.; Pratama, M.Y.; Omstead, A.N.; Gorbonova, A.; Mansoor, R.; Melton-Kreft, R.; Jobe, B.A.; Wagner, P.L.; Kelly, R.J.; Goel, A. A blood-based circulating microbial metagenomic panel for early diagnosis and prognosis of esophageal adenocarcinoma. Br. J. Cancer 2022, 127, 2016–2024. [Google Scholar] [CrossRef]
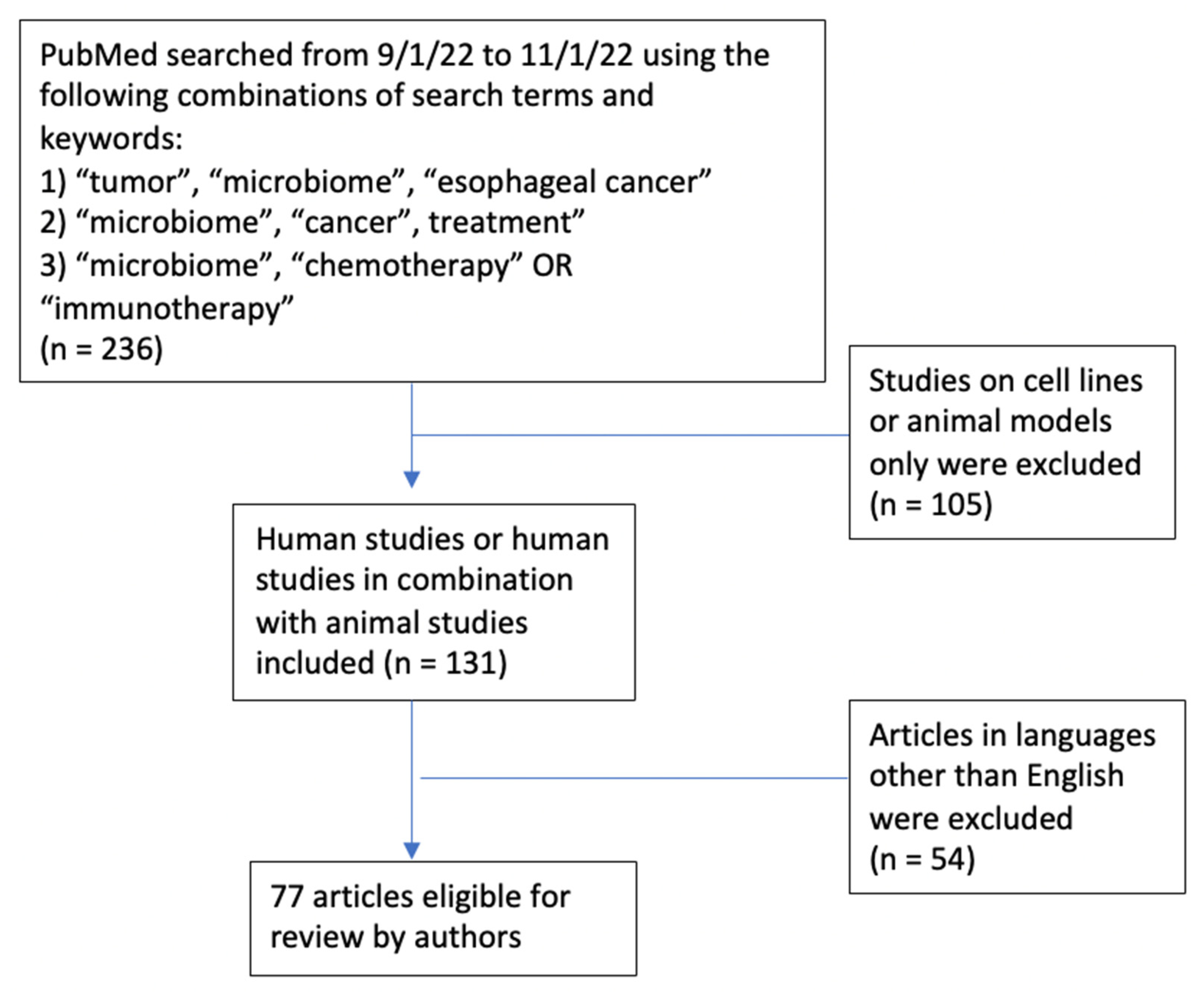
| Study | Tumor | Subjects | Immunotherapy | Methods | Findings |
|---|---|---|---|---|---|
| Chaput (2017) [65] | Metastatic Melanoma | Human | Anti-CTLA-4 | Fecal 16S rRNA, Peripheral blood immunophenotyping | Responders enriched for Faecalibacterium and Firmicutes, and with lower baseline peripheral Tregs and alpha-beta CD4 T-cells |
| Frankel (2017) [66] | Metastatic Melanoma | Human | Anti-CTLA-4, Anti-PD-1, Anti-CTLA-4 + Anti-PD-1 | Fecal metagenomics, fecal metabolomics | Responders enriched for Bacteroides and Streptococcus; MB enriched for fatty acid synthesis |
| Gopalakrishnan (2018) [67] | Metastatic Melanoma | Human, mouse | Anti-PD-1 | Oral and fecal metagenomics; tumor mutation burden; tumor-infiltrating lymphocytes, peripheral blood immunophenotypes | Responders enriched for Ruminococcaceae and Faecalibacterium, and with greater CD8+ TIL density; Bacteroides abundance associated with non-response and fewer CD8+ TILs |
| Matson (2018) [68] | Metastatic Melanoma | Human, mouse | Anti-PD-1, Anti-CTLA-4 | Fecal 16S rRNA, fecal metagenomics, fecal qPCR; FMT | Responders enriched for E. faecium, C. aerofaciens, B. adolescentis, B. lonus, K. pneumoniae, V. parvula, P. merdae; FMT from responders to germ-free mice with improved tumor control with anti-PD-1 therapy |
| Routy (2018) [69] | Non-small-cell lung carcinoma, Renal cell carcinoma/urothelial carcinoma | Human, mouse | Anti-PD-1/PD-L1 | Fecal metagenomics; FMT | ICI responders enriched for Firmicutes, Akkermansia, Alistipes; germ-free mice with better anti-tumor response after human responder FMT than non-responder FMT |
| Tanoue (2019) [70] | MC38 adenocarcinoma mouse model | Human, mouse | Anti-PD-1 | FMT of pre-specified mix of 11 bacterial strains | FMT-treated mice with better spontaneous (without ICI) and anti-PD-1 tumor responses |
| Baruch (2021) [71] | Metastatic Melanoma | Human | Anti-PD-1 | Human-to-human, ICI-responsive-to-non-responsive FMT | 3/10 non-responders salvaged anti-PD-1 tumor response |
| Davar (2021) [72] | Metastatic Melanoma | Human | Anti-PD-1 | Human-to-human, ICI-responsive-to-non-responsive FMT | 6/15 non-responders salvaged anti-PD-1 tumor response |
| MB Intervention | Cancer Treatment | Tumor Type | Study Phase | Trial ID | Status |
|---|---|---|---|---|---|
| FMT | ICI | Renal cell Carcinoma/bladder | 1 | NCT04038619 | Active, recruiting |
| FMT | ICI | Melanoma, non-small-cell lung cancer | 1 | NCT03819296 | Active, recruiting |
| FMT | ICI | Melanoma, non-small-cell lung cancer | 2 | NCT04951583 | Active, recruiting |
| FMT | ICI | Prostate | 2 | NCT04116775 | Active, recruiting |
| FMT | ICI | Renal cell carcinoma | 2 | NCT04758507 | Active, recruiting |
| FMT | Allogeneic stem cell transplant | Hematologic malignancies | 2 | NCT04935684 | Active, recruiting |
| Probiotic | ICI + chemotherapy | Non-small-cell lung cancer | 2 | NCT04699721 | Active |
| Probiotic | Chemotherapy | Colorectal cancer | 2 | NCT04131803 | Active |
| Prebiotic/Fiber | ICI | Melanoma | 2 | NCT04645680 | Active, recruiting |
| Prebiotic/Fiber | Radiotherapy | Colorectal cancer, bladder cancer, prostate cancer | 3 | NCT04534075 | Active, recruiting |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, A.; Lieu, C.H.; Kim, S.S. The Local Microbiome in Esophageal Cancer and Treatment Response: A Review of Emerging Data and Future Directions. Cancers 2023, 15, 3562. https://doi.org/10.3390/cancers15143562
Pandey A, Lieu CH, Kim SS. The Local Microbiome in Esophageal Cancer and Treatment Response: A Review of Emerging Data and Future Directions. Cancers. 2023; 15(14):3562. https://doi.org/10.3390/cancers15143562
Chicago/Turabian StylePandey, Abhishek, Christopher H. Lieu, and Sunnie S. Kim. 2023. "The Local Microbiome in Esophageal Cancer and Treatment Response: A Review of Emerging Data and Future Directions" Cancers 15, no. 14: 3562. https://doi.org/10.3390/cancers15143562
APA StylePandey, A., Lieu, C. H., & Kim, S. S. (2023). The Local Microbiome in Esophageal Cancer and Treatment Response: A Review of Emerging Data and Future Directions. Cancers, 15(14), 3562. https://doi.org/10.3390/cancers15143562






