Validation of In Vitro Trained Transcriptomic Radiosensitivity Signatures in Clinical Cohorts
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
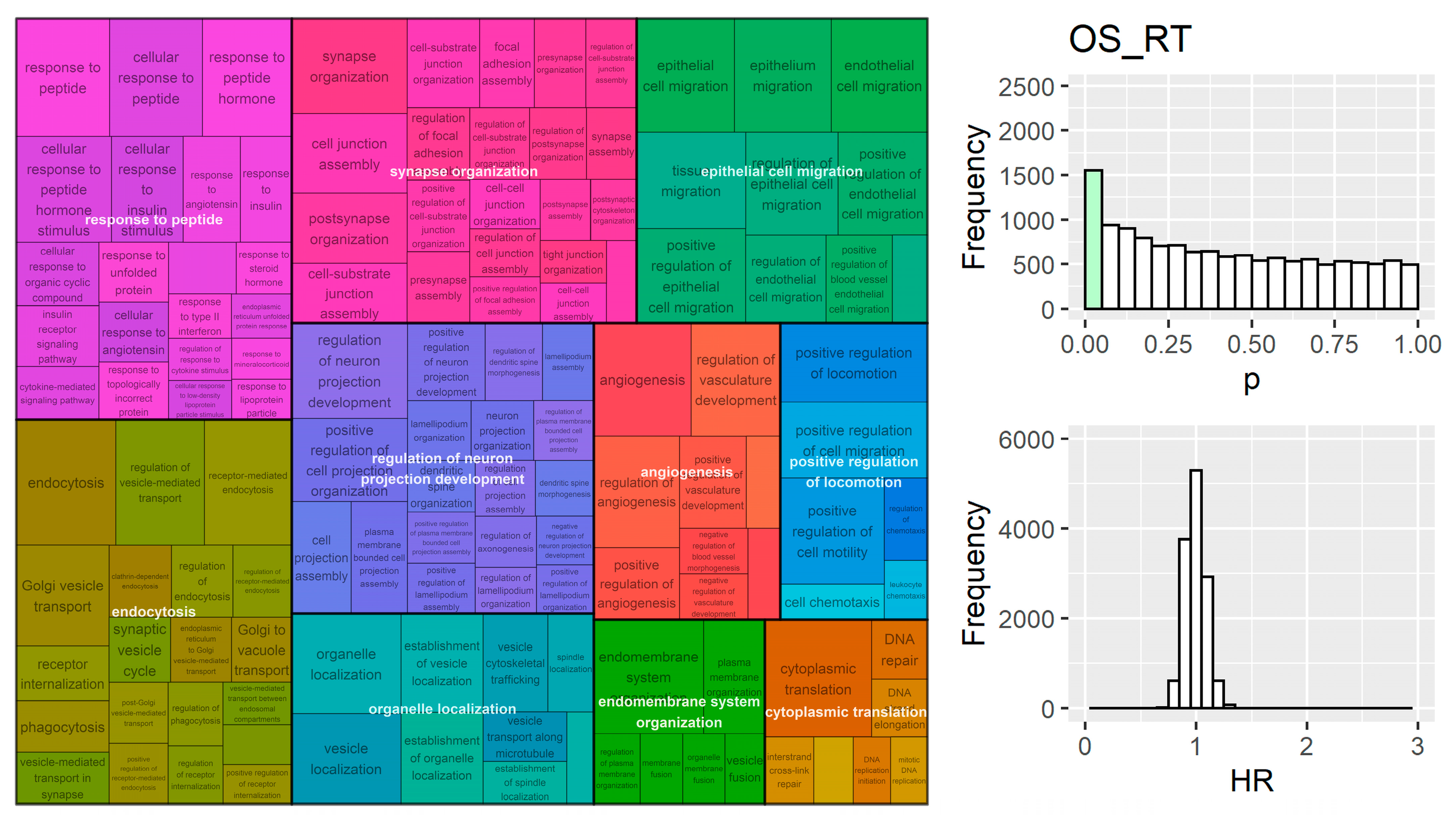
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lederman, M. The Early History of Radiotherapy: 1895–1939. Int. J. Radiat. Oncol. Biol. Phys. 1981, 7, 639–648. [Google Scholar] [CrossRef]
- Holsti, L.R. Development of Clinical Radiotherapy since 1896. Acta Oncol. 1995, 34, 995–1003. [Google Scholar] [CrossRef]
- Pitter, K.L.; Casey, D.L.; Lu, Y.C.; Hannum, M.; Zhang, Z.; Song, X.; Pecorari, I.; Mcmillan, B.; Ma, J.; Samstein, R.M.; et al. Pathogenic ATM Mutations in Cancer and a Genetic Basis for Radiotherapeutic Efficacy. J. Natl. Cancer Inst. 2021, 113, 266–273. [Google Scholar] [CrossRef]
- Sjöström, M.; Chang, S.L.; Fishbane, N.; Davicioni, E.; Zhao, S.G.; Hartman, L.; Holmberg, E.; Feng, F.Y.; Speers, C.W.; Pierce, L.J.; et al. Clinicogenomic Radiotherapy Classifier Predicting the Need for Intensified Locoregional Treatment After Breast-Conserving Surgery for Early-Stage Breast Cancer. J. Clin. Oncol. 2019, 37, 3340–3349. [Google Scholar] [CrossRef]
- Speers, C.; Zhao, S.; Liu, M.; Bartelink, H.; Pierce, L.J.; Feng, F.Y. Development and Validation of a Novel Radiosensitivity Signature in Human Breast Cancer. Clin. Cancer Res. 2015, 21, 3667–3677. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.G.; Chang, S.L.; Spratt, D.E.; Erho, N.; Yu, M.; Ashab, H.A.D.; Alshalalfa, M.; Speers, C.; Tomlins, S.A.; Davicioni, E.; et al. Development and Validation of a 24-Gene Predictor of Response to Postoperative Radiotherapy in Prostate Cancer: A Matched, Retrospective Analysis. Lancet Oncol. 2016, 17, 1612–1620. [Google Scholar] [CrossRef] [PubMed]
- Eschrich, S.; Zhang, H.; Zhao, H.; Boulware, D.; Lee, J.H.; Bloom, G.; Torres-Roca, J.F. Systems Biology Modeling of the Radiation Sensitivity Network: A Biomarker Discovery Platform. Int. J. Radiat. Oncol. Biol. Phys. 2009, 75, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Hall, J.S.; Iype, R.; Senra, J.; Taylor, J.; Armenoult, L.; Oguejiofor, K.; Li, Y.; Stratford, I.; Stern, P.L.; O’Connor, M.J.; et al. Investigation of Radiosensitivity Gene Signatures in Cancer Cell Lines. PLoS ONE 2014, 9, e86329. [Google Scholar] [CrossRef]
- O’Connor, J.D.; Overton, I.M.; McMahon, S.J. RadSigBench: A Framework for Benchmarking Functional Genomics Signatures of Cancer Cell Radiosensitivity. Brief. Bioinform. 2022, 23, bbab561. [Google Scholar] [CrossRef]
- Eschrich, S.A.; Fulp, W.J.; Pawitan, Y.; Foekens, J.A.; Smid, M.; Martens, J.W.M.; Echevarria, M.; Kamath, V.; Lee, J.H.; Harris, E.E.; et al. Validation of a Radiosensitivity Molecular Signature in Breast Cancer. Clin. Cancer Res. 2012, 18, 5134–5143. [Google Scholar] [CrossRef]
- Ahmed, K.A.; Chinnaiyan, P.; Fulp, W.J.; Eschrich, S.; Torres-Roca, J.F.; Caudell, J.J. The Radiosensitivity Index Predicts for Overall Survival in Glioblastoma. Oncotarget 2015, 6, 34414–34422. [Google Scholar] [CrossRef]
- Strom, T.; Hoffe, S.E.; Fulp, W.; Frakes, J.; Coppola, D.; Springett, G.M.; Malafa, M.P.; Harris, C.L.; Eschrich, S.A.; Torres-Roca, J.F.; et al. Radiosensitivity Index Predicts for Survival with Adjuvant Radiation in Resectable Pancreatic Cancer. Radiother. Oncol. 2015, 117, 159–164. [Google Scholar] [CrossRef]
- Strom, T.; Torres-Roca, J.F.; Parekh, A.; Naghavi, A.O.; Caudell, J.J.; Oliver, D.E.; Messina, J.L.; Khushalani, N.I.; Zager, J.S.; Sarnaik, A.; et al. Regional Radiation Therapy Impacts Outcome for Node-Positive Cutaneous Melanoma. J. Natl. Compr. Cancer Netw. 2017, 15, 473–482. [Google Scholar] [CrossRef]
- Torres-Roca, J.F.; Fulp, W.J.; Caudell, J.J.; Servant, N.; Bollet, M.A.; van de Vijver, M.; Naghavi, A.O.; Harris, E.E.; Eschrich, S.A. Integration of a Radiosensitivity Molecular Signature Into the Assessment of Local Recurrence Risk in Breast Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2015, 93, 631–638. [Google Scholar] [CrossRef]
- Scott, J.G.; Berglund, A.; Schell, M.J.; Mihaylov, I.; Fulp, W.J.; Yue, B.; Welsh, E.; Caudell, J.J.; Ahmed, K.; Strom, T.S.; et al. A Genome-Based Model for Adjusting Radiotherapy Dose (GARD): A Retrospective, Cohort-Based Study. Lancet Oncol. 2017, 18, 202–211. [Google Scholar] [CrossRef]
- Scott, J.G.; Sedor, G.; Ellsworth, P.; Scarborough, J.A.; Ahmed, K.A.; Oliver, D.E.; Eschrich, S.A.; Kattan, M.W.; Torres-Roca, J.F. Pan-Cancer Prediction of Radiotherapy Benefit Using Genomic-Adjusted Radiation Dose (GARD): A Cohort-Based Pooled Analysis. Lancet Oncol. 2021, 22, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.S.; Kim, S.C.; Kim, S.J.; Park, C.H.; Jeung, H.-C.; Kim, Y.B.; Ahn, J.B.; Chung, H.C.; Rha, S.Y. Identification of a Radiosensitivity Signature Using Integrative Metaanalysis of Published Microarray Data for NCI-60 Cancer Cells. BMC Genom. 2012, 13, 348. [Google Scholar] [CrossRef]
- Venet, D.; Dumont, J.E.; Detours, V. Most Random Gene Expression Signatures Are Significantly Associated with Breast Cancer Outcome. PLoS Comput. Biol. 2011, 7, e1002240. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Bolstad, B.M.; Collin, F.; Cope, L.M.; Hobbs, B.; Speed, T.P. Summaries of Affymetrix GeneChip Probe Level Data. Nucleic Acids Res. 2003, 31, e15. [Google Scholar] [CrossRef] [PubMed]
- McLendon, R.; Friedman, A.; Bigner, D.; Van Meir, E.G.; Brat, D.J.; Mastrogianakis, G.M.; Olson, J.J.; Mikkelsen, T.; Lehman, N.; Aldape, K.; et al. Comprehensive Genomic Characterization Defines Human Glioblastoma Genes and Core Pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Shedden, K.; Taylor, J.M.G.; Enkemann, S.A.; Tsao, M.S.; Yeatman, T.J.; Gerald, W.L.; Eschrich, S.; Jurisica, I.; Giordano, T.J.; Misek, D.E.; et al. Gene Expression-Based Survival Prediction in Lung Adenocarcinoma: A Multi-Site, Blinded Validation Study. Nat. Med. 2008, 14, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Klijn, J.G.; Zhang, Y.; Sieuwerts, A.M.; Look, M.P.; Yang, F.; Talantov, D.; Timmermans, M.; Meijer-van Gelder, M.E.; Yu, J.; et al. Gene-Expression Profiles to Predict Distant Metastasis of Lymph-Node-Negative Primary Breast Cancer. Lancet 2005, 365, 671–679. [Google Scholar] [CrossRef]
- Pawitan, Y.; Bjöhle, J.; Amler, L.; Borg, A.L.; Egyhazi, S.; Hall, P.; Han, X.; Holmberg, L.; Huang, F.; Klaar, S.; et al. Gene Expression Profiling Spares Early Breast Cancer Patients from Adjuvant Therapy: Derived and Validated in Two Population-Based Cohorts. Breast Cancer Res. 2005, 7, R953–R964. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.r-project.org/ (accessed on 23 June 2022).
- Sean, D.; Meltzer, P.S. GEOquery: A Bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [Google Scholar] [CrossRef]
- Colaprico, A.; Silva, T.C.; Olsen, C.; Garofano, L.; Cava, C.; Garolini, D.; Sabedot, T.S.; Malta, T.M.; Pagnotta, S.M.; Castiglioni, I.; et al. TCGAbiolinks: An R/Bioconductor Package for Integrative Analysis of TCGA Data. Nucleic Acids Res. 2016, 44, e71. [Google Scholar] [CrossRef]
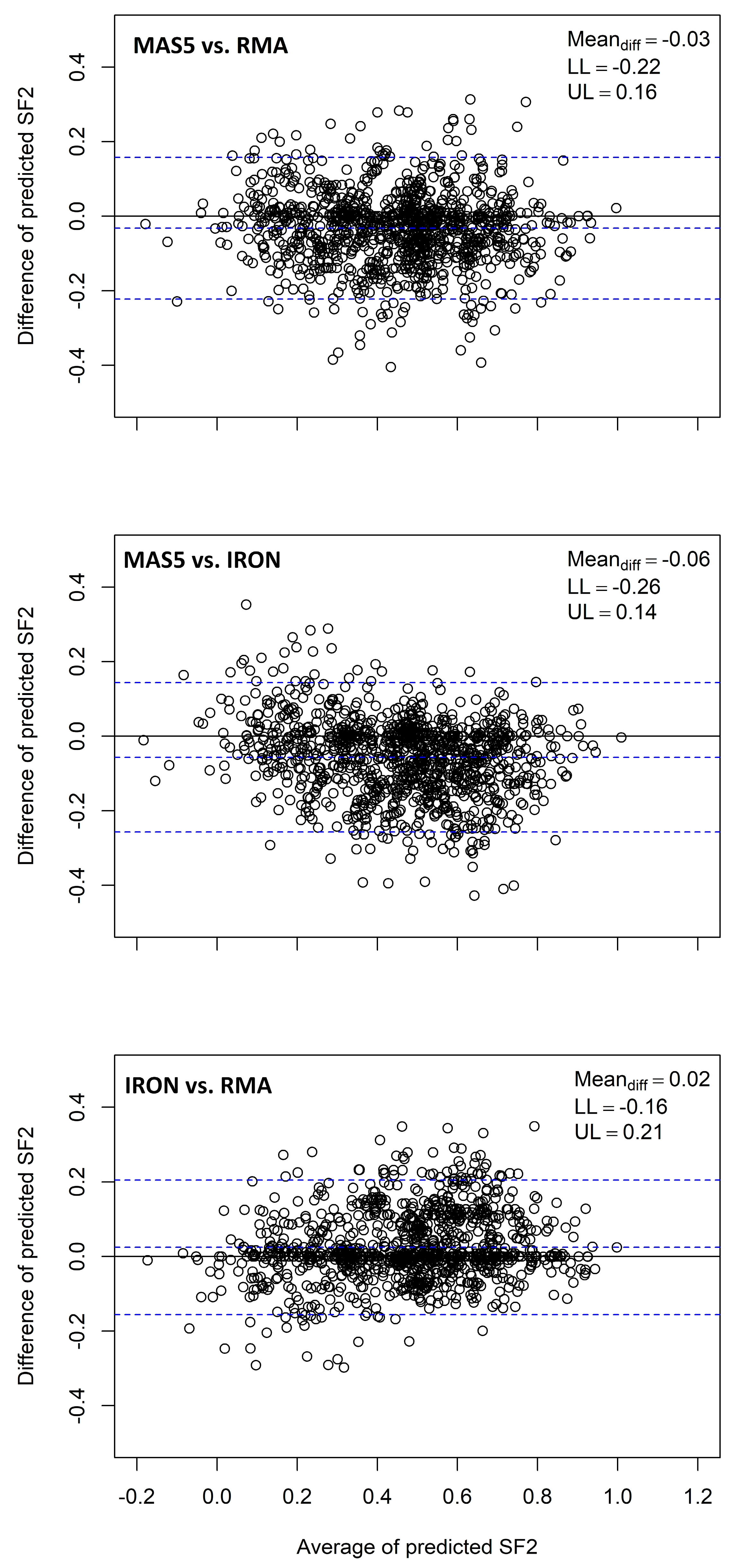
- Bland, M.J.; Altman, D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 327, 307–310. [Google Scholar] [CrossRef]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy—Analysis of Affymetrix GeneChip Data at the Probe Level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef]
- Welsh, E.A.; Eschrich, S.A.; Berglund, A.E.; Fenstermacher, D.A. Iterative Rank-Order Normalization of Gene Expression Microarray Data. BMC Bioinform. 2013, 14, 153. [Google Scholar] [CrossRef]
- Harrell, F. Rms 6.3-0: Regression Modeling Strategies. Available online: https://cran.r-project.org/package=rms (accessed on 12 September 2022).
- Klammer, A.A.; Park, C.Y.; Noble, W.S. Statistical Calibration of the SEQUEST Xcorr Function. J. Proteome Res. 2009, 8, 2106–2113. [Google Scholar] [CrossRef]
- Arnold, T.; Emerson, J. Nonparametric Goodness-of-Fit Tests for Discrete Null Distributions. R J. 2011, 3, 34. [Google Scholar] [CrossRef]
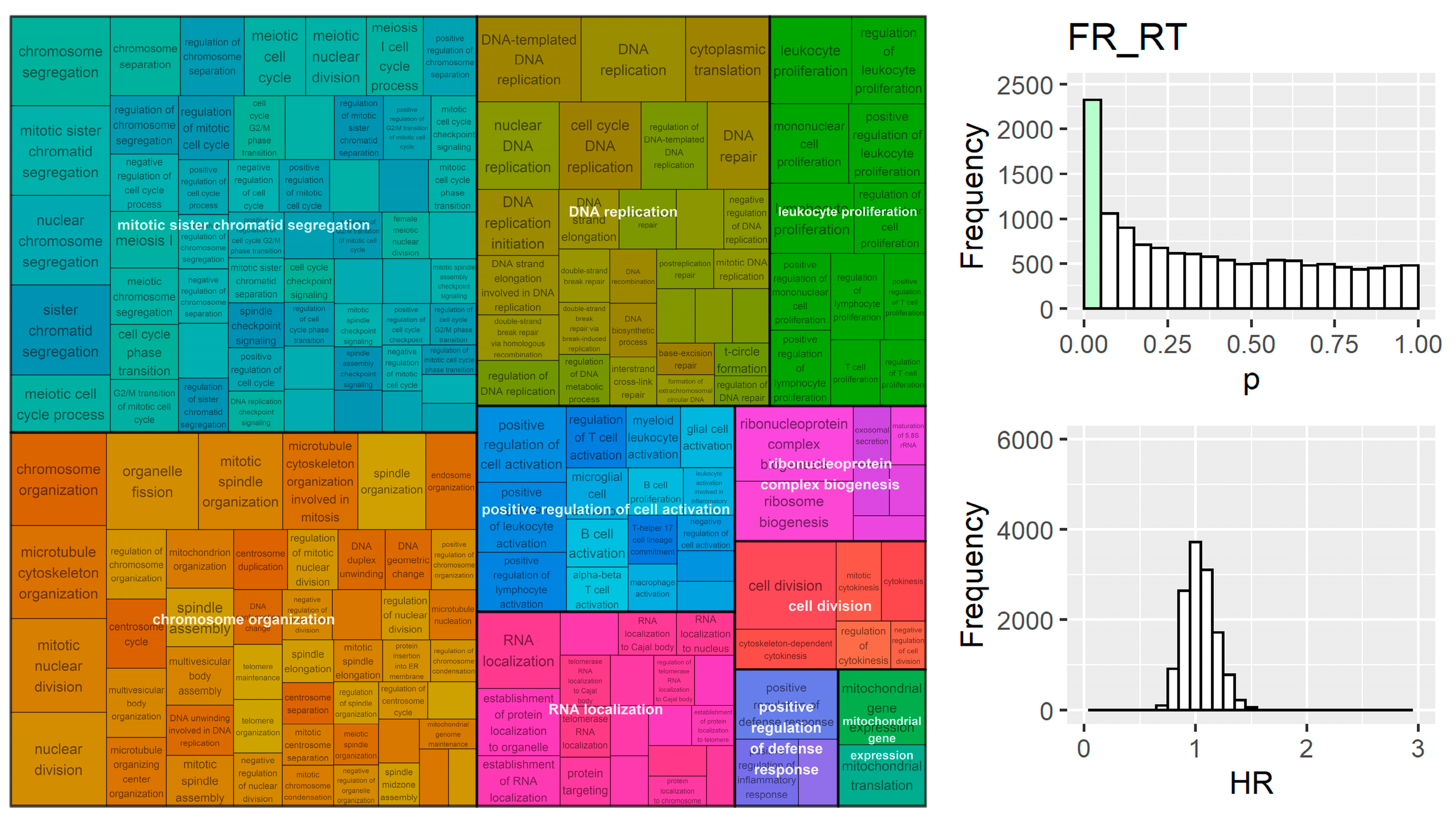
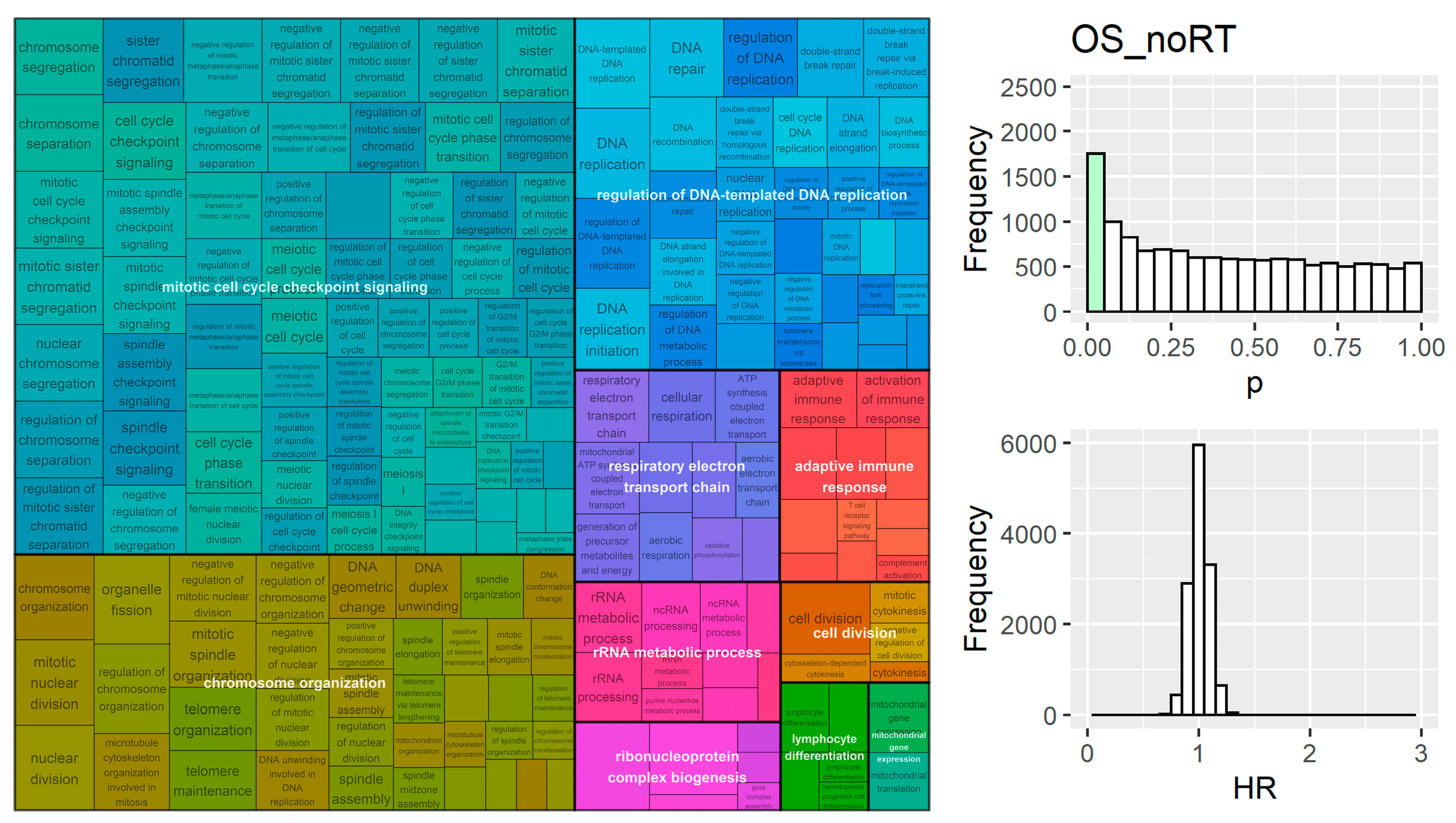
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing Yoav Benjamini and Yosef Hochberg. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar]
- Sayols, S. rrvgo: A Bioconductor package for interpreting lists of Gene Ontology terms. Micropubl. Biol. 2023, 2023. [Google Scholar] [CrossRef]
- Yu, G.; Li, F.; Qin, Y.; Bo, X.; Wu, Y.; Wang, S. GOSemSim: An R Package for Measuring Semantic Similarity among GO Terms and Gene Products. Bioinformatics 2010, 26, 976–978. [Google Scholar] [CrossRef]
- Grass, G.D.; Alfonso, J.C.L.; Welsh, E.; Ahmed, K.A.; Teer, J.K.; Pilon-Thomas, S.; Harrison, L.B.; Cleveland, J.L.; Mulé, J.J.; Eschrich, S.A.; et al. The Radiosensitivity Index Gene Signature Identifies Distinct Tumor Immune Microenvironment Characteristics Associated With Susceptibility to Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2022, 113, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Finn, O.J. Immuno-oncology: Understanding the function and dysfunction of the immune system in cancer. Ann. Oncol. 2012, 23, viii6–viii9. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, T.M.; Keyomarsi, K. Role of Cell Cycle in Mediating Sensitivity to Radiotherapy. Int. J. Radiat. Oncol. Biol. Phys. 2004, 59, 928–942. [Google Scholar] [CrossRef]
- Shoemaker, R.H. The NCI60 Human Tumour Cell Line Anticancer Drug Screen. Nat. Rev. Cancer 2006, 6, 813–823. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia Enables Predictive Modelling of Anticancer Drug Sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Nusinow, D.P.; Szpyt, J.; Ghandi, M.; Rose, C.M.; McDonald, E.R.; Kalocsay, M.; Jané-Valbuena, J.; Gelfand, E.; Schweppe, D.K.; Jedrychowski, M.; et al. Quantitative Proteomics of the Cancer Cell Line Encyclopedia. Cell 2020, 180, 387–402.e16. [Google Scholar] [CrossRef]
- Amundson, S.A.; Do, K.T.; Vinikoor, L.C.; Lee, R.A.; Koch-Paiz, C.A.; Ahn, J.; Reimers, M.; Chen, Y.; Scudiero, D.A.; Weinstein, J.N.; et al. Integrating Global Gene Expression and Radiation Survival Parameters across the 60 Cell Lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res. 2008, 68, 415–424. [Google Scholar] [CrossRef]
- Lewis, J.E.; Kemp, M.L. Integration of Machine Learning and Genome-Scale Metabolic Modeling Identifies Multi-Omics Biomarkers for Radiation Resistance. Nat. Commun. 2021, 12, 2700. [Google Scholar] [CrossRef] [PubMed]
- Manem, V.S.K.; Lambie, M.; Smith, I.; Smirnov, P.; Kofia, V.; Freeman, M.; Koritzinsky, M.; Abazeed, M.E.; Haibe-Kains, B.; Bratman, S.V. Modeling Cellular Response in Large-Scale Radiogenomic Databases to Advance Precision Radiotherapy. Cancer Res. 2019, 79, 6227–6237. [Google Scholar] [CrossRef]
- Abazeed, M.E.; Adams, D.J.; Hurov, K.E.; Tamayo, P.; Creighton, C.J.; Sonkin, D.; Giacomelli, A.O.; Du, C.; Fries, D.F.; Wong, K.K.; et al. Integrative Radiogenomic Profiling of Squamous Cell Lung Cancer. Cancer Res. 2013, 73, 6289–6298. [Google Scholar] [CrossRef] [PubMed]
- West, C.M.L.; Davidson, S.E.; Roberts, S.A.; Hunter, R.D. Intrinsic Radiosensitivity and Prediction of Patient Response to Radiotherapy for Carcinoma of the Cervix. Br. J. Cancer 1993, 68, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Stausbøl-Grøn, B.; Bentzen, S.M.; Jørgensen, K.E.; Nielsen, O.S.; Bundgaard, T.; Overgaard, J. In Vitro Radiosensitivity of Tumour Cells and Fibroblasts Derived from Head and Neck Carcinomas: Mutual Relationship and Correlation with Clinical Data. Br. J. Cancer 1999, 79, 1074–1084. [Google Scholar] [CrossRef]
- Nuryadi, E.; Permata, T.B.M.; Komatsu, S.; Oike, T.; Nakano, T. Inter-Assay Precision of Clonogenic Assays for Radiosensitivity in Cancer Cell Line A549. Oncotarget 2018, 9, 13706–13712. [Google Scholar] [CrossRef] [PubMed]
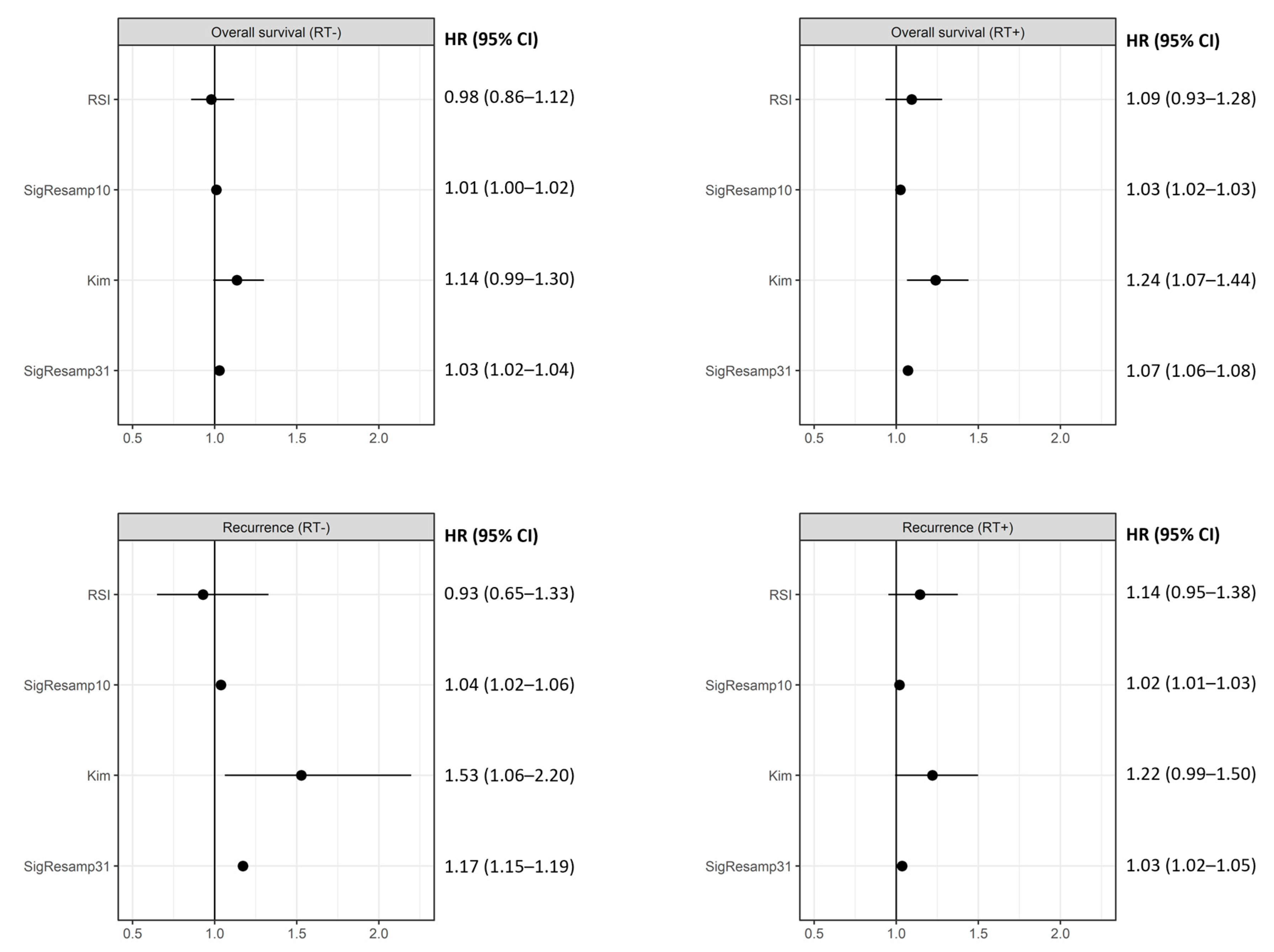


| Group | Datasets | n | Events |
|---|---|---|---|
| Overall Survival (RT+) [OS_RT] | TCGA glioblastoma | 186 | 133 |
| Karolinska breast cancer cohort | 77 | 17 | |
| LUAD cohort | 65 | 51 | |
| Overall Survival (RT-) [OS_noRT] | TCGA glioblastoma | 55 | 55 |
| Karolinska breast cancer cohort | 82 | 23 | |
| LUAD cohort | 364 | 174 | |
| Recurrence (RT+) [FR_RT] | Erasmus breast cancer cohort | 282 | 91 |
| Karolinska breast cancer cohort | 77 | 19 | |
| Recurrence (RT-) [FR_noRT] | Erasmus breast cancer cohort | 62 | 12 |
| Karolinska breast cancer cohort | 82 | 21 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
O’Connor, J.D.; Overton, I.M.; McMahon, S.J. Validation of In Vitro Trained Transcriptomic Radiosensitivity Signatures in Clinical Cohorts. Cancers 2023, 15, 3504. https://doi.org/10.3390/cancers15133504
O’Connor JD, Overton IM, McMahon SJ. Validation of In Vitro Trained Transcriptomic Radiosensitivity Signatures in Clinical Cohorts. Cancers. 2023; 15(13):3504. https://doi.org/10.3390/cancers15133504
Chicago/Turabian StyleO’Connor, John D., Ian M. Overton, and Stephen J. McMahon. 2023. "Validation of In Vitro Trained Transcriptomic Radiosensitivity Signatures in Clinical Cohorts" Cancers 15, no. 13: 3504. https://doi.org/10.3390/cancers15133504
APA StyleO’Connor, J. D., Overton, I. M., & McMahon, S. J. (2023). Validation of In Vitro Trained Transcriptomic Radiosensitivity Signatures in Clinical Cohorts. Cancers, 15(13), 3504. https://doi.org/10.3390/cancers15133504






