Bioinformatics Screen Reveals Gli-Mediated Hedgehog Signaling as an Associated Pathway to Poor Immune Infiltration of Dedifferentiated Liposarcoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
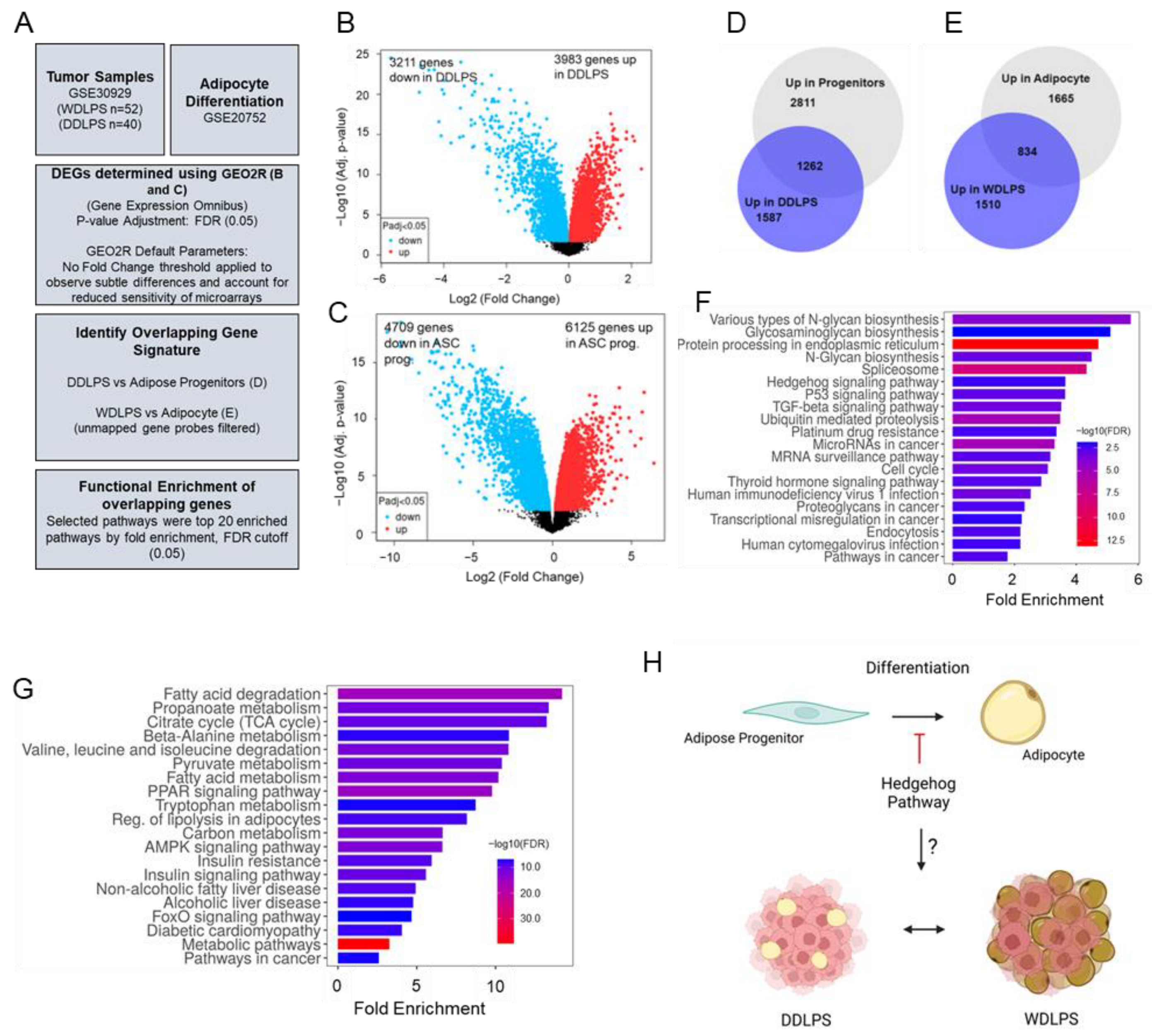
2.1. Hedgehog Signaling Is Enriched in Dedifferentiated Adipose Progenitors and DDLPS Tumors
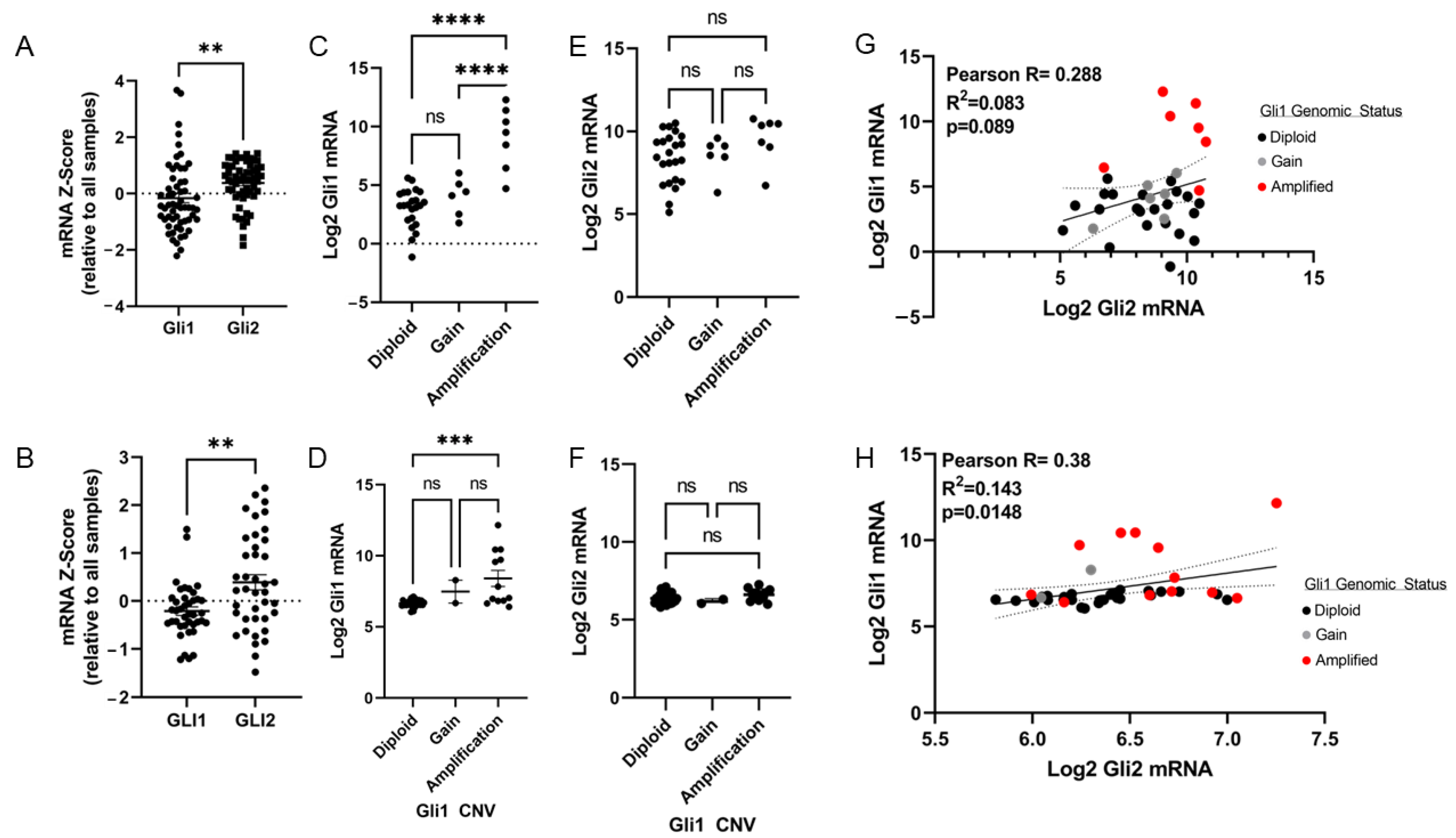
2.2. Chromosome 12q13-12q15 Amplification Increases Gli1 Expression, but Not Gli2
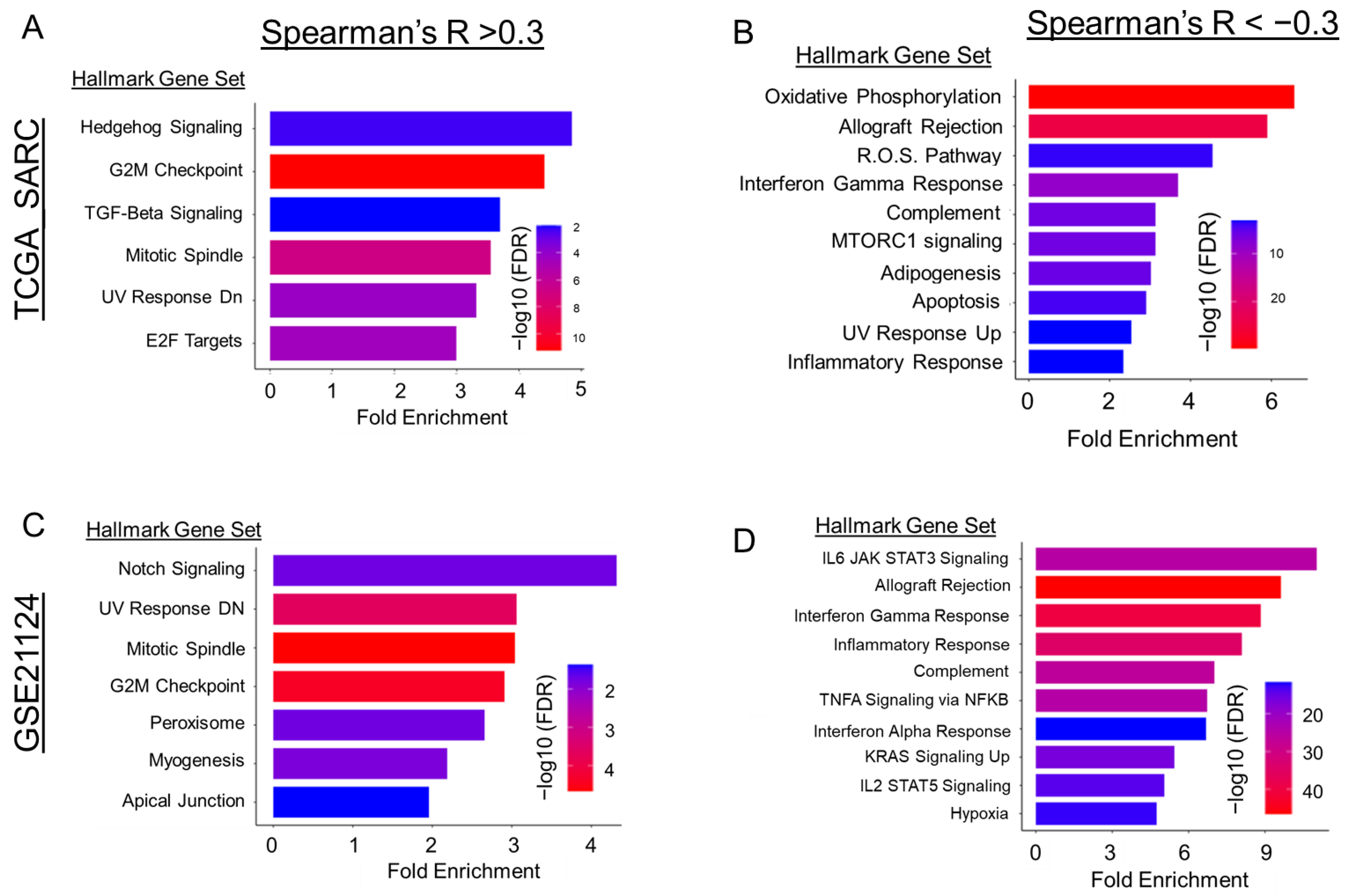
2.3. Gli2 Expression and Downstream Hedgehog Signaling Are Elevated in DDLPS Tumors
2.4. Gli2-Focused Co-Expression Analysis Reveals an Inverse Relationship between Gli2 and Immune Cell Population Markers While Establishing a Positive Correlation to Fibroblasts and ECM Gene Markers
3. Discussion
4. Limitations
5. Methods
5.1. Software, Databases, and Computational Tools
5.2. Dataset Accession and Descriptions
5.3. Genomics Profiling
5.4. Transcriptional Profiling, Differentially Expressed Genes, and Co-Expression Analysis
5.5. Microarrays, Tissue RNA Sequencing, and Single-Cell RNA Sequencing
5.6. Gene Set Enrichment Analysis (GSEA) and Gene Ontology
5.7. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singer, S.; Socci, N.D.; Ambrosini, G.; Sambol, E.; Decarolis, P.; Wu, Y.; O’Connor, R.; Maki, R.; Viale, A.; Sander, C.; et al. Gene Expression Profiling of Liposarcoma Identifies Distinct Biological Types/Subtypes and Potential Therapeutic Targets in Well-Differentiated and Dedifferentiated Liposarcoma. Cancer Res. 2007, 67, 6626–6636. [Google Scholar] [CrossRef]
- Jones, R.L.; Lee, A.T.J.; Thway, K.; Huang, P.H. Clinical and Molecular Spectrum of Liposarcoma. J. Clin. Oncol. 2018, 36, 151–159. [Google Scholar] [CrossRef]
- Osuna-Soto, J.; Caro Cuenca, T.; Sanz-Zorrilla, A.; Torrecilla-Martínez, A.; Ortega Salas, R.; Leiva-Cepas, F. Prognosis and Survival of Patients Diagnosed with Well-Differentiated and Dedifferentiated Retroperitoneal Liposarcoma. Cir. Esp. (Engl. Ed.) 2022, 100, 622–628. [Google Scholar] [CrossRef]
- Serguienko, A.; Braadland, P.; Meza-Zepeda, L.A.; Bjerkehagen, B.; Myklebost, O. Accurate 3-Gene-Signature for Early Diagnosis of Liposarcoma Progression. Clin. Sarcoma Res. 2020, 10, 4. [Google Scholar] [CrossRef]
- Mulita, F.; Verras, G.I.; Liolis, E.; Tchabashvili, L.; Kehagias, D.; Kaplanis, C.; Perdikaris, I.; Kehagias, I. Recurrent Retroperitoneal Liposarcoma: A Case Report and Literature Review. Clin. Case Rep. 2021, 9, e04717. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for Functional Genomics Data Sets—Update. Nucleic Acids Res. 2013, 41, 991–995. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The CBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the CBioPortal Complementary Data Sources and Analysis Options. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Creytens, D.; Van Gorp, J.; Speel, E.J.; Ferdinande, L. Characterization of the 12q Amplicons in Lipomatous Soft Tissue Tumors by Multiplex Ligation-Dependent Probe Amplification-Based Copy Number Analysis. Anticancer Res. 2015, 35, 1835–1842. [Google Scholar]
- Creytens, D. What’s New in Adipocytic Neoplasia? Virchows Arch. 2020, 476, 29–39. [Google Scholar] [CrossRef]
- Matushansky, I.; Hernando, E.; Socci, N.D.; Matos, T.; Mills, J.; Edgar, M.A.; Schwartz, G.K.; Singer, S.; Cordon-cardo, C.; Maki, R.G. A Developmental Model of Sarcomagenesis Defines a Differentiation-Based Classification for Liposarcomas. Am. J. Pathol. 2008, 172, 1069–1080. [Google Scholar] [CrossRef]
- Abeshouse, A.; Adebamowo, C.; Adebamowo, S.N.; Akbani, R.; Akeredolu, T.; Ally, A.; Anderson, M.L.; Anur, P.; Appelbaum, E.L.; Armenia, J.; et al. Comprehensive and Integrated Genomic Characterization of Adult Soft Tissue Sarcomas. Cell 2017, 171, 950–965.e28. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, T.S.; Xu, Z.; Zhang, X.; Wang, L.; Gimble, J.M.; Lander, E.S.; Rosen, E.D. Comparative Epigenomic Analysis of Murine and Human Adipogenesis. Cell 2010, 143, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Gobble, R.M.; Qin, L.X.; Brill, E.R.; Angeles, C.V.; Ugras, S.; O’Connor, R.B.; Moraco, N.H.; DeCarolis, P.L.; Antonescu, C.; Singer, S. Expression Profiling of Liposarcoma Yields a Multigene Predictor of Patient Outcome and Identifies Genes That Contribute to Liposarcomagenesis. Cancer Res. 2011, 71, 2697–2705. [Google Scholar] [CrossRef]
- Hulsen, T.; de Vlieg, J.; Alkema, W. BioVenn—A Web Application for the Comparison and Visualization of Biological Lists Using Area-Proportional Venn Diagrams. BMC Genom. 2008, 9, 488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, X.X.; Liu, Z.F.; Guo, X.R.; Cui, X.W.; Ji, C.B.; Zhong, H.; Chi, X. Inhibition of Hedgehog Signaling Promotes White Adipose Tissue Browning. Mol. Cell. Endocrinol. 2020, 518, 110970. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.H.; Dong, J.C.; Bian, Q. Small Molecules for Mesenchymal Stem Cell Fate Determination. World J. Stem Cells 2019, 11, 1084–1103. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.; Jiang, K.; Jia, J. Hedgehog Signaling Promotes Lipolysis in Adipose Tissue through Directly Regulating Bmm/ATGL Lipase. Dev. Biol. 2020, 457, 128–139. [Google Scholar] [CrossRef]
- Braune, J.; Weyer, U.; Matz-Soja, M.; Hobusch, C.; Kern, M.; Kunath, A.; Klöting, N.; Kralisch, S.; Blüher, M.; Gebhardt, R.; et al. Hedgehog Signalling in Myeloid Cells Impacts on Body Weight, Adipose Tissue Inflammation and Glucose Metabolism. Diabetologia 2017, 60, 889–899. [Google Scholar] [CrossRef]
- Todoric, J.; Strobl, B.; Jais, A.; Boucheron, N.; Bayer, M.; Amann, S.; Lindroos, J.; Teperino, R.; Prager, G.; Bilban, M.; et al. Cross-Talk between Interferon-γ and Hedgehog Signaling Regulates Adipogenesis. Diabetes 2011, 60, 1668–1676. [Google Scholar] [CrossRef]
- Fontaine, C.; Cousin, W.; Plaisant, M.; Dani, C.; Peraldi, P. Hedgehog Signaling Alters Adipocyte Maturation of Human Mesenchymal Stem Cells. Stem Cells 2008, 26, 1037–1046. [Google Scholar] [CrossRef]
- Teicher, B.A. Searching for Molecular Targets in Sarcoma. Biochem. Pharmacol. 2012, 84, 1–10. [Google Scholar] [CrossRef]
- Lucchesi, C.; Khalifa, E.; Laizet, Y.; Soubeyran, I.; Mathoulin-Pelissier, S.; Chomienne, C.; Italiano, A. Targetable Alterations in Adult Patients with Soft-Tissue Sarcomas: Insights for Personalized Therapy. JAMA Oncol. 2018, 4, 1398–1404. [Google Scholar] [CrossRef]
- Liu, J.; Mao, R.; Lao, I.W.; Yu, L.; Bai, Q.; Zhou, X.; Wang, J. GLI1-Altered Mesenchymal Tumor: A Clinicopathological and Molecular Analysis of Ten Additional Cases of an Emerging Entity. Virchows Arch. 2022, 480, 1087–1099. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sun, C.; Jiang, J.; Xie, Y.; Li, B.; Cui, X.; Chen, Y.; Liu, C.; Li, F. GLI1 Expression Is an Important Prognostic Factor That Contributes to the Poor Prognosis of Rhabdomyosarcoma. Histol. Histopathol. 2016, 31, 329–337. [Google Scholar] [CrossRef]
- Nacev, B.A.; Sanchez-Vega, F.; Smith, S.A.; Antonescu, C.R.; Rosenbaum, E.; Shi, H.; Tang, C.; Socci, N.D.; Rana, S.; Gularte-Mérida, R.; et al. Clinical Sequencing of Soft Tissue and Bone Sarcomas Delineates Diverse Genomic Landscapes and Potential Therapeutic Targets. Nat. Commun. 2022, 13, 3405. [Google Scholar] [CrossRef]
- Barretina, J.; Taylor, B.S.; Banerji, S.; Ramos, A.H.; Lagos-Quintana, M.; Decarolis, P.L.; Shah, K.; Socci, N.D.; Weir, B.A.; Ho, A.; et al. Subtype-Specific Genomic Alterations Define New Targets for Soft-Tissue Sarcoma Therapy. Nat. Genet. 2010, 42, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Frischauf, A.-M.; Plamberger, S.; Kaser, A.; Regl, G.; Schmid, C.; Schnidar, H.; Aberger, F.; Kasper, M.; Eichberger, T.; Sander, V. Overlapping and Distinct Transcriptional Regulator Properties of the GLI1 and GLI2 Oncogenes. Genomics 2006, 87, 616–632. [Google Scholar] [CrossRef]
- Ali, S.A.; Niu, B.; Cheah, K.S.E.; Alman, B. Unique and Overlapping GLI1 and GLI2 Transcriptional Targets in Neoplastic Chondrocytes. PLoS ONE 2019, 14, e0211333. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A Single–Cell Type Transcriptomics Map of Human Tissues. Sci. Adv. 2021, 7, abh2169. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a Knowledge-Based Human Protein Atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- Norreen-Thorsen, M.; Struck, E.C.; Öling, S.; Zwahlen, M.; Von Feilitzen, K.; Odeberg, J.; Lindskog, C.; Pontén, F.; Uhlén, M.; Dusart, P.J.; et al. A Human Adipose Tissue Cell-Type Transcriptome Atlas. Cell Rep. 2022, 40, 111046. [Google Scholar] [CrossRef] [PubMed]
- Hildreth, A.D.; Ma, F.; Wong, Y.Y.; Sun, R.; Pellegrini, M.; O’Sullivan, T.E. Single-Cell Sequencing of Human White Adipose Tissue Identifies New Cell States in Health and Obesity. Nat. Immunol. 2021, 22, 639–653. [Google Scholar] [CrossRef]
- Johnson, R.W.; Merkel, A.R.; Page, J.M.; Ruppender, N.S.; Guelcher, S.A.; Sterling, J.A. Wnt Signaling Induces Gene Expression of Factors Associated with Bone Destruction in Lung and Breast Cancer. Clin. Exp. Metastasis 2014, 31, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.W.; Nguyen, M.P.; Padalecki, S.S.; Grubbs, B.G.; Merkel, A.R.; Oyajobi, B.O.; Matrisian, L.M.; Mundy, G.R.; Sterling, J.A. TGF-β Promotion of Gli2-Induced Expression of Parathyroid Hormone-Related Protein, an Important Osteolytic Factor in Bone Metastasis, Is Independent of Canonical Hedgehog Signaling. Cancer Res. 2011, 71, 822–831. [Google Scholar] [CrossRef] [PubMed]
- Sterling, J.A.; Oyajobi, B.O.; Grubbs, B.; Padalecki, S.S.; Munoz, S.A.; Gupta, A.; Story, B.; Zhao, M.; Mundy, G.R. The Hedgehog Signaling Molecule Gli2 Induces Parathyroid Hormone-Related Peptide Expression and Osteolysis in Metastatic Human Breast Cancer Cells. Cancer Res. 2006, 66, 7548–7553. [Google Scholar] [CrossRef]
- Han, D.; Rodriguez-bravo, V.; Charytonowicz, E.; Demicco, E.; Domingo-domenech, J.; Maki, R.G.; Cordon-cardo, C. Targeting Sarcoma Tumor-Initiating Cells through Differentiation Therapy. Stem Cell Res. 2017, 21, 117–123. [Google Scholar] [CrossRef]
- Domingo-Domenech, J.; Vidal, S.J.; Rodriguez-Bravo, V.; Castillo-Martin, M.; Quinn, S.A.; Rodriguez-Barrueco, R.; Bonal, D.M.; Charytonowicz, E.; Gladoun, N.; de la Iglesia-Vicente, J.; et al. Suppression of Acquired Docetaxel Resistance in Prostate Cancer through Depletion of Notch- and Hedgehog-Dependent Tumor-Initiating Cells. Cancer Cell 2012, 22, 373–388. [Google Scholar] [CrossRef]
- Grund-Gröschke, S.; Stockmaier, G.; Aberger, F. Hedgehog/GLI Signaling in Tumor Immunity—New Therapeutic Opportunities and Clinical Implications. Cell Commun. Signal. 2019, 17, 172. [Google Scholar] [CrossRef]
- Jiang, J.; Ding, Y.; Chen, Y.; Lu, J.; Chen, Y.; Wu, G.; Xu, N.; Wang, H.; Teng, L. Pan-Cancer Analyses Reveal That Increased Hedgehog Activity Correlates with Tumor Immunosuppression and Resistance to Immune Checkpoint Inhibitors. Cancer Med. 2022, 11, 847–863. [Google Scholar] [CrossRef]
- Petty, A.J.; Li, A.; Wang, X.; Dai, R.; Heyman, B.; Hsu, D.; Huang, X.; Yang, Y. Hedgehog Signaling Promotes Tumor-Associated Macrophage Polarization to Suppress Intratumoral CD8+ T Cell Recruitment. J. Clin. Investig. 2019, 129, 5151–5162. [Google Scholar] [CrossRef] [PubMed]
- Cascio, S.; Chandler, C.; Zhang, L.; Sinno, S.; Gao, B.; Onkar, S.; Bruno, T.C.; Vignali, D.A.A.; Mahdi, H.; Osmanbeyoglu, H.U.; et al. Cancer-Associated MSC Drive Tumor Immune Exclusion and Resistance to Immunotherapy, Which Can Be Overcome by Hedgehog Inhibition. Sci. Adv. 2021, 7, abi5790. [Google Scholar] [CrossRef] [PubMed]
- Hinshaw, D.C.; Hanna, A.; Lama-Sherpa, T.; Metge, B.; Kammerud, S.C.; Benavides, G.A.; Kumar, A.; Alsheikh, H.A.; Mota, M.; Chen, D.; et al. Hedgehog Signaling Regulates Metabolism and Polarization of Mammary Tumor-Associated Macrophages. Cancer Res. 2021, 81, 5425–5437. [Google Scholar] [CrossRef]
- Scales, M.K.; Velez-Delgado, A.; Steele, N.G.; Schrader, H.E.; Stabnick, A.M.; Yan, W.; Mercado Soto, N.M.; Nwosu, Z.C.; Johnson, C.; Zhang, Y.; et al. Combinatorial Gli Activity Directs Immune Infiltration and Tumor Growth in Pancreatic Cancer. PLoS Genet. 2022, 18, e1010315. [Google Scholar] [CrossRef]
- Cazet, A.S.; Hui, M.N.; Elsworth, B.L.; Wu, S.Z.; Roden, D.; Chan, C.L.; Skhinas, J.N.; Collot, R.; Yang, J.; Harvey, K.; et al. Targeting Stromal Remodeling and Cancer Stem Cell Plasticity Overcomes Chemoresistance in Triple Negative Breast Cancer. Nat. Commun. 2018, 9, 2897. [Google Scholar] [CrossRef] [PubMed]
- Peiris-Pagès, M.; Sotgia, F.; Lisanti, M. Chemotherapy Induces the Cancer-Associated Fibroblast Phenotype, Activating Paracrine Hedgehog-GLI Signalling in Breast Cancer Cells. Oncotarget 2015, 6, 10728–10745. [Google Scholar] [CrossRef]
- Tang, Y.; Chen, Y.; Bao, Y.; Mahara, S.; Maryam, S.; Yatim, J.M.; Oguz, G.; Leng, P. Hypoxic Tumor Microenvironment Activates GLI2 via HIF-1{alpha} and TGF-{beta}2 to Promote Chemoresistance in Colorectal Cancer [Medical Sciences]. Proc. Natl. Acad. Sci. USA 2018, 115, E5990–E5999. [Google Scholar] [CrossRef]
- Steele, N.G.; Biffi, G.; Kemp, S.B.; Zhang, Y.; Drouillard, D.; Syu, L.J.; Hao, Y.; Oni, T.E.; Brosnan, E.; Elyada, E.; et al. Inhibition of Hedgehog Signaling Alters Fibroblast Composition in Pancreatic Cancer. Clin. Cancer Res. 2021, 27, 2023–2037. [Google Scholar] [CrossRef]
- Nagao, H.; Ijiri, K.; Hirotsu, M.; Ishidou, Y.; Yamamoto, T.; Nagano, S.; Takizawa, T.; Nakashima, K.; Komiya, S.; Setoguchi, T. Role of GLI2 in the Growth of Human Osteosarcoma. J. Pathol. 2011, 224, 169–179. [Google Scholar] [CrossRef]
- Nagao-Kitamoto, H.; Setoguchi, T.; Kitamoto, S.; Nakamura, S.; Tsuru, A.; Nagata, M.; Nagano, S.; Ishidou, Y.; Yokouchi, M.; Kitajima, S.; et al. Ribosomal Protein S3 Regulates GLI2-Mediated Osteosarcoma Invasion. Cancer Lett. 2015, 356, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Hirotsu, M.; Setoguchi, T.; Sasaki, H.; Matsunoshita, Y.; Gao, H.; Nagao, H.; Kunigou, O.; Komiya, S. Smoothened as a New Therapeutic Target for Human Osteosarcoma. Mol. Cancer 2010, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Joo, J.; Christensen, L.; Warner, K.; States, L.; Kang, H.G.; Vo, K.; Lawlor, E.R.; May, W.A. GLI1 Is a Central Mediator of EWS/FLI1 Signaling in Ewing Tumors. PLoS ONE 2009, 4, e0007608. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Liu, A. Spop Promotes Skeletal Development and Homeostasis by Positively Regulating Ihh Signaling. Proc. Natl. Acad. Sci. USA 2016, 113, 14751–14756. [Google Scholar] [CrossRef]
- Polychronidou, G.; Karavasilis, V.; Pollack, S.M.; Huang, P.H.; Lee, A.; Jones, R.L. Novel Therapeutic Approaches in Chondrosarcoma. Futur. Oncol. 2017, 13, 637–648. [Google Scholar] [CrossRef]
- Beauchamp, E.; Bulut, G.; Abaan, O.; Chen, K.; Merchant, A.; Matsui, W.; Endo, Y.; Rubin, J.S.; Toretsky, J.; Üren, A. GLI1 Is a Direct Transcriptional Target of EWS-FLI1 Oncoprotein. J. Biol. Chem. 2009, 284, 9074–9082. [Google Scholar] [CrossRef]
- Yao, Z.; Han, L.; Chen, Y.; He, F.; Sun, B.; Kamar, S.; Zhang, Y.; Yang, Y.; Wang, C.; Yang, Z. Hedgehog Signalling in the Tumourigenesis and Metastasis of Osteosarcoma, and Its Potential Value in the Clinical Therapy of Osteosarcoma Review. Cell Death Dis. 2018, 9, 701. [Google Scholar] [CrossRef]
- Fleming, J.T.; Brignola, E.; Chen, L.; Guo, Y.; Zhao, S.; Wang, Q.; Li, B.; Correa, H.; Ermilov, A.N.; Dlugosz, A.A.; et al. Insight into the Etiology of Undifferentiated Soft Tissue Sarcomas from a Novel Mouse Model. Mol. Cancer Res. 2019, 17, 1024–1035. [Google Scholar] [CrossRef]
- Campbell, V.T.; Nadesan, P.; Ali, S.A.; Wang, C.Y.Y.; Whetstone, H.; Poon, R.; Wei, Q.; Keilty, J.; Proctor, J.; Wang, L.W.; et al. Hedgehog Pathway Inhibition in Chondrosarcoma Using the Smoothened Inhibitor IPI-926 Directly Inhibits Sarcoma Cell Growth. Mol. Cancer Ther. 2014, 13, 1259–1269. [Google Scholar] [CrossRef]
- Srivastava, R.K.; Kaylani, S.Z.; Edrees, N.; Li, C.; Talwelkar, S.S.; Xu, J.; Palle, K.; Pressey, J.G.; Athar, M. GLI Inhibitor GANT-61 Diminishes Embryonal and Alveolar Rhabdomyosarcoma Growth by Inhibiting Shh/AKT-MTOR Axis. Oncotarget 2014, 5, 12151–12165. [Google Scholar] [CrossRef]
- Kelleher, F.C.; Cain, J.E.; Healy, J.M.; Watkins, D.N.; Thomas, D.M. Prevailing Importance of the Hedgehog Signaling Pathway and the Potential for Treatment Advancement in Sarcoma. Pharmacol. Ther. 2012, 136, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Dai, D.; Xie, L.; Shui, Y.; Li, J.; Wei, Q. Identification of Tumor Prognostic Genes in Sarcoma. Front. Genet. 2021, 12, 620705. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.Y.; Qu, W.Q.; Peng, H.H.; Yu, Y.F.; Jiang, Z.Z.; Qi, B.W.; Yu, A.X. Stemness Subtypes and Scoring System Predict Prognosis and Efficacy of Immunotherapy in Soft Tissue Sarcoma. Front. Immunol. 2022, 13, 796606. [Google Scholar] [CrossRef] [PubMed]
- Mulita, F.; Verras, G.I.; Anagnostopoulos, C.N.; Kotis, K. A Smarter Health through the Internet of Surgical Things. Sensors 2022, 22, 4577. [Google Scholar] [CrossRef]



| Software/Database | Description | Location |
|---|---|---|
| Gene Expression Omnibus (GEO) |
| https://www.ncbi.nlm.nih.gov/geo/ (Accessed on 12 August 2022) |
| GEO2R |
| https://www.ncbi.nlm.nih.gov/geo/geo2r/ (Accessed on 12 August 2022) |
| ShinyGO (0.77) |
| http://bioinformatics.sdstate.edu/go/ (Accessed on 9 December 2022). |
| CBioPortal |
| https://www.cbioportal.org/ (Accessed on 10 June 2021) |
| GSEA |
| http://www.gsea-sigdb.org/gsea/index.jsp (Accessed on 17 April 2022) |
| GraphPad Prism (9.5.0) |
| |
| Biorender |
| https://www.biorender.com/ (Accessed on 19 February 2023) |
| Human Protein Atlas |
| https://www.proteinatlas.org/ENSG00000074047-GLI2/single+cell+type (Accessed on 13 June 2022) |
| Dataset | PMID/URL | Accession Location | Description |
|---|---|---|---|
| GSE30929 | 21335544 | Gene Expression Omnibus (GEO) GSE30929 | Expression analysis by UG133A array WDLPS (n = 52) DDLPS (n = 40) |
| GSE20697 | 20887899 | Gene Expression Omnibus (GEO) GSE20697 | Whole-transcript expression data by UG133A array Time course of adipose stem cell differentiation |
| MSK-IMPACT | 35705560 | CBioPortal Sarcoma_MSKCC_2022 | Genomics profiling by sequencing DDLPS (n = 167) |
| TCGA-SARC | 29100075 | CBioPortal sarc_tcga_pan_can_atlas_2018 | Genomics and transcriptome profiling by sequencing DDLPS (n = 59) |
| GSE21124 | 20601955 | CBioPortal, Sarc_mskcc (2010) Gene Expression Omnibus (GEO) GSE21122/GSE21123 | Expression and genomics analysis by UG133A array DDLPS (n = 50) Matched Normal (n = 50) Normal Fat (n = 9) |
| Human Protein Atlas | 25613900 28495876 | https://www.proteinatlas.org/ENSG00000074047-GLI2 (Accessed on 13 June 2022) | Bulk RNA sequencing and single-cell sequencing of subcutaneous adipose tissue |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beadle, E.P.; Bennett, N.E.; Rhoades, J.A. Bioinformatics Screen Reveals Gli-Mediated Hedgehog Signaling as an Associated Pathway to Poor Immune Infiltration of Dedifferentiated Liposarcoma. Cancers 2023, 15, 3360. https://doi.org/10.3390/cancers15133360
Beadle EP, Bennett NE, Rhoades JA. Bioinformatics Screen Reveals Gli-Mediated Hedgehog Signaling as an Associated Pathway to Poor Immune Infiltration of Dedifferentiated Liposarcoma. Cancers. 2023; 15(13):3360. https://doi.org/10.3390/cancers15133360
Chicago/Turabian StyleBeadle, Erik P., Natalie E. Bennett, and Julie A. Rhoades. 2023. "Bioinformatics Screen Reveals Gli-Mediated Hedgehog Signaling as an Associated Pathway to Poor Immune Infiltration of Dedifferentiated Liposarcoma" Cancers 15, no. 13: 3360. https://doi.org/10.3390/cancers15133360
APA StyleBeadle, E. P., Bennett, N. E., & Rhoades, J. A. (2023). Bioinformatics Screen Reveals Gli-Mediated Hedgehog Signaling as an Associated Pathway to Poor Immune Infiltration of Dedifferentiated Liposarcoma. Cancers, 15(13), 3360. https://doi.org/10.3390/cancers15133360






