Simple Summary
Neuroblastoma is a pediatric tumor originating from the precursors of sympathetic nerves. The disease is known for its high heterogeneity. Hence, developing adequate preclinical models reflecting the complex biology of neuroblastoma is particularly challenging. This paper describes the current status of the available neuroblastoma models with their strengths and limitations, and demonstrates the future perspectives for preclinical neuroblastoma research.
Abstract
Preclinical in vitro and in vivo models remain indispensable tools in cancer research. These classic models, including two- and three-dimensional cell culture techniques and animal models, are crucial for basic and translational studies. However, each model has its own limitations and typically does not fully recapitulate the course of the human disease. Therefore, there is an urgent need for the development of novel, advanced systems that can allow for efficient evaluation of the mechanisms underlying cancer development and progression, more accurately reflect the disease pathophysiology and complexity, and effectively inform therapeutic decisions for patients. Preclinical models are especially important for rare cancers, such as neuroblastoma, where the availability of patient-derived specimens that could be used for potential therapy evaluation and screening is limited. Neuroblastoma modeling is further complicated by the disease heterogeneity. In this review, we present the current status of preclinical models for neuroblastoma research, discuss their development and characteristics emphasizing strengths and limitations, and describe the necessity of the development of novel, more advanced and clinically relevant approaches.
1. Introduction
Preclinical disease models play crucial roles in all fields of biomedical research, including drug discovery and development, the implementation of drug screening, diagnostic tests, prophylactic and therapeutic vaccines, evaluation of disease mechanisms, and discovering biochemical pathways. Despite their limitations, these models have been indispensable in cancer research conducted before clinical trials. However, none of the preclinical cancer models are ideal. The extent of the clinical relevance of the existing models has been a long-standing problem in cancer research [1,2,3,4,5].
Classic models, such as in vitro two-dimensional cell cultures and laboratory animals, have been extensively used in cancer research for decades [6]. Various established cell lines, including HeLa, are most commonly used, since they are publicly available, inexpensive, easy to handle, and typically provide highly replicable results. Unfortunately, the erroneous use of cells in laboratories around the world is surprisingly frequent—the cross-contamination rate can be as high as 25%, and numerous cell lines can be misidentified and mislabeled [7,8,9,10], which jeopardizes the research quality. However, even if used under their true identity, after many years of passaging the established cancer cell lines often no longer resemble the original cancers. They undergo changes that make them different from the source tumor: genetically, morphologically, metabolically, and physiologically. One of the reasons for this phenotypic drift is culture on plastic, in 2D setting, lacking the structure and microenvironment of the tissue of origin [5,11]. Moreover, while tumor cell lines can be grown in laboratories for a long time, the same cannot be conducted with corresponding normal cells, derived—as a necessary control—from patients’ normal tissues [12].
Mouse models of human cancer are not without problems as well. Ideally, they should recapitulate the events occurring in a patient and mimic the pathology, genetics, and therapeutic response of human disease. Neither classic mouse models, such as transgenic mice and conventional knockouts, nor more advanced ones (e.g., conditional knockouts and mice with regulated expression of oncogenes) fulfill all the requirements for a perfect model. The mouse organism resembles its human counterpart in many aspects, but there are also important differences (e.g., the control of telomeres and telomerase) that affect and limit potential research and the relevance of the obtained results to clinical practice [13,14,15,16].
Because of the limitations of classic preclinical models, new advanced models are being developed and extensively utilized. Various in vitro techniques such as three-dimensional cell cultures including gel embedding, scaffold culture, hanging drop culture, microfluidic devices, 3D bioprinting [7,17,18,19,20], induced pluripotent stem cells [21,22], and patient-derived 2D cell culture methods [23,24,25,26,27]. Recent years also brought significant advances in animal models, including humanized mouse models and patient-derived xenografts (PDXs) [28,29]. Importantly, none of these techniques fully recapitulate the process of carcinogenesis and cancer progression. Hence, they can be optimized and combined to complement each other, and thereby provide reliable tissue and organ models [30,31,32,33,34]. Currently, morphologically and genetically accurate complex in vitro models (CIVMs) are being developed to enable studies on particular cancer types [35].
Neuroblastoma is a rare solid cancer of the sympathetic nervous system, derived from neural crest cells. It is the most common childhood malignancy; the median age of diagnosis is 18 months and most of the cases occur in children below 10 years of age. Neuroblastoma is a very heterogenous disease, with a diverse clinical presentation and molecular complexity [36,37,38,39,40,41]. Despite an advancement in therapy in recent years, the prognosis for high-risk patients remains poor [42]. Most high-risk neuroblastomas initially respond to the therapy, but eventually relapse; the fatality rate of the disease is 50%. These unfavorable outcomes are commonly associated with amplification of the MYCN protooncogene (encoding N-MYC transcription factor), mutations of the anaplastic lymphoma kinase (ALK), and segmental chromosomal alterations (e.g., loss of chromosome arm 1p and 11q, gain of 17q) [37,43,44]. Moreover, patients with neuroblastoma may experience various neurological symptoms, metabolic syndrome, growth and puberty impairment, secondary neoplasms, and other syndromes, depending on the location of the primary tumor and metastases [40,45]. Hallmarks of neuroblastoma—heterogenous and diverse clinical manifestations, poor prognosis of advanced disease and high metastatic frequency—require new strategies in drug discovery and administration [46,47]. Advanced preclinical models are indispensable in that approach, as well as in attempts for a better understanding of the mechanisms and biology of the disease [36,48]. Our contemporary review summarizes the current status as well as recent developments in preclinical models used in various fields of neuroblastoma research, both in vitro and in vivo. We emphasize the importance of the clinically relevant models, especially for a rare disease such as neuroblastoma, discuss the strengths and limitations of each model, and describe the future advances in this field.
2. In Vitro Models of Neuroblastoma
2.1. Conventional 2D Cell Cultures
Currently, neuroblastoma research relies largely on cell lines cultured in vitro in two-dimensional settings. Numerous established neuroblastoma cell lines of human or rodent origin are available commercially (Table 1) [49,50]. Despite the disadvantages, the cells maintained using this technique are easy to use, and the method is highly productive and inexpensive. They preserve the distinct chromosomal aberrations that are characteristic for neuroblastoma and contribute to the disease prognosis and patient outcome [51,52]. Recent advances in CRISPR/Cas9 technology allow for further genetic modification aiming at recapitulating such genomic changes and thereby testing their functions [53]. Moreover, neuroblastoma cell lines recapitulate heterogeneity in differentiation stages observed in human tumors, ranging from undifferentiated mesenchymal to committed adrenergic cells [54,55]. For these reasons, the established cell lines are broadly used in various fields of neuroblastoma research, including basic tumor biology and interactions with the tumor microenvironment [56,57,58], as well as the development of novel treatment strategies, treatment screening, and monitoring [47,59,60,61]. Moreover, the neuroblastoma cell lines have been used in a wide spectrum of studies on genetic, biochemical, functional, and structural neuroblastoma characteristics, for example on the interaction of neuroblastoma cells with Schwann cells, the synthesis of neurotransmitters, differentiation and transdifferentiation processes, chromosomal structure, the role of MYCN expression, the importance of neuropeptide Y and hypoxia, and many others [57,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83]. A recent CRISPR/Cas9 screen performed in a panel of neuroblastoma cell lines has led to the identification of EZH2 as a molecular driver of the MYCN-amplified neuroblastomas, paving the way for similar studies in the future [84], importantly, in 2D cell lines, potential chemotherapy [85,86,87,88,89], radiation [90], and immunotherapy [91,92,93,94] screening. Furthermore, they are utilized to determine the role of genetic and chromosomal alterations in tumor growth, development, and the possible prognosis of the disease [95,96,97,98].
Established neuroblastoma cell lines have also been used for validating the mechanistic models used for the prediction of neuroblastoma progression based on the analysis of molecular networks [99]. Due to their neuronal features, neuroblastoma cell lines are also used as models in neuroscience, e.g., in studies on Alzheimer’s disease [100], Parkinson’s disease [101], and in virology research [102,103].
Despite the rarity of the disease, the field of neuroblastoma research is equipped in a vast number of established cell lines. This includes unique sets of cell lines developed from the same patient before diagnosis and post-treatment, rarely available for other cancer types [104]. This includes SK-N-BE(1) and SK-N-BE(2), SMS-KAN and SMS-KANR, SMS-KCN and SMS-KCNR, as well as CHLA15 and CHLA20 cell lines. Moreover, recent years brought significant progress in the development of new neuroblastoma research tools. Thanks to collaborative efforts led by the Children’s Oncology group, a wide range of cell lines developed from different biological materials (tumors, bone marrow, or blood), at different stages of the disease (at diagnosis, post-treatment, and post-mortem) is now available through the Childhood Cancer Repository [105]. This collection of well-characterized neuroblastoma cell lines reflecting neuroblastoma progression and representing its most aggressive, therapy-resistant, and metastatic features provides an excellent experimental tool for designing and testing novel therapeutic strategies.
Although most commonly used in cancer research, the conventional 2D cell culture is associated with significant disadvantages. Even though freshly obtained neuroblastoma tumor initiating cells retain certain tumor characteristics in cell culture [106], it has been known for years now that in vitro cultivation causes an accumulation of numerous alterations in cells [107]. The cells may undergo rapid epigenetic and transcriptional changes [108], differentiate [109], lose the expression of certain genes [110], change the biophysical properties of the cell membrane [111], lose sensitivity to oxidative stress [112], and undergo many other alterations. Conventional 2D cell lines lack polarity and their contact with tumor environment is affected [4], they are chromosomally unstable [3], and they do not preserve the heterogeneity and complexity of the tissue of origin. The heterogeneity of the tumor is currently described not only as a difference between the cancer cells in morphology, transcriptional profiles, and metabolism, but also includes the differences between the tumor microenvironment, which plays an important role in tumor resistance to therapy, the higher level of metastasis, and aggressiveness [113,114,115]. With two-dimensional cell lines, we are not able to properly reproduce these conditions in vitro [2,5,11,26,116].
In general, neuroblastoma cell lines, similar to other conventional cancer cell lines, do not properly recapitulate the original tumor properties and do not fully reflect the complexity and heterogeneity of the malignancy [117]. Thus, they may not be the ideal model for preclinical studies as the results obtained with them may not be transferable to patients. Therefore, various approaches to solve the problem with conventional 2D cell cultures in preclinical studies have been attempted. These include various three-dimensional cultures, co-cultures with additional cells, and complex models with microfluidic perfusion systems [118,119].

Table 1.
Examples of widely used neuroblastoma cell lines commercially available from American Type Culture Collection (ATCC).
Table 1.
Examples of widely used neuroblastoma cell lines commercially available from American Type Culture Collection (ATCC).
| Name | Origin | Stage (INSS *) | Treatment | MYCN Status | ALK Status | P53 Status | Differentiation Status | References |
|---|---|---|---|---|---|---|---|---|
| IMR-32 | Human | none | amplified | wt | wt | adrenergic | [49,54,85,120,121] | |
| SK-N-SH | Human | 4 | CT/RT | non-amplified | mut | wt | adrenergic | [54,85,105,122,123,124] |
| SK-SY5Y | Human; thrice cloned (SK-N-SH -> SH-SY -> SH-SY5 -> SH-SY5Y) subline of SK-N-SH | 4 | CT/RT | non-amplified | mut | wt | adrenergic | [50,54,85,124,125,126] |
| SK-N-BE(2) | Human | 4 | CT/RT | amplified | wt | wt | [49,85,125,127] | |
| BE(2)-C | Human; clone of SK-N-BE(2) | 4 | amplified | adrenergic | [54,125,128] | |||
| BE(2)-M17 | Human; clone of SK-N-BE(2) | 4 | [125] | |||||
| Neuro-2a | Mouse | - | [79] | |||||
| SK-N-FI | Human | 4 | CT | non-amplified | wt | mut | adrenergic | [54,77,85,105] |
| SK-N-DZ | Human | 4 | amplified | wt | wt | [78,85,123] | ||
| B35 | Rat | - | [66] | |||||
| SK-N-AS | Human | 4 | non-amplified | wt | wt | mesenchymal | [50,54,77,123,124] | |
| N1E-115 | Mouse | - | [65] | |||||
| NBFL | Human | 4 | [64] | |||||
| CHP-212 | Human | amplified | [49,128] | |||||
| NB41A3 | Mouse | - | [62] |
* International Neuroblastoma Staging System [129]; CT: chemotherapy; RT: radiotherapy; wt: wild type; mut: mutant.
2.2. Conditionally Reprogrammed Cells
Conditional reprogramming as the novel method of cell culture has been described in 2012 [130]. This method allows growing the epithelial cells, both cancerous and normal, for an indefinite time in a 2D in vitro setting without the transduction with any exogenous genes. The proliferative and adult stem-like state of the conditionally reprogrammed cells is maintained by a co-culture with irradiated mouse fibroblasts as the feeder cells, and by the presence of the Rho-associated kinase (ROCK) inhibitor, Y-27632 [25]. These cells are karyotype-stable, the induction of conditional reprogramming is rapid and reversible [131]. Conditional reprogramming is the epitome of personalized medicine; the cells can be directly obtained from a patient, cultured, and used for potential therapeutic approaches [23]. They can be also used as models for certain cancer types, including rare diseases, e.g., neuroendocrine cervical carcinoma [24,27,132], as well as biobanked for future basic and preclinical applications, for example genetic and chemosensitivity testing [133,134,135,136]. Conditional reprogramming is an inexpensive, easy to use, and robust method [25]. It can be used for growing normal cells [26,137], expanding cells generated from PDXs [30], as well as growing animal cells [138]. It has been shown that conditionally reprogrammed cells obtained from mammary tumors retain the genetic characteristics of the source malignancy [139] and maintain the expression of the estrogen receptor ERα; a well-known challenge in breast cancer research [140]. Undoubtedly, the main disadvantage of conditional reprogramming is that being a two-dimensional setting, it does not preserve the structural organization of a tumor.
We have shown that conditional reprogramming can be used to generate, culture, and biobank neuroblastoma cell lines [141]. Murine neuroblastoma conditionally reprogrammed cells collected from tumors arising in TH-MYCN mice retain the characteristic heterogenic neuroblastoma phenotype including a mesenchymal and neuronal component [54] (Figure 1A), and are useful as a neuroblastoma cell model in basic research [67]. These cells may also be grown in 3D cell culture settings (Figure 1B–D), thus serving as an example of the combination of various preclinical models. We propose that conditional reprogramming—when applied to human neuroblastoma samples—may serve as a novel reliable model for basic and translational research, including personalized drug testing.
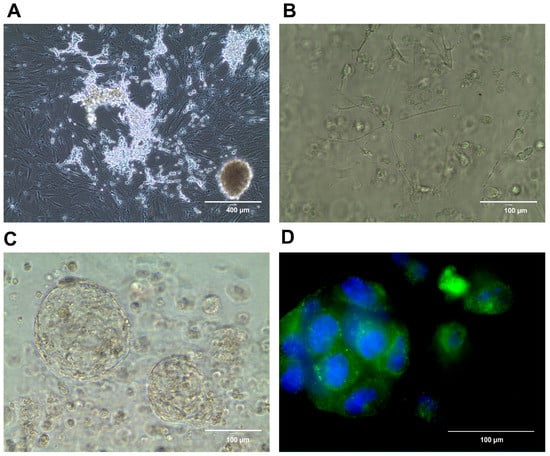
Figure 1.
Conditionally reprogrammed murine neuroblastoma cells (CR-NB). Cells growing in a 2D cell culture, demonstrating an adrenergic cell population growing atop a mesenchymal cell population, as shown before [141] (A). The same CR-NB cell line in Matrigel 1 day (B) and 28 days of a 3D culture (C). The localization of neuropeptide Y in the 3D CR-NB structure (green); DNA (blue) stained using Hoechst 33258 (D).
2.3. 3D Cell Cultures
Cancer is a complex, multidimensional disease, so it cannot be adequately represented in a 2D cell culture setting. Classic 2D cultures lack the microenvironment of the tumor and the complex structure of the tissue as whole. On the other hand, when animals are used as preclinical models, their genetic and pathophysiological characteristics are different from the human body in many ways. Moreover, animal research is associated with ethical issues, as the number of animals used for biomedical experimentation should be reduced as much as possible [142]. Therefore, in an attempt to create better preclinical cancer models, three-dimensional cell cultures are becoming increasingly utilized. This includes spheroids and organoids in various matrices, as well as more complex settings, such as microfluidic systems or material engineering for the scaffolds [33,143,144]. In cancer research, 3D in vitro models are particularly useful in representing certain cancer-specific hallmarks, including the replication potential, tissue invasion, and response to growth signals [116,145,146].
Spheroids are maintained in a scaffold-free and gel-free setting: they are grown in the hanging drop culture or on low-attachment plastic surfaces. The cells aggregate spontaneously, and the aggregates are composed of different subpopulations of cells, thus expressing the structural heterogeneity important for studies aimed for discovering anticancer therapy or the identification of pathways involved in tumor biology and cancer biomarkers [18,147,148,149]. The 3D multicellular cultures in vitro have many advantages over traditional cells maintained in the 2D setting, as they recapitulate the cellular complexity and heterogeneity of a tumor and the well-defined structural organization of the colony [150]. Spheroid models are also utilized in various aspects of neuroblastoma research, including the development and monitoring of combination chemotherapy [59,151,152,153], stem cell research [154], radiobiological studies [155], and the exploration of tumor growth determinants and their complexity [156]. Special microgravity-assay bioreactors, promoting spontaneous cellular interactions and aggregations and thus permitting formations of spherical 3D cell cultures, have also been useful for propagating neuroblastoma cells [157].
The local microenvironment, specifically its main component, the extracellular matrix (ECM), plays a crucial role in cancer development. It controls almost every aspect of cellular behavior, being involved in the regulation of processes such as cancer invasion, tumor-associated inflammation, cell polarity, and cancer stem cell niches [158]. Therefore, gel-embedded 3D cultures are thought to more accurately mimic in vivo tumor conditions, and they are arguably considered a better in vitro tumor model than growth in suspension. Matrigel, collagen, alginate, and other emerging gel techniques, such as engineered scaffolding structures [159,160,161], provide adequate structural support for cells, which allows for more precise modeling interactions between tumor cells and ECM. This, in turn, results in clinically relevant outcomes, such as testing the response to anticancer drugs [20,145]. These organoid models have been used for breast cancer cells [17], urological cancers [162], lung cancer [163], brain tissue [164], and many others [20,145,165,166]. Their strengths include the applicability to numerous research fields, from basic science to modelling diseases, including development and infectious diseases, drug discovery and screening, as well as personalized medicine [33,167]. They are versatile, expandable, genetically stable, and can be adapted for gene-modification techniques. However, they are costly and time-consuming.
The 3D gel-embedded cell cultures, utilizing for example the collagen-based scaffolds supplemented with nanohydroxyapatite or glycosaminoglycans [168] or composite hydrogels based on gelatin or alginate [118,169,170], are tested or used for neuroblastoma as well [171], to study the heterogeneity of the tumor [98], immunotyping [172], and chemosensitivity [173,174]. They are expected to improve the correlation between the results obtained in vitro and the outcomes in patients.
2.4. Complex In Vitro Cultures
Recently, a novel 3D bioprinting technique that recapitulates the tumor microenvironment and the spatial distribution of the cells as well as allows for the addition of other types of cells, has been applied to neuroblastoma cells [118,119,175]. To evaluate the role of vascularization and angiogenesis in neuroblastomas, an advanced model mimicking neuroblastoma vasculature was established [176,177,178,179]. These models may allow for advanced studies on neuroblastoma structure, progression, angiogenesis, and metastasis. In the near future, they may prove to be useful, providing a precision medicine platform for the assessment of potential therapeutic options.
2.5. Models of Neuroblastoma Initiation
The culture of neuroblastoma cells derived from human and mouse tumors serves as a useful tool to test the disease phenotypes and response to treatment; however, it does not recapitulate the early events leading to the disease initiation during sympathoadrenal development. To fill this gap, researchers have utilized in vitro techniques mimicking normal sympathetic differentiation, in the presence or absence of genetic aberrations observed in human neuroblastomas. These approaches include the differentiation of human and mouse embryonic stem cells, as well as human induced pluripotent cells into the neural crest, sympathoadrenal progenitors, or sympathetic nerves [180,181,182,183]. Others propose the use of human neural progenitor cells obtained by the direct reprogramming of the somatic cells, as well as the isolation of sympathetic neuroblasts from chick embryos or sympathoadrenal progenitor cells from postnatal murine adrenal glands [90,180,184]. Thus far, the number of available models of neuroblastoma initiation is limited and the methodology is still being developed. However, as the interest in the dysregulation of developmental processes leading to malignant transformation is growing, the above techniques become more commonly used in research on neuroblastomas and other neuronal tumors [182].
3. Animal Models of Neuroblastoma
Rodent cancer models, especially murine models, have been utilized in neuroblastoma research for decades to explore the genetics and mechanisms of the disease, as well as treatment and diagnostic options. The ability to recapitulate multiple processes involved in cancer progression and interactions between tumor cells and their environment are advantages of animal models. However, such experiments are typically costly, time-consuming, and not suitable for high-throughput studies. Commonly used murine models of neuroblastoma are classified into syngeneic, transgenic, xenograft, and humanized animal models [117]. The strengths and limitations of the most commonly used in vivo neuroblastoma models are summarized in Table 2.
3.1. Genetically Engineered Mouse Models
The best characterized alteration in human neuroblastoma and the most important predictor of its poor prognosis is the amplification of a Myc-related gene, MYCN. Moreover, MYCN directly influences the genetic and epigenetic changes observed in neuroblastoma [185]. Because of that, MYCN has been extensively used for modelling neuroblastoma in mice. In 1997, Weiss and colleagues created transgenic mice that overexpress N-myc in neural crest lineage cells under the control of rat tyrosine hydroxylase gene (TH) promoter [186]. Transgenic TH-MYCN mice demonstrate a high rate of neuroblastoma tumors that closely resemble human disease, based on their histology, pathology, molecular biology, and location in the paraspinal sympathetic ganglia [187,188]. The tumors also exhibit the heterogeneity observed in human neuroblastomas, with both adrenergic and mesenchymal populations present within the tumor tissue [141]. However, no tumors are observed in the adrenal glands of TH-MYCN mice, which is a common location of human neuroblastoma. Moreover, despite a high MYCN expression, which in humans correlates with metastatic disease, no overt metastases are seen in the TH-MYCN model. While disseminated tumor cells are observed in the lungs, no metastases in typical neuroblastoma locations, i.e., bones, bone marrow, or liver, are observed [189]. Lastly, the experiments on these mice are complicated by a long time for tumor development and the lack of the full penetrance, with the reported tumor frequency in hemizygous mice between 30% and 50% [186]. Nevertheless, the TH-MYCN murine model—as well as derived modified models (e.g., TH-MYCN/Mdm2+/−, TH-MYCN/TH-Cre/Casp8flox/flox, LSL-MYCN; Dbh-iCre, TH-MYCN/Trp53KI/KI), which can overcome some issues associated with the original TH-MYCN mice—are extensively utilized in various fields of neuroblastoma research. These include testing the potential chemotherapeutics [187,190,191,192,193,194,195,196], the identification of biomarkers [197,198], and a study on various aspects and mechanisms of tumorigenesis, and metastasis, as well as the development and progression of neuroblastomas [43,195,199,200,201,202,203,204,205].
In addition to MYCN amplification, aggressive neuroblastoma is also associated with activating mutations of the ALK oncogene and the overexpression of the epigenetic regulator involved in development, LIN28B. While some attempts of creating a mouse model expressing either the most common ALK mutation in neuroblastoma, ALK F1174L [206], or overexpressing LIN28B [207] in sympathetic lineage resulted in the formation of neuroblastoma tumors, other studies indicated that these genetic lesions alone are not capable of triggering a neuroblastoma. Instead, in these models, the expression of ALK mutants [208,209] or LIN28B [210] resulted in increased tumorigenicity, when combined with MYCN overexpression, as seen in TH-MYCN/ALKF1174 mice. Nevertheless, the genetic changes detected in the above models correlate well with the genetic landscape of human neuroblastomas, making them an adequate model to investigate the perturbations of developmental processes leading to neuroblastoma initiation and the therapy response of these tumors [196,208,211,212]. Unfortunately, similarly to the original TH-MYCN mice, none of these models accurately recapitulate the metastatic processes observed in neuroblastoma patients, as the tumors developing in the transgenic mice spread rarely and do not show tropism to the metastatic niches most common in neuroblastoma patients. Recently, a new model with c-MYC overexpression has been reported (Dbh-iCre/CAG-C-MYC mice); however, the details on the tumor frequency and disease phenotype are not yet available [212].
In addition to the animal models mimicking the genetic changes observed in neuroblastomas, other transgenic murine models rely on the overexpression of virus-derived transforming genes [213]. They include mice overexpressing human adenovirus type 12E1A and E1B under the regulatory control of the mouse mammary tumor virus long terminal repeat [214], the early region of JC virus [215], and the hybrid of the metallothionein promoter–enhancer and the ret oncogene [216]. However, these models are less relevant to human disease and are not broadly used.
3.2. Syngeneic Murine Models
Syngeneic models of neuroblastoma utilize the mouse neuroblastoma cell line C1300 and its derivatives, such as Neuro-2A. These tumors possess immune and histological characteristics similar to human neuroblastomas. Because of that, they have been used successfully for testing chemotherapeutic strategies and immune-mediated approaches [188,217]. These models are inexpensive, easy to handle, and reproducible; however, they do not recapitulate the human cancer genetically, they demonstrate low cellular heterogeneity, and human immune cells are absent from them [16,218]. Consequently, they are considered as having a low relevance to human biology [117].
3.3. Xenograft Murine Models
Mouse xenografts are the most commonly used animal models of neuroblastoma [219,220,221]. This approach introduces human neuroblastoma cells into immunocompromised mice—typically athymic nude or severe combined immunodeficiency disease (SCID) mice. While these mice are excellent recipients of xenografts, the absence or partial absence of the immune system may restrict their use, e.g., for studies on immunotherapy [222]. The human cells can be introduced subcutaneously or orthotopically to the adrenal gland fat pad. While the first approach is easier to perform, it does not recapitulate the neuroblastoma tumor environment. Orthotopic injections, on the other hand, are technically challenging and require surgery to reach the adrenal gland.
Mouse xenografts are most commonly derived from established neuroblastoma cell lines, as they are widely available and easy to use. However, as described above, these cell lines are characterized by their own limitations, which may impact the results of the experiments in vivo. To overcome the issues related to the cell lines, patient-derived tissue fragments (or cells) are used for grafting [117,223] Using intact patient-derived tumors is preferable since it may bypass the alterations acquired by in vitro cultivation [224]. These tumors and cells are transplanted orthotopically or heterotopically into animals [58,225,226]. Importantly, to overcome a problem of scarcely available neuroblastoma patient-derived samples, cryopreserved implants may be used for generating xenografts [224]. The intact tumor patient-derived xenografts (PDXs) accurately recapitulate the human cancer microenvironment complexity, retain high-risk neuroblastoma features, such as high vascularization, as well as the presence of tumor-associated macrophages and cancer-associated fibroblast infiltration [227,228]. Overall, PDXs are considered to be of high relevance to human pathology [117] and can serve as in vivo models for the screening of potential therapeutic options, including immunotherapy [229] and virotherapy [230]. However, there are numerous limitations associated with these models, such as a high cost and the time-consuming labor involved in their maintenance, the limited availability of fresh tissues or PDXs, the modifications of the phenotype in cells derived from PDXs upon cell culture, and only around 50% of the engraftment success rate [109,225,231]. Nevertheless, the number of available neuroblastoma PDXs is growing. A large collection of well-characterized patient-derived tumors is now available through the Pediatric Preclinical Testing Consortium and Childhood Cancer Repository [232].
3.4. Humanized Mice and Other Future Murine Neuroblastoma Models
Importantly, xenograft models rely on immunocompromised animals that lack a functional immune system. To circumvent this issue, mice with a humanized immune system are being investigated as potential cancer models. These models require either an injection of human peripheral blood cells into mice, or the simultaneous injection of stromal tissue with tumor tissue [16,28,48,233]. However, thus far, no successful attempt with humanized mice models has been reported for neuroblastoma.
Mouse-human neural crest chimeras as neuroblastoma models were described in 2020 by Cohen et al. In this model, the human neural crest cells carrying genetic lesions observed in neuroblastomas were injected into a mouse embryo. This experimental approach resulted in the formation of human neuroblastomas in immunocompetent mice, accompanied by the potent infiltration by mouse immune cells [234]. Thus, in the future this approach may be considered a powerful model to test the immune aspects of neuroblastoma, potentially overcoming some issues related to traditional murine xenograft models developed in immunocompromised mice.
Other interesting attempts include personalized approaches, e.g., by generating a couple of different in vitro and in vivo models from the same patient-derived cells [235] that, taken together, may more closely represent the complexity of human disease. In another approach, treatment guided by patient personalized tumor grafts has been demonstrated [222]. However, the latter has not been tested for neuroblastomas.

Table 2.
Preclinical in vivo models for neuroblastoma research, their major strengths, limitations, and human relevance.
Table 2.
Preclinical in vivo models for neuroblastoma research, their major strengths, limitations, and human relevance.
| Model | Strengths | Limitations | Human Relevance | References |
|---|---|---|---|---|
| Syngeneic mouse models |
|
| low | [16,117] |
| Transgenic mouse models |
|
| intermediate | [186,193,195,199,209] |
| Human xenograft murine models |
|
| low | [219,220,221] |
| PDX models |
|
| high | [223,224,225,228] |
| Zebrafish models |
|
| low | [117,236] |
3.5. Other Animal Models Proposed for Neuroblastoma Research
Except nude mice, nude rats have been utilized as potential models for neuroblastomas. However, these models did not exactly recapitulate the course of human malignancy [237]. Alternatively, some neural crest pathologies, including neuroblastoma, involving ALK activity and LIN28B gene expression were studied in the Xenopus model [238,239,240]. Zebrafish (Danio rerio) have been recently proposed as a promising platform to study genetics, pathogenesis, and progression processes in neuroblastoma [236,240,241,242,243]. Due to technical advantages over the mouse models, such as lower cost, shorter time to tumor development, and easy imaging, this model has become widely used in the neuroblastoma field. In addition, a recently established avian embryonic model involving grafting human neuroblastoma cells into chick embryos at the sympatho–adrenal crest level has been proven to be a valuable tool in recapitulating the interactions between normal developmental processes, the local microenvironment, and neuroblastoma progression [244,245]. The growth, differentiation, and potential drug sensitivity of neuroblastoma tumor can be also analyzed on the chick embryo chorioallantoic membrane [246,247,248]. However, even though the chick chorioallantoic membrane model is highly reproducible and easy to handle, it represents a low relevance to human pathophysiology [117].
3.6. Models of Neuroblastoma Metastasis
Modeling a metastatic disease has been a long-standing challenge in the field of neuroblastoma research. As described above, the tumors developing in transgenic mice typically do not form overt metastases. Similarly, the metastases from xenografts, including orthotopic tumors, are scarce and rarely seen in the niches relevant to human disease. Recapitulating osseous dissemination has proven to be particularly difficult. To overcome this problem, systemic injections to the tail vein are widely used [249,250]. Alternatively, intracardiac injections that facilitate bone colonization can be employed [251]. However, both models omit the initial stages of the metastatic process, which involve local invasion, intravasation, and escape from the primary tumor. On the other hand, direct injections of neuroblastoma cells into the bone cavity may be useful in investigations into the interactions between tumor cells and the bone environment, yet they do not recapitulate their dissemination [58]. This gap in our ability to model metastatic processes in neuroblastoma may be in the future filled by the use of PDXs. Recent years brought some reports of orthotopic PDXs metastasizing to the clinically relevant niches, such as bone, bone marrow, and liver [224]. However, as described above, the utility of these models is still hindered by the limited availability of neuroblastoma PDXs. Interestingly, a similar metastatic pattern has been shown upon orthotopic co-injection of established neuroblastoma cell lines with human mesenchymal stem cells [252]. Lastly, zebrafish models are useful to track neuroblastoma invasiveness; however, obtaining metastatic niche-specific information in this model is challenging.
4. Conclusions and Future Directions
Neuroblastoma research relies on preclinical models; however, no existing model is free of limitations. Therefore, it is of crucial importance to develop novel, advanced models that may serve as platforms for basic and translational research, especially for new treatment approaches. Conventional preclinical models in vitro, such as two-dimensional cell cultures, lack adequate representation of a tumor and do not recapitulate properly its biology, 3D architecture, topology, and many other features. Animal models, despite their importance in translational medicine and medical research fields, including neuroblastoma, also have serious disadvantages. They do not accurately recapitulate many aspects of human disease, such as its heterogeneity, the role of the immune system, and the complexity of the tumor microenvironment. The results of preclinical studies quite often do not predict the outcome in cancer patients; thus, the research on potential chemical agents does not proceed to clinical development. The gap between 2D cell cultures and animal models can be bridged by complex three-dimensional systems that closely recapitulate the course of human cancer [2,116,144,156,171]. Recent developments in three-dimensional, microfluidic, and multicellular systems used for neuroblastoma research [150] are promising, especially for testing new therapeutic approaches [118,160,253,254], including natural product research [177], immunotherapy [172,255,256], and new multimodal strategies for high-risk neuroblastoma [149]. A heterogenous disease such as neuroblastoma may largely benefit from comprehensive approaches involving a combination of various biological models supplemented by mathematical simulations [146,235,257,258]. Such strategies give promise for a better understanding of the malignancy, which may facilitate the development of effective therapies and improve the outcome for patients. However, it is important to note that currently available neuroblastoma models aim mainly at recapitulating the aggressive disease and do not reflect a full clinical spectrum of the disease. While this is justified by the need to test novel therapies for high-grade neuroblastoma, the basic biology of low-grade tumors and spontaneous regression, which is characteristic for these malignancies, remains understudied.
Author Contributions
E.K. and J.K. equally participated in manuscript design, writing, and revision. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by NIH grants: 1RO1CA123211, R01CA197964, and 1R21CA198698, as well as grants from the Children’s Cancer Foundation to J.K. In addition, internal funds from the Center for Cell Reprogramming at Georgetown University Medical Center were used to support the work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kamb, A. What’s Wrong with Our Cancer Models? Nat. Rev. Drug Discov. 2005, 4, 161–165. [Google Scholar] [CrossRef]
- Krüger, M.; Kopp, S. Tumor Models and Drug Targeting In Vitro—Where Are We Today? Where Do We Go from Here? Cancers 2023, 15, 1768. [Google Scholar] [CrossRef]
- He, Z.; Wilson, A.; Rich, F.; Kenwright, D.; Stevens, A.; Low, Y.S.; Thunders, M. Chromosomal Instability and Its Effect on Cell Lines. Cancer Rep. 2023, 6, e1822. [Google Scholar] [CrossRef] [PubMed]
- Movia, D.; Prina-Mello, A. Cancer Cell Culture Methods and Protocols Methods in Molecular Biology; Movia, D., Prina-Mello, A., Eds.; Humana Press: Totowa, NJ, USA, 2023. [Google Scholar]
- Wilding, J.L.; Bodmer, W.F. Cancer Cell Lines for Drug Discovery and Development. Cancer Res. 2014, 74, 2377–2384. [Google Scholar] [CrossRef] [PubMed]
- Voskoglou-Nomikos, T.; Pater, J.L.; Seymour, L. Clinical Predictive Value of the In Vitro Cell Line, Human Xenograft, and Mouse Allograft Preclinical Cancer Models. Clin. Cancer Res. 2003, 9, 4227–4239. [Google Scholar] [PubMed]
- Biju, T.S.; Priya, V.V.; Francis, A.P. Role of Three-Dimensional Cell Culture in Therapeutics and Diagnostics: An Updated Review. Drug Deliv. Transl. Res. 2023. online ahead of print. [Google Scholar] [CrossRef]
- Ye, F.; Chen, C.; Qin, J.; Liu, J.; Zheng, A.C. Genetic Profiling Reveals an Alarming Rate of Cross-Contamination among Human Cell Lines Used in China. FASEB J. 2015, 29, 4268–4272. [Google Scholar] [CrossRef]
- Drexler, H.G.; Dirks, W.G.; Matsuo, Y.; MacLeod, R.A.F. False Leukemia-Lymphoma Cell Lines: An Update on over 500 Cell Lines. Leukemia 2003, 17, 416–426. [Google Scholar] [CrossRef]
- Lacroix, M. Persistent Use of “False” Cell Lines. Int. J. Cancer 2008, 122, 1–4. [Google Scholar] [CrossRef]
- Abbot, A. Cell Culture: Biology’s New Dimension. Nature 2003, 424, 870–872. [Google Scholar] [CrossRef]
- Shay, J.; Wright, W. Hayflick, His Limit, and Cellular Ageing. Nat. Rev. Mol. Cell Biol. 2000, 1, 72–76. [Google Scholar] [CrossRef] [PubMed]
- Balmain, A.; Harris, C.C. Carcinogenesis in Mouse and Human Cells: Parallels and Paradoxes. Carcinogenesis 2000, 21, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Hann, B.; Balmain, A. Building “validated” Mouse Models of Human Cancer. Curr. Opin. Cell Biol. 2001, 13, 778–784. [Google Scholar] [CrossRef]
- Jonkers, J.; Berns, A. Conditional Mouse Models of Sporadic Cancer. Nat. Rev. Cancer 2002, 2, 251–265. [Google Scholar] [CrossRef]
- Seitz, G.; Armeanu-Ebinger, S.; Warmann, S.; Fuchs, J. Animal Models of Extracranial Pediatric Solid Tumors (Review). Oncol. Lett. 2012, 4, 859–864. [Google Scholar] [CrossRef]
- Dhiman, H.K.; Ray, A.R.; Panda, A.K. Three-Dimensional Chitosan Scaffold-Based MCF-7 Cell Culture for the Determination of the Cytotoxicity of Tamoxifen. Biomaterials 2005, 26, 979–986. [Google Scholar] [CrossRef]
- Weigelt, B.; Ghajar, C.M.; Bissell, M.J. The Need for Complex 3D Culture Models to Unravel Novel Pathways and Identify Accurate Biomarkers in Breast Cancer. Adv. Drug Deliv. Rev 2014, 69–70, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Gao, Y.; Hao, Y.; Li, E.; Wang, Y.; Zhang, J.; Wang, W.; Gao, Z.; Wang, Q. Application of a Microfluidic Chip-Based 3D Co-Culture to Test Drug Sensitivity for Individualized Treatment of Lung Cancer. Biomaterials 2013, 34, 4109–4117. [Google Scholar] [CrossRef]
- Lv, D.; Hu, Z.; Lu, L.; Lu, H.; Xu, X. Three-Dimensional Cell Culture: A Powerful Tool in Tumor Research and Drug Discovery. Oncol. Lett. 2017, 14, 6999–7010. [Google Scholar] [CrossRef]
- Ito, T.; Kawai, Y.; Yasui, Y.; Iriguchi, S.; Minagawa, A.; Ishii, T.; Miyoshi, H.; Taketo, M.M.; Kawada, K.; Obama, K.; et al. The Therapeutic Potential of Multiclonal Tumoricidal T Cells Derived from Tumor Infiltrating Lymphocyte-1derived IPS Cells. Commun. Biol. 2021, 4, 694. [Google Scholar] [CrossRef]
- Grskovic, M.; Javaherian, A.; Strulovici, B.; Daley, G.Q. Induced Pluripotent Stem Cells-Opportunities for Disease Modelling and Drug Discovery. Nat. Rev. Drug Discov. 2011, 10, 915–929. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Myers, S.; Wang, J.; Zhou, D.; Woo, J.A.; Kallakury, B.; Ju, A.; Bazylewicz, M.; Carter, Y.M.; Albanese, C.; et al. Use of Reprogrammed Cells to Identify Therapy for Respiratory Papillomatosis. N. Engl. J. Med. 2012, 367, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Yuan, H.; Krawczyk, E.; Blancato, J.; Albanese, C.; Zhou, D.; Wang, N.; Paul, S.; Alkhilaiwi, F.; Palechor-Ceron, N.; Dakic, A.; et al. HPV Positive Neuroendocrine Cervical Cancer Cells Are Dependent on Myc but Not E6/E7 Viral Oncogenes. Sci. Rep. 2017, 7, srep45617. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Krawczyk, E.; Suprynowicz, F.A.; Palechor-Ceron, N.; Yuan, H.; Dakic, A.; Simic, V.; Zheng, Y.L.; Sripadhan, P.; Chen, C.; et al. Conditional Reprogramming and Long-Term Expansion of Normal and Tumor Cells from Human Biospecimens. Nat. Protoc. 2017, 12, 439–451. [Google Scholar] [CrossRef]
- Palechor-Ceron, N.; Krawczyk, E.; Dakic, A.; Simic, V.; Yuan, H.; Blancato, J.; Wang, W.; Hubbard, F.; Zheng, Y.L.; Dan, H.; et al. Conditional Reprogramming for Patient-Derived Cancer Models and next-Generation Living Biobanks. Cells 2019, 8, 1327. [Google Scholar] [CrossRef]
- Timofeeva, O.A.; Palechor-Ceron, N.; Li, G.; Yuan, H.; Krawczyk, E.; Zhong, X.; Liu, G.; Upadhyay, G.; Dakic, A.; Yu, S.; et al. Conditionally Reprogrammed Normal and Primary Tumor Prostate Epithelial Cells: A Novel Patient-Derived Cell Model for Studies of Human Prostate Cancer. Oncotarget 2017, 8, 22741–22758. [Google Scholar] [CrossRef]
- De La Rochere, P.; Guil-Luna, S.; Decaudin, D.; Azar, G.; Sidhu, S.S.; Piaggio, E. Humanized Mice for the Study of Immuno-Oncology. Trends Immunol. 2018, 39, 748–763. [Google Scholar] [CrossRef]
- Zhou, Y.; Xia, J.; Xu, S.; She, T.; Zhang, Y.; Sun, Y.; Wen, M.; Jiang, T.; Xiong, Y.; Lei, J. Experimental Mouse Models for Translational Human Cancer Research. Front. Immunol. 2023, 14, 1095388. [Google Scholar] [CrossRef]
- Mondal, A.M.; Ma, A.H.; Li, G.; Krawczyk, E.; Yuan, R.; Lu, J.; Schlegel, R.; Stamatakis, L.; Kowalczyk, K.J.; Philips, G.K.; et al. Fidelity of a PDX-CR Model for Bladder Cancer. Biochem. Biophys. Res. Commun. 2019, 517, 49–56. [Google Scholar] [CrossRef]
- Khamis, Z.I.; Sarker, D.B.; Xue, Y.; Al-Akkary, N.; James, V.D.; Zeng, C.; Li, Y.; Sang, Q.X.A. Modeling Human Brain Tumors and the Microenvironment Using Induced Pluripotent Stem Cells. Cancers 2023, 15, 1253. [Google Scholar] [CrossRef]
- Luo, J.; Li, P. Human Pluripotent Stem Cell-Derived Brain Organoids as In Vitro Models for Studying Neural Disorders and Cancer. Cell Biosci. 2021, 11, 99. [Google Scholar] [CrossRef] [PubMed]
- Novelli, G.; Spitalieri, P.; Murdocca, M.; Centanini, E.; Sangiuolo, F. Organoid Factory: The Recent Role of the Human Induced Pluripotent Stem Cells (HiPSCs) in Precision Medicine. Front. Cell Dev. Biol. 2023, 10, 1059579. [Google Scholar] [CrossRef] [PubMed]
- Jung, O.; Song, M.J.; Ferrer, M. Operationalizing the Use of Biofabricated Tissue Models as Preclinical Screening Platforms for Drug Discovery and Development. SLAS Discov. 2021, 26, 1164–1176. [Google Scholar] [CrossRef] [PubMed]
- Browning, J.R.; Derr, P.; Derr, K.; Doudican, N.; Michael, S.; Lish, S.R.; Taylor, N.A.; Krueger, J.G.; Ferrer, M.; Carucci, J.A.; et al. A 3D Biofabricated Cutaneous Squamous Cell Carcinoma Tissue Model with Multi-Channel Confocal Microscopy Imaging Biomarkers to Quantify Antitumor Effects of Chemotherapeutics in Tissue. Oncotarget 2020, 11, 2587–2596. [Google Scholar] [CrossRef]
- Pinto, N.R.; Applebaum, M.A.; Volchenboum, S.L.; Matthay, K.K.; London, W.B.; Ambros, P.F.; Nakagawara, A.; Berthold, F.; Schleiermacher, G.; Park, J.R.; et al. Advances in Risk Classification and Treatment Strategies for Neuroblastoma. J. Clin. Oncol. 2015, 33, 3008–3017. [Google Scholar] [CrossRef] [PubMed]
- Kuzyk, A.; Gartner, J.; Mai, S. Identification of Neuroblastoma Subgroups Based on Three-Dimensional Telomere Organization. Transl. Oncol. 2016, 9, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Gheytanchi, E.; Mehrazma, M.; Madjd, Z. Expression of Ki-67, P53 and VEGF in Pediatric Neuroblastoma. Asian Pac. J. Cancer Prev. 2014, 15, 3065–3070. [Google Scholar] [CrossRef]
- Maris, J.M. Recent Advances in Neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef]
- Matthay, K.K.; Maris, J.M.; Schleiermacher, G.; Nakagawara, A.; Mackall, C.L.; Diller, L.; Weiss, W.A. Neuroblastoma. Nat. Rev. Dis. Primers 2016, 2, 16078. [Google Scholar] [CrossRef]
- Ackermann, S.; Cartolano, M.; Hero, B.; Welte, A.; Kahlert, Y.; Roderwieser, A.; Bartenhagen, C.; Walter, E.; Gecht, J.; Kerschke, L.; et al. A Mechanistic Classification of Clinical Phenotypes in Neuroblastoma. Science 2018, 362, 1165–1170. [Google Scholar] [CrossRef]
- DuBois, S.G.; Macy, M.E.; Henderson, T.O. High-Risk and Relapsed Neuroblastoma: Toward More Cures and Better Outcomes. Am. Soc. Clin. Oncol. Educ. Book 2022, 42, 768–780. [Google Scholar] [CrossRef]
- Cheung, N.K.V.; Dyer, M.A. Neuroblastoma: Developmental Biology, Cancer Genomics and Immunotherapy. Nat. Rev. Cancer 2013, 13, 397–411. [Google Scholar] [CrossRef]
- Brodeur, G.M. Neuroblastoma: Biological Insights into a Clinical Enigma. Nat. Rev. Cancer 2003, 3, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Speleman, F.; Park, J.R.; Henderson, T.O. Neuroblastoma: A Tough Nut to Crack. ASCO Educ. Book 2016, 35, e548–e557. [Google Scholar] [CrossRef] [PubMed]
- Matthay, K.K.; George, R.E.; Yu, A.L. Promising Therapeutic Targets in Neuroblastoma. Clin. Cancer Res. 2012, 18, 2740–2753. [Google Scholar] [CrossRef]
- Castel, V.; Segura, V.; Berlanga, P. Emerging Drugs for Neuroblastoma. Expert Opin. Emerg. Drugs 2013, 18, 155–171. [Google Scholar] [CrossRef]
- Ornell, K.J.; Coburn, J.M. Developing Preclinical Models of Neuroblastoma: Driving Therapeutic Testing. BMC Biomed. Eng. 2019, 1, 33. [Google Scholar] [CrossRef]
- Thiele, C.J. Neuroblastoma Cell Lines. In Human Cell Culture Volume I: Cancer Cell Lines Part 1: Chapter 2 Neuroblastoma; Masters, J.R.W., Palsson, B.O., Eds.; Kluwer Academic Publishers: New York, NY, USA, 1999. [Google Scholar]
- Campos Cogo, S.; Gradowski Farias da Costa do Nascimento, T.; de Almeida Brehm Pinhatti, F.; de França Junior, N.; Santos Rodrigues, B.; Regina Cavalli, L.; Elifio-Esposito, S. An Overview of Neuroblastoma Cell Lineage Phenotypes and In Vitro Models. Exp. Biol. Med. 2020, 245, 1637–1647. [Google Scholar] [CrossRef]
- Paolini, L.; Hussain, S.; Galardy, P.J. Chromosome Instability in Neuroblastoma: A Pathway to Aggressive Disease. Front. Oncol. 2022, 12, 988972. [Google Scholar] [CrossRef]
- Attiyeh, E.F.; London, W.B.; Mossé, Y.P.; Wang, Q.; Winter, C.; Khazi, D.; McGrady, P.W.; Seeger, R.C.; Thomas Look, A.; Shimada, H.; et al. Chromosome 1p and 11q Deletions and Outcome in Neuroblastoma. N. Engl. J. Med. 2005, 24, 2243–2253. [Google Scholar] [CrossRef]
- Parvin, S.; Akter, J.; Takenobu, H.; Katai, Y.; Satoh, S.; Okada, R.; Haruta, M.; Mukae, K.; Wada, T.; Ohira, M.; et al. ATM Depletion Induces Proteasomal Degradation of FANCD2 and Sensitizes Neuroblastoma Cells to PARP Inhibitors. BMC Cancer 2023, 23, 313. [Google Scholar] [CrossRef]
- van Groningen, T.; Koster, J.; Valentijn, L.J.; Zwijnenburg, D.A.; Akogul, N.; Hasselt, N.E.; Broekmans, M.; Haneveld, F.; Nowakowska, N.E.; Bras, J.; et al. Neuroblastoma Is Composed of Two Super-Enhancer-Associated Differentiation States. Nat. Genet. 2017, 49, 1261–1266. [Google Scholar] [CrossRef]
- van Groningen, T.; Akogul, N.; Westerhout, E.M.; Chan, A.; Hasselt, N.E.; Zwijnenburg, D.A.; Broekmans, M.; Stroeken, P.; Haneveld, F.; Hooijer, G.K.J.; et al. A NOTCH Feed-Forward Loop Drives Reprogramming from Adrenergic to Mesenchymal State in Neuroblastoma. Nat. Commun. 2019, 10, 1530. [Google Scholar] [CrossRef] [PubMed]
- Shmakova, A.A.; Klimovich, P.S.; Rysenkova, K.D.; Popov, V.S.; Gorbunova, A.S.; Karpukhina, A.A.; Karagyaur, M.N.; Rubina, K.A.; Tkachuk, V.A.; Semina, E.V. Urokinase Receptor UPAR Downregulation in Neuroblastoma Leads to Dormancy, Chemoresistance and Metastasis. Cancers 2022, 14, 994. [Google Scholar] [CrossRef]
- Huertas-Castaño, C.; Gómez-Muñoz, M.A.; Pardal, R.; Vega, F.M. Hypoxia in the Initiation and Progression of Neuroblastoma Tumours. Int. J. Mol. Sci. 2020, 21, 39. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Cai, W.; Li, S.; Da, Z.; Sun, H.; Ma, L.; Lin, Y.; Zhi, D. Characterization of Neuroblastoma Bone Invasion/Metastasis in Established Bone Metastatic Model of SY5Y and KCNR Cell Lines. Child’s Nerv. Syst. 2013, 29, 1097–1105. [Google Scholar] [CrossRef] [PubMed]
- Cuperus, R.; Tytgat, G.A.M.; Leen, R.; Brites, P.; Bras, J.; Caron, H.N.; Van Kuilenburg, A.P.B. Pleiotropic Effects of Fenretinide in Neuroblastoma Cell Lines and Multicellular Tumor Spheroids. Int. J. Oncol. 2008, 32, 1011–1019. [Google Scholar] [CrossRef] [PubMed]
- Roomi, M.W.; Kalinovsky, T.; Roomi, N.W.; Niedzwiecki, A.; Rath, M. Inhibition of the SK-N-MC Human Neuroblastoma Cell Line In Vivo and In Vitro by a Novel Nutrient Mixture. Oncol. Rep. 2013, 29, 1714–1720. [Google Scholar] [CrossRef]
- Lu, C.; Everhart, L.; Tilan, J.; Kuo, L.; Sun, C.C.J.; Munivenkatappa, R.B.; Jönsson-Rylander, A.C.; Sun, J.; Kuan-Celarier, A.; Li, L.; et al. Neuropeptide y and Its Y2 Receptor: Potential Targets in Neuroblastoma Therapy. Oncogene 2010, 29, 5630–5642. [Google Scholar] [CrossRef]
- Thompson, J.M.; London, E.D.; Johnson, J.E. Ultrastructural, Functional and Biochemical Characteristics of Mouse and Human Neuroblastoma Cell Lines. Neuroscience 1982, 7, 1807–1815. [Google Scholar] [CrossRef]
- Gazitt, Y.; He, Y.J.; Chang, L.; Koza, S.; Fisk, D.; Graham-Pole, J. Expression of N-Myc, c-Myc, and MDR-1 Proteins in Newly Established Neuroblastoma Cell Lines: A Study by Immunofluorescence Staining and Flow Cytometry. Cancer Res. 1992, 52, 2957–2965. [Google Scholar]
- Symes, A.J.; Rao, M.S.; Lewis, S.E.; Landis, S.C.; Hyman, S.E.; Stephen Fink, J. Ciliary Neurotrophic Factor Coordinately Activates Transcription of Neuropeptide Genes in a Neuroblastoma Cell Line. Proc. Natl. Acad. Sci. USA 1993, 90, 572–576. [Google Scholar] [CrossRef]
- Amano, T.; Richelson, E.; Nirenberg, M. Neurotransmitter Synthesis by Neuroblastoma Clones (Neuroblast Differentiation/Cell Culture/Choline Acetyltransferase/Acetylcholinesterase/Tyrosine Hydroxylase/Axons-Dendrites). Proc. Natl. Acad. Sci. USA 1972, 69, 258–263. [Google Scholar] [CrossRef]
- Schubert, D.; Heinemann, S.; Carlisle, W.; Tarikas, H.; Kimes, B.; Patrick, J.; Steinbach, J.H.; Culp, W.; Brandt, B.L. Clonal Cell Lines from the Rat Central Nervous System. Nature 1974, 2, 224–227. [Google Scholar] [CrossRef] [PubMed]
- Abualsaud, N.; Caprio, L.; Galli, S.; Krawczyk, E.; Alamri, L.; Zhu, S.; Gallicano, G.I.; Kitlinska, J. Neuropeptide Y/Y5 Receptor Pathway Stimulates Neuroblastoma Cell Motility Through RhoA Activation. Front. Cell Dev. Biol. 2021, 8, 627090. [Google Scholar] [CrossRef] [PubMed]
- Rupniak, H.T.; Rein, G.; Powell, J.F.; Ryder, T.A.; Carson, S.; Povey, S.; Hill, B.T. Characteristics of a New Human Neuroblastoma Cell Line Which Differentiates in Response to Cyclic Adenosine 3′:5′-Monophosphate1. Cancer Res. 1984, 44, 2600–2607. [Google Scholar] [PubMed]
- West, G.J.; Uki, J.; Herschman, H.R.; Seeger Robert, C. Adrenergic, Cholinergic, and Inactive Human Neuroblastoma Cell Lines with the Action-Potential Na+ Ionophore. Cancer Res. 1977, 37, 1372–1376. [Google Scholar] [PubMed]
- Schlesinger, H.R.; Gerson, J.M.; Moorhead, P.S.; Maguire, H.; Hummeler, K. Establishment and Characterization of Human Neuroblastoma Cell Lines. Cancer Res. 1976, 36, 3094–3100. [Google Scholar]
- Gilbert, F.; Balaban, G.; Moorhead, P.; Bianchi, D.; Schlesinger, H. Abnormalities of Chromosome Lp in Human Neuroblastoma Tumors and Cell Lines. Cancer Genet. Cytogenet. 1982, 7, 33–42. [Google Scholar] [CrossRef]
- Naiditch, J.A.; Jie, C.; Lautz, T.B.; Yu, S.; Clark, S.; Voronov, D.; Chu, F.; Madonna, M.B. Mesenchymal Change and Drug Resistance in Neuroblastoma. J. Surg. Res. 2015, 193, 279–288. [Google Scholar] [CrossRef]
- Seeger, R.C.; Rayner, S.A.; Banerjee, A.; Chung, H.; Laug, E.; Neustein, H.B.; Benedict, W.F. Morphology, Growth, Chromosomal Pattern, and Fibrinolytic of Two New Human Neuroblastoma Cell Lines. Cancer Res. 1977, 37, 1364–1371. [Google Scholar]
- Pajtler, K.W.; Mahlow, E.; Odersky, A.; Lindner, S.; Stephan, H.; Bendix, I.; Eggert, A.; Schramm, A.; Schulte, J.H. Neuroblastoma in Dialog with Its Stroma: NTRK1 Is a Regulator of Cellular Cross-Talk with Schwann Cells. Oncotarget 2014, 5, 11180–11192. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Tsubota, S.; Nishio, N.; Takahashi, Y.; Kadomatsu, K. Combination of Tumor Necrosis Factor-α and Epidermal Growth Factor Induces the Adrenergic-to-Mesenchymal Transdifferentiation in SH-SY5Y Neuroblastoma Cells. Cancer Sci. 2021, 112, 715–724. [Google Scholar] [CrossRef]
- Wolpaw, A.J.; Grossmann, L.D.; Dessau, J.L.; Dong, M.M.; Aaron, B.J.; Brafford, P.A.; Volgina, D.; Pascual-Pasto, G.; Rodriguez-Garcia, A.; Uzun, Y.; et al. Epigenetic State Determines Inflammatory Sensing in Neuroblastoma. Proc. Natl. Acad. Sci. USA 2022, 119, e2102358119. [Google Scholar] [CrossRef]
- Iavarone, A.; Lasorella, A.; Servidei, T.; Riccardi, R.; Mastrangelo, R. Uptake and Storage of M-Lodobenzylguanidine Are Frequent Neuronal Functions of Human Neuroblastoma Cell Lines. Cancer Res. 1993, 53, 304–309. [Google Scholar] [PubMed]
- Matsumoto, M.; Akiyama, T.; Miyatake, S.; Kikuchi, H.; Hanaoka, M.; Namba, Y. Expression of Proto-Oncogene Products during Drug-Induced Differentiation of a Neuroblastoma Cell Line SK-N-DZ. Acta Neuropathol. 1989, 79, 217–221. [Google Scholar] [CrossRef]
- Olmsted, J.B.; Carlson, K.; Klebe, R.; Ruddle, F.; Rosenbaum, J. Isolation of Microtubule Protein from Cultured Mouse Neuroblastoma Cells. Proc. Natl. Acad. Sci. USA 1970, 65, 129–136. [Google Scholar] [CrossRef]
- Ross, R.A.; Walton, J.D.; Han, D.; Guo, H.F.; Cheung, N.K.V. A Distinct Gene Expression Signature Characterizes Human Neuroblastoma Cancer Stem Cells. Stem Cell Res. 2015, 15, 419–426. [Google Scholar] [CrossRef]
- Spengler, B.A.; Lazarova, D.L.; Ross, R.A.; Biedler, J.L. Cell Lineage and Differentiation State Are Primary Determinants of MYCN Gene Expression and Malignant Potential in Human Neuroblastoma Cells. Oncol. Res. 1997, 9, 467–476. [Google Scholar]
- Foley, J.; Cohn, S.L.; Salwen, H.R.; Parysek, L.M. Differential Expression of N-Myc in Phenotypically Distinct Subclones of a Human Neuroblastoma Cell Line. Cancer Res. 1991, 51, 6338–6345. [Google Scholar]
- Ciccarone, V.; Spengler, B.A.; Meyers, M.B.; Biedler, J.L.; Ross, R.A. Phenotypic Diversification in Human Neuroblastoma Cells: Expression of Distinct Neural Crest Lineages. Cancer Res. 1989, 49, 219–225. [Google Scholar]
- Chen, L.; Alexe, G.; Dharia, N.V.; Ross, L.; Iniguez, A.B.; Conway, A.S.; Wang, E.J.; Veschi, V.; Lam, N.; Qi, J.; et al. CRISPR-Cas9 Screen Reveals a MYCN-Amplified Neuroblastoma Dependency on EZH2. J. Clin. Investig. 2018, 128, 446–462. [Google Scholar] [CrossRef]
- Blanco-Luquin, I.; Lázcoz, P.; Celay, J.; Castresana, J.S.; Encío, I.J. In Vitro Assessment of the Role of P53 on Chemotherapy Treatments in Neuroblastoma Cell Lines. Pharmaceuticals 2021, 14, 1184. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia Enables Predictive Modelling of Anticancer Drug Sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef]
- Park, J.H.; Szemes, M.; Vieira, G.C.; Melegh, Z.; Malik, S.; Heesom, K.J.; Von Wallwitz-Freitas, L.; Greenhough, A.; Brown, K.W.; Zheng, Y.G.; et al. Protein Arginine Methyltransferase 5 Is a Key Regulator of the MYCN Oncoprotein in Neuroblastoma Cells. Mol. Oncol. 2015, 9, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.K.; Mallepalli, S.; Damu, A.; Vadde, R. Neuroblastoma and Stem Cell Therapy: An Updated Review. CNS Neurol. Disord. Drug Targets 2020, 20, 625–643. [Google Scholar] [CrossRef] [PubMed]
- Saintas, E.; Abrahams, L.; Ahmad, G.T.; Ajakaiye, A.O.M.; Alhumaidi, A.S.H.A.M.; Ashmore-Harris, C.; Clark, I.; Dura, U.K.; Fixmer, C.N.; Ike-Morris, C.; et al. Acquired Resistance to Oxaliplatin Is Not Directly Associated with Increased Resistance to DNA Damage in SK-N-ASrOXALI4000, a Newly Established Oxaliplatin-Resistant Sub-Line of the Neuroblastoma Cell Line SK-N-AS. PLoS ONE 2017, 12, e0172140. [Google Scholar] [CrossRef]
- Sitnikov, D.; Revkova, V.; Ilina, I.; Shatalova, R.; Komarov, P.; Struleva, E.; Konoplyannikov, M.; Kalsin, V.; Baklaushev, V. Sensitivity of Neuroblastoma and Induced Neural Progenitor Cells to High-Intensity THz Radiation. Int. J. Mol. Sci. 2023, 24, 6558. [Google Scholar] [CrossRef]
- Cornel, A.M.; Dunnebach, E.; Hofman, D.A.; Das, S.; Sengupta, S.; Van Den Ham, F.; Wienke, J.; Strijker, J.G.M.; Van Den Beemt, D.A.M.H.; Essing, A.H.W.; et al. Epigenetic Modulation of Neuroblastoma Enhances T Cell and NK Cell Immunogenicity by Inducing a Tumor-Cell Lineage Switch. J. Immunother. Cancer 2022, 10, e005002. [Google Scholar] [CrossRef]
- Jiménez, C.; Moreno, L.; Segura, M.F. Epigenetic Therapies for Neuroblastoma: Immunogenicity Awakens. Mol. Oncol. 2023, 17, 718–721. [Google Scholar] [CrossRef]
- Iguchi, M.; Yagyu, S.; Kambe, K.; Higashi, M.; Fumino, S.; Kishida, T.; Iehara, T.; Mazda, O.; Tajiri, T. Development of Anti-GD2 Antibody-Producing Mesenchymal Stem Cells as Cellular Immunotherapy. Anticancer Res. 2023, 43, 2417–2424. [Google Scholar] [CrossRef] [PubMed]
- Karapurkar, J.K.; Kim, M.S.; Colaco, J.C.; Suresh, B.; Sarodaya, N.; Kim, D.H.; Park, C.H.; Hong, S.H.; Kim, K.S.; Ramakrishna, S. CRISPR/Cas9-Based Genome-Wide Screening of the Deubiquitinase Subfamily Identifies USP3 as a Protein Stabilizer of REST Blocking Neuronal Differentiation and Promotes Neuroblastoma Tumorigenesis. J. Exp. Clin. Cancer Res. 2023, 42, 121. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Monleon, A.; Gaarder, J.; Djos, A.; Kogner, P.; Fransson, S. Identification of Recurrent 3q13.31 Chromosomal Rearrangement Indicates LSAMP as a Tumor Suppressor Gene in Neuroblastoma. Int. J. Oncol. 2023, 62, 27. [Google Scholar] [CrossRef]
- Eleveld, T.F.; Bakali, C.; Eijk, P.P.; Stathi, P.; Vriend, L.E.; Poddighe, P.J.; Ylstra, B. Engineering Large-Scale Chromosomal Deletions by CRISPR-Cas9. Nucleic Acids Res. 2021, 49, 12007–12016. [Google Scholar] [CrossRef]
- Sanmartín, E.; Muñoz, L.; Piqueras, M.; Sirerol, J.A.; Berlanga, P.; Cañete, A.; Castel, V.; Font De Mora, J. Deletion of 11q in Neuroblastomas Drives Sensitivity to PARP Inhibition. Clin. Cancer Res. 2017, 23, 6875–6887. [Google Scholar] [CrossRef]
- López-Carrasco, A.; Martín-Vañó, S.; Burgos-Panadero, R.; Monferrer, E.; Berbegall, A.P.; Fernández-Blanco, B.; Navarro, S.; Noguera, R. Impact of Extracellular Matrix Stiffness on Genomic Heterogeneity in MYCN-Amplified Neuroblastoma Cell Line. J. Exp. Clin. Cancer Res. 2020, 39, 226. [Google Scholar] [CrossRef] [PubMed]
- Kasemeier-Kulesa, J.C.; Schnell, S.; Woolley, T.; Spengler, J.A.; Morrison, J.A.; McKinney, M.C.; Pushel, I.; Wolfe, L.A.; Kulesa, P.M. Predicting Neuroblastoma Using Developmental Signals and a Logic-Based Model. Biophys. Chem. 2018, 238, 30–38. [Google Scholar] [CrossRef]
- de Medeiros, L.M.; De Bastiani, M.A.; Rico, E.P.; Schonhofen, P.; Pfaffenseller, B.; Wollenhaupt-Aguiar, B.; Grun, L.; Barbé-Tuana, F.; Zimmer, E.R.; Castro, M.A.A.; et al. Cholinergic Differentiation of Human Neuroblastoma SH-SY5Y Cell Line and Its Potential Use as an In Vitro Model for Alzheimer’s Disease Studies. Mol. Neurobiol. 2019, 56, 7355–7367. [Google Scholar] [CrossRef]
- Xie, H.-R.; Hu, L.-S.; Li, G.-Y. SH-SY5Y Human Neuroblastoma Cell Line: In Vitro Cell Model of Dopaminergic Neurons in Parkinson’s Disease. Chin. Med. J. 2010, 123, 1086–1092. [Google Scholar]
- Pandhare, J.; Dash, S.; Jones, B.; Villalta, F.; Dash, C. A Novel Role of Proline Oxidase in HIV-1 Envelope Glycoprotein-Induced Neuronal Autophagy. J. Biol. Chem. 2015, 290, 25439–25451. [Google Scholar] [CrossRef]
- Shastry, P.; Basu, A.; Rajadhyaksha, M.S. Neuroblastoma Cell Lines—A Versatile In Vitro Model in Neurobiology. Int. J. Neurosci. 2001, 108, 109–126. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.P.; Biedler, J.L.; Spengler, B.A.; Reynolds, D.A.; Ross, R.A.; Frenkel, E.P.; Smith, R.G. Characterization of Human Neuroblastoma Cell Lines Established Before and After Therapy. J. Natl. Cancer Inst. 1986, 76, 375–387. [Google Scholar] [PubMed]
- Harenza, J.L.; DIamond, M.A.; Adams, R.N.; Song, M.M.; Davidson, H.L.; Hart, L.S.; Dent, M.H.; Fortina, P.; Reynolds, C.P.; Maris, J.M. Transcriptomic Profiling of 39 Commonly-Used Neuroblastoma Cell Lines. Sci. Data 2017, 4, 170033. [Google Scholar] [CrossRef]
- Bate-Eya, L.T.; Ebus, M.E.; Koster, J.; Den Hartog, I.J.M.; Zwijnenburg, D.A.; Schild, L.; Van Der Ploeg, I.; Dolman, M.E.M.; Caron, H.N.; Versteeg, R.; et al. Newly-Derived Neuroblastoma Cell Lines Propagated in Serum-Free Media Recapitulate the Genotype and Phenotype of Primary Neuroblastoma Tumours. Eur. J. Cancer 2014, 50, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Siranosian, B.; Ha, G.; Tang, H.; Oren, Y.; Hinohara, K.; Strathdee, C.A.; Dempster, J.; Lyons, N.J.; Burns, R.; et al. Genetic and Transcriptional Evolution Alters Cancer Cell Line Drug Response. Nature 2018, 560, 325–330. [Google Scholar] [CrossRef]
- Nestor, C.E.; Ottaviano, R.; Reinhardt, D.; Cruickshanks, H.A.; Mjoseng, H.K.; McPherson, R.C.; Lentini, A.; Thomson, J.P.; Dunican, D.S.; Pennings, S.; et al. Rapid Reprogramming of Epigenetic and Transcriptional Profiles in Mammalian Culture Systems. Genome Biol. 2015, 16, 11. [Google Scholar] [CrossRef]
- Persson, C.U.; Von Stedingk, K.; Bexell, D.; Merselius, M.; Braekeveldt, N.; Gisselsson, D.; Arsenian-Henriksson, M.; Påhlman, S.; Wigerup, C. Neuroblastoma Patient-Derived Xenograft Cells Cultured in Stem-Cell Promoting Medium Retain Tumorigenic and Metastatic Capacities but Differentiate in Serum. Sci. Rep. 2017, 7, 10274. [Google Scholar] [CrossRef]
- McNerney, K.O.; Karageorgos, S.; Ferry, G.M.; Wolpaw, A.J.; Burudpakdee, C.; Khurana, P.; Toland, C.N.; Vemu, R.; Vu, A.; Hogarty, M.D.; et al. TH-MYCN Tumors, but Not Tumor-Derived Cell Lines, Are Adrenergic Lineage, GD2+, and Responsive to Anti-GD2 Antibody Therapy. Oncoimmunology 2022, 11, 2075204. [Google Scholar] [CrossRef]
- Santillo, S. Changes in Biophysical Properties of Undifferentiated SH-SY5Y Cells During Long-Term Cultures. Neuroscience 2022, 482, 143–158. [Google Scholar] [CrossRef]
- Kelner, M.J.; Diccianni, M.B.; Yu, A.L.; Rutherford, M.R.; Estes, L.A.; Morgenstern, R. Absence of MGST1 MRNA and Protein Expression in Human Neuroblastoma Cell Lines and Primary Tissue. Free Radic. Biol. Med. 2014, 69, 167–171. [Google Scholar] [CrossRef]
- Grainger, D.W. Cell-Based Drug Testing; This World Is Not Flat. Adv. Drug Deliv. Rev. 2014, 69–70, 7–11. [Google Scholar] [CrossRef]
- Proietto, M.; Crippa, M.; Damiani, C.; Pasquale, V.; Sacco, E.; Vanoni, M.; Gilardi, M. Tumor Heterogeneity: Preclinical Models, Emerging Technologies, and Future Applications. Front. Oncol. 2023, 13, 1164535. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Zhou, J.; Xiao, Q.; Fujiwara, K.; Zhang, M.; Mo, G.; Gong, W.; Zheng, L. Cancer-Associated Fibroblast Heterogeneity Is Associated with Organ-Specific Metastasis in Pancreatic Ductal Adenocarcinoma. J. Hematol. Oncol. 2021, 14, 184. [Google Scholar] [CrossRef] [PubMed]
- Hickman, J.A.; Graeser, R.; de Hoogt, R.; Vidic, S.; Brito, C.; Gutekunst, M.; van der Kuip, H. Imi Predect consortium Three-Dimensional Models of Cancer for Pharmacology and Cancer Cell Biology: Capturing Tumor Complexity In Vitro/Ex Vivo. Biotechnol. J. 2014, 9, 1115–1128. [Google Scholar] [CrossRef]
- Nolan, J.C.; Frawley, T.; Tighe, J.; Soh, H.; Curtin, C.; Piskareva, O. Preclinical Models for Neuroblastoma: Advances and Challenges. Cancer Lett. 2020, 474, 53–62. [Google Scholar] [CrossRef]
- Corallo, D.; Frabetti, S.; Candini, O.; Gregianin, E.; Dominici, M.; Fischer, H.; Aveic, S. Emerging Neuroblastoma 3D In Vitro Models for Pre-Clinical Assessments. Front. Immunol. 2020, 11, 584214. [Google Scholar] [CrossRef] [PubMed]
- Quinn, C.H.; Beierle, A.M.; Beierle, E.A. Artificial Tumor Microenvironments in Neuroblastoma. Cancers 2021, 13, 1629. [Google Scholar] [CrossRef]
- Tumilowicz, J.J.; Nichols, W.W.; Cholon, J.J.; Greene, A.E. Definition of a Continuous Human Cell Line Derived from Neuroblastoma. Cancer Res. 1970, 30, 2110–2118. [Google Scholar]
- Reynolds, C.P.; Regino Perez-Polo, J. Human neuroblastoma: Glial induced morphological differentiation. Neurosci. Lett. 1975, 1, 91–97. [Google Scholar] [CrossRef]
- Biedler, J.L.; Helson, L.; Spengler, B.A. Morphology and Growth, Tumorigenicity, and Cytogenetics of Human Neuroblastoma Cells in Continuous Culture. Cancer Res. 1973, 33, 2643–2652. [Google Scholar]
- Mossé, Y.P.; Laudenslager, M.; Longo, L.; Cole, K.A.; Wood, A.; Attiyeh, E.F.; Laquaglia, M.J.; Sennett, R.; Lynch, J.E.; Perri, P.; et al. Identification of ALK as a Major Familial Neuroblastoma Predisposition Gene. Nature 2008, 455, 930–935. [Google Scholar] [CrossRef] [PubMed]
- Levesque, A.A.; Pappalardo, R.M.; Puli, P.; Enzor, L.A.; Angeles, C. P53 Oligomerization Status as an Indicator of Sensitivity of P53-Wildtype Neuroblastomas to the Combination of DNA Damaging Agent and Chk1 Inhibitor. PLoS ONE 2022, 17, e0263463. [Google Scholar] [CrossRef] [PubMed]
- Biedler, J.L.; Roffler-Tarlov, S.; Schachner, M.; Freedman, L.S. Multiple Neurotransmitter Synthesis by Human Neuroblastoma Cell Lines and Clones. Cancer Res. 1978, 38, 3751–3757. [Google Scholar]
- Qin Wang, H.; Halilovic, E.; Li, X.; Liang, J.; Cao, Y.; Rakiec, D.P.; Ruddy, D.A.; Jeay, S.; Wuerthner, J.U.; Timple, N.; et al. Combined ALK and MDM2 Inhibition Increases Antitumor Activity and Overcomes Resistance in Human ALK Mutant Neuroblastoma Cell Lines and Xenograft Models. eLife 2017, 6, e17137. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, D.; Ambrogio, C.; Pastorino, F.; Brignole, C.; Martinengo, C.; Carosio, R.; Loi, M.; Pagnan, G.; Emionite, L.; Cilli, M.; et al. Selective Therapeutic Targeting of the Anaplastic Lymphoma Kinase with Liposomal SiRNA Induces Apoptosis and Inhibits Angiogenesis in Neuroblastoma. Mol. Ther. 2011, 19, 2201–2212. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Chu, P.; Lingeman, R.; McDaniel, H.; Kechichian, S.; Hickey, R.J.; Liu, Z.; Yuan, Y.C.; Sandoval, J.A.; Fields, G.B.; et al. The Mechanism by Which MYCN Amplification Confers an Enhanced Sensitivity to a PCNA-Derived Cell Permeable Peptide in Neuroblastoma Cells. Ebiomedicine 2015, 2, 1923–1931. [Google Scholar] [CrossRef]
- Brodeur, G.M.; Pritchard, J.; Berthold, F.; Carlsen, N.L.T.; Castel, V.; Castleberry, R.P.; De Bernardi, B.; Evans, A.E.; Favrot, M.; Hedborg, F.; et al. Revisions of the International Criteria for Neuroblastoma Diagnosis, Staging, and Response to Treatment Purpose and Methods: Based on Preliminary Expe-Rience, There Was a Need for Modifications and Clarifi-Cations in the International Neuroblastoma Staging Sys-Tem (INSS) and International Neuroblastoma Response Criteria (INRC). J. Clin. Oncol. 1993, 11, 1466–1477. [Google Scholar] [PubMed]
- Liu, X.; Ory, V.; Chapman, S.; Yuan, H.; Albanese, C.; Kallakury, B.; Timofeeva, O.A.; Nealon, C.; Dakic, A.; Simic, V.; et al. ROCK Inhibitor and Feeder Cells Induce the Conditional Reprogramming of Epithelial Cells. Am. J. Pathol. 2012, 180, 599–607. [Google Scholar] [CrossRef] [PubMed]
- Suprynowicz, F.A.; Upadhyay, G.; Krawczyk, E.; Kramer, S.C.; Hebert, J.D.; Liu, X.; Yuan, H.; Cheluvaraju, C.; Clapp, P.W.; Boucher, R.C.; et al. Conditionally Reprogrammed Cells Represent a Stem-like State of Adult Epithelial Cells. Proc. Natl. Acad. Sci. USA 2012, 109, 20035–20040. [Google Scholar] [CrossRef]
- Saeed, K.; Rahkama, V.; Eldfors, S.; Bychkov, D.; Mpindi, J.P.; Yadav, B.; Paavolainen, L.; Aittokallio, T.; Heckman, C.; Wennerberg, K.; et al. Comprehensive Drug Testing of Patient-Derived Conditionally Reprogrammed Cells from Castration-Resistant Prostate Cancer. Eur. Urol. 2017, 71, 319–327. [Google Scholar] [CrossRef]
- Alamri, A.M.; Liu, X.; Blancato, J.K.; Haddad, B.R.; Wang, W.; Zhong, X.; Choudhary, S.; Krawczyk, E.; Kallakury, B.V.; Davidson, B.J.; et al. Expanding Primary Cells from Mucoepidermoid and Other Salivary Gland Neoplasms for Genetic and Chemosensitivity Testing. DMM Dis. Model. Mech. 2018, 11, dmm031716. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Thomas, C.; Shrivastava, N.; Gersten, A.; Gadsden, N.; Schlecht, N.; Kawachi, N.; Schiff, B.A.; Smith, R.V.; Rosenblatt, G.; et al. Establishment of a Diverse Head and Neck Squamous Cancer Cell Bank Using Conditional Reprogramming Culture Methods. J. Med. Virol. 2023, 95, e28388. [Google Scholar] [CrossRef] [PubMed]
- Alothman, S.J.; Kang, K.; Liu, X.; Krawczyk, E.; Azhar, R.I.; Hu, R.; Goerlitz, D.; Kallakury, B.V.; Furth, P.A. Characterization of Transcriptome Diversity and In Vitro Behavior of Primary Human High-Risk Breast Cells. Sci. Rep. 2022, 12, 6159. [Google Scholar] [CrossRef]
- Su, S.; Di Poto, C.; Roy, R.; Liu, X.; Cui, W.; Kroemer, A.; Ressom, H.W. Highlight Article: Long-Term Culture and Characterization of Patient-Derived Primary Hepatocytes Using Conditional Reprogramming. Exp. Biol. Med. 2019, 244, 857–864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Bai, Q.; Chen, Y.; Ye, L.; Wu, X.; Long, X.; Ye, L.; Liu, J.; Li, H. Conditionally Reprogrammed Human Normal Bronchial Epithelial Cells Express Comparable Levels of Cytochromes P450 and Are Sensitive to BaP Induction. Biochem. Biophys. Res. Commun. 2018, 503, 2132–2138. [Google Scholar] [CrossRef] [PubMed]
- Moorefield, E.C.; Blue, R.E.; Quinney, N.L.; Gentzsch, M.; Ding, S. Generation of Renewable Mouse Intestinal Epithelial Cell Monolayers and Organoids for Functional Analyses. BMC Cell Biol. 2018, 19, 15. [Google Scholar] [CrossRef]
- Mahajan, A.S.; Sugita, B.M.; Duttargi, A.N.; Saenz, F.; Krawczyk, E.; McCutcheon, J.N.; Fonseca, A.S.; Kallakury, B.; Pohlmann, P.; Gusev, Y.; et al. Genomic Comparison of Early-Passage Conditionally Reprogrammed Breast Cancer Cells to Their Corresponding Primary Tumors. PLoS ONE 2017, 12, e0186190. [Google Scholar] [CrossRef]
- Jin, L.; Qu, Y.; Gomez, L.J.; Chung, S.; Han, B.; Gao, B.; Yue, Y.; Gong, Y.; Liu, X.; Amersi, F.; et al. Characterization of Primary Human Mammary Epithelial Cells Isolated and Propagated by Conditional Reprogrammed Cell Culture. Oncotarget 2018, 9, 11503. [Google Scholar] [CrossRef]
- Krawczyk, E.; Hong, S.H.; Galli, S.; Trinh, E.; Wietlisbach, L.; Misiukiewicz, S.F.; Tilan, J.U.; Chen, Y.S.; Schlegel, R.; Kitlinska, J. Murine Neuroblastoma Cell Lines Developed by Conditional Reprogramming Preserve Heterogeneous Phenotypes Observed In Vivo. Lab. Investig. 2020, 100, 38–51. [Google Scholar] [CrossRef]
- Festing, S.; Wilkinson, R. The Ethics of Animal Research. Talking Point on the Use of Animals in Scientific Research. EMBO Rep. 2007, 8, 526–530. [Google Scholar] [CrossRef]
- Miserocchi, G.; Bocchini, M.; Cortesi, M.; Arienti, C.; De Vita, A.; Liverani, C.; Mercatali, L.; Bravaccini, S.; Ulivi, P.; Zanoni, M. Combining Preclinical Tools and Models to Unravel Tumor Complexity: Jump into the next Dimension. Front. Immunol. 2023, 14, 1171141. [Google Scholar] [CrossRef] [PubMed]
- Tosca, E.M.; Ronchi, D.; Facciolo, D.; Magni, P. Replacement, Reduction, and Refinement of Animal Experiments in Anticancer Drug Development: The Contribution of 3D In Vitro Cancer Models in the Drug Efficacy Assessment. Biomedicines 2023, 11, 1058. [Google Scholar] [CrossRef] [PubMed]
- Kimlin, L.C.; Casagrande, G.; Virador, V.M. In Vitro Three-Dimensional (3D) Models in Cancer Research: An Update. Mol. Carcinog. 2013, 52, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Restan Perez, M.; da Silva, V.A.; Thomsen, J.; Bhardwaj, L.; Andrade, T.; Alhussan, A.; Willerth, S.M. 3D Bioprinting Complex Models of Cancer. Biomater. Sci. 2023, 11, 3414–3430. [Google Scholar] [CrossRef]
- Hirschhaeuser, F.; Menne, H.; Dittfeld, C.; West, J.; Mueller-Klieser, W.; Kunz-Schughart, L.A. Multicellular Tumor Spheroids: An Underestimated Tool Is Catching up Again. J. Biotechnol. 2010, 148, 3–15. [Google Scholar] [CrossRef]
- Zanoni, M.; Piccinini, F.; Arienti, C.; Zamagni, A.; Santi, S.; Polico, R.; Bevilacqua, A.; Tesei, A. 3D Tumor Spheroid Models for in Vitro Therapeutic Screening: A Systematic Approach to Enhance the Biological Relevance of Data Obtained. Sci. Rep. 2016, 6, srep19103. [Google Scholar] [CrossRef]
- Kaess, C.; Matthes, M.; Gross, J.; Waetzig, R.; Heise, T.; Corbacioglu, S.; Sommer, G. Evaluating the RIST Molecular-Targeted Regimen in a Three-Dimensional Neuroblastoma Spheroid Cell Culture Model. Cancers 2023, 15, 1749. [Google Scholar] [CrossRef]
- Nasehi, R.; Abdallah, A.T.; Pantile, M.; Zanon, C.; Vogt, M.; Rütten, S.; Fischer, H.; Aveic, S. 3D Geometry Orchestrates the Transcriptional Landscape of Metastatic Neuroblastoma Cells in a Multicellular In Vitro Bone Model. Mater. Today Bio. 2023, 19, 100596. [Google Scholar] [CrossRef]
- Kock, A.; Bergqvist, F.; Steinmetz, J.; Elfman, L.H.M.; Korotkova, M.; Johnsen, J.I.; Jakobsson, P.J.; Kogner, P.; Larsson, K. Establishment of an In Vitro 3D Model for Neuroblastoma Enables Preclinical Investigation of Combined Tumor-Stroma Drug Targeting. FASEB J. 2020, 34, 11101–11114. [Google Scholar] [CrossRef]
- Baek, N.H.; Seo, O.W.; Kim, M.S.; Hulme, J.; An, S.S.A. Monitoring the Effects of Doxorubicin on 3D-Spheroid Tumor Cells in Real-Time. Oncol. Targets Ther. 2016, 9, 7207–7218. [Google Scholar] [CrossRef]
- Besançon, O.G.; Tytgat, G.A.M.; Meinsma, R.; Leen, R.; Hoebink, J.; Kalayda, G.V.; Jaehde, U.; Caron, H.N.; van Kuilenburg, A.B.P. Synergistic Interaction between Cisplatin and Gemcitabine in Neuroblastoma Cell Lines and Multicellular Tumor Spheroids. Cancer Lett. 2012, 319, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Craig, B.T.; Rellinger, E.J.; Alvarez, A.L.; Dusek, H.L.; Qiao, J.; Chung, D.H. Induced Differentiation Inhibits Sphere Formation in Neuroblastoma. Biochem. Biophys. Res. Commun. 2016, 477, 255–259. [Google Scholar] [CrossRef] [PubMed]
- Tesei, A.; Sarnelli, A.; Arienti, C.; Menghi, E.; Medri, L.; Gabucci, E.; Pignatta, S.; Falconi, M.; Silvestrini, R.; Zoli, W.; et al. In Vitro Irradiation System for Radiobiological Experiments. Radiat. Oncol. 2013, 8, 257. [Google Scholar] [CrossRef] [PubMed]
- Thoma, C.R.; Zimmermann, M.; Agarkova, I.; Kelm, J.M.; Krek, W. 3D Cell Culture Systems Modeling Tumor Growth Determinants in Cancer Target Discovery. Adv. Drug Deliv. Rev. 2014, 69–70, 29–41. [Google Scholar] [CrossRef]
- Redden, R.A.; Doolin, E.J. Microgravity Assay of Neuroblastoma: In Vitro Aggregation Kinetics and Organoid Morphology Correlate with MYCN Expression. Vitr. Cell Dev. Biol. Anim. 2011, 47, 312–317. [Google Scholar] [CrossRef]
- Lu, P.; Weaver, V.M.; Werb, Z. The Extracellular Matrix: A Dynamic Niche in Cancer Progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef]
- Aljitawi, O.S.; Li, D.; Xiao, Y.; Zhang, D.; Ramachandran, K.; Stehno-Bittel, L.; Van Veldhuizen, P.; Lin, T.L.; Kambhampati, S.; Garimella, R. A Novel Three-Dimensional Stromal-Based Model for In Vitro Chemotherapy Sensitivity Testing of Leukemia Cells. Leuk. Lymphoma 2014, 55, 378–391. [Google Scholar] [CrossRef]
- Innala, M.; Riebe, I.; Kuzmenko, V.; Sundberg, J.; Gatenholm, P.; Hanse, E.; Johannesson, S. 3D Culturing and Differentiation of SH-SY5Y Neuroblastoma Cells on Bacterial Nanocellulose Scaffolds. Artif. Cells Nanomed. Biotechnol. 2014, 42, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Almici, E.; Caballero, D.; Montero, J.; Samitier, J. 3D Neuroblastoma In Vitro Models Using Engineered Cell-Derived Matrices. In Biomaterials for 3D Tumor Modeling; Elsevier: Amsterdam, The Netherlands, 2020; pp. 107–130. ISBN 9780128181287. [Google Scholar]
- Neuhaus, J.; Rabien, A.; Reinhold, A.; Koehler, L.; Berndt-Paetz, M. 3D Tumor Models in Urology. Int. J. Mol. Sci. 2023, 24, 6232. [Google Scholar] [CrossRef]
- Yokota, E.; Iwai, M.; Yukawa, T.; Yoshida, M.; Naomoto, Y.; Haisa, M.; Monobe, Y.; Takigawa, N.; Guo, M.; Maeda, Y.; et al. Clinical Application of a Lung Cancer Organoid (Tumoroid) Culture System. NPJ Precis. Oncol. 2021, 5, 29. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral Organoids Model Human Brain Development and Microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Huch, M. Disease Modelling in Human Organoids. DMM Dis. Model. Mech. 2019, 12, dmm039347. [Google Scholar] [CrossRef]
- Magré, L.; Verstegen, M.M.A.; Buschow, S.; van der Laan, L.J.W.; Peppelenbosch, M.; Desai, J. Emerging Organoid-Immune Co-Culture Models for Cancer Research: From Oncoimmunology to Personalized Immunotherapies. J. Immunother. Cancer 2023, 11, e006290. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Cong, L.; Cong, X. Patient-Derived Organoids in Precision Medicine: Drug Screening, Organoid-on-a-Chip and Living Organoid Biobank. Front. Oncol. 2021, 11, 762184. [Google Scholar] [CrossRef] [PubMed]
- Gallagher, C.; Murphy, C.; Kelly, G.; O’brien, F.J.; Piskareva, O. Three-Dimensional In Vitro Biomimetic Model of Neuroblastoma Using Collagen-Based Scaffolds. J. Vis. Exp. 2021, 2021, e62627. [Google Scholar] [CrossRef]
- Bordoni, M.; Karabulut, E.; Kuzmenko, V.; Fantini, V.; Pansarasa, O.; Cereda, C.; Gatenholm, P. 3D Printed Conductive Nanocellulose Scaffolds for the Differentiation of Human Neuroblastoma Cells. Cells 2020, 9, 682. [Google Scholar] [CrossRef]
- Desai, A.; Kisaalita, W.S.; Keith, C.; Wu, Z.Z. Human Neuroblastoma (SH-SY5Y) Cell Culture and Differentiation in 3-D Collagen Hydrogels for Cell-Based Biosensing. Biosens. Bioelectron. 2006, 21, 1483–1492. [Google Scholar] [CrossRef]
- Fabbri, R.; Cacopardo, L.; Ahluwalia, A.; Magliaro, C. Advanced 3D Models of Human Brain Tissue Using Neural Cell Lines: State-of-the-Art and Future Prospects. Cells 2023, 12, 1181. [Google Scholar] [CrossRef]
- Marrella, A.; Dondero, A.; Aiello, M.; Casu, B.; Olive, D.; Regis, S.; Bottino, C.; Pende, D.; Meazza, R.; Caluori, G.; et al. Cell-Laden Hydrogel as a Clinical-Relevant 3D Model for Analyzing Neuroblastoma Growth, Immunophenotype, and Susceptibility to Therapies. Front. Immunol. 2019, 10, 1876. [Google Scholar] [CrossRef]
- Curtin, C.; Nolan, J.C.; Conlon, R.; Deneweth, L.; Gallagher, C.; Tan, Y.J.; Cavanagh, B.L.; Asraf, A.Z.; Harvey, H.; Miller-Delaney, S.; et al. A Physiologically Relevant 3D Collagen-Based Scaffold–Neuroblastoma Cell System Exhibits Chemosensitivity Similar to Orthotopic Xenograft Models. Acta Biomater. 2018, 70, 84–97. [Google Scholar] [CrossRef]
- Monferrer, E.; Dobre, O.; Trujillo, S.; González Oliva, M.A.; Trubert-Paneli, A.; Acevedo-León, D.; Noguera, R.; Salmeron-Sanchez, M. Vitronectin-Based Hydrogels Recapitulate Neuroblastoma Growth Conditions. Front. Cell Dev. Biol. 2022, 10, 988699. [Google Scholar] [CrossRef]
- Nothdurfter, D.; Ploner, C.; Coraça-Huber, D.C.; Wilflingseder, D.; Müller, T.; Hermann, M.; Hagenbuchner, J.; Ausserlechner, M.J. 3D Bioprinted, Vascularized Neuroblastoma Tumor Environment in Fluidic Chip Devices for Precision Medicine Drug Testing. Biofabrication 2022, 14, 035002. [Google Scholar] [CrossRef] [PubMed]
- Villasante, A.; Sakaguchi, K.; Kim, J.; Cheung, N.K.; Nakayama, M.; Parsa1, H.; Okano, T.; Shimizu, T.; Vunjak-Novakovic, G. Vascularized Tissue-Engineered Model for Studying Drug Resistance in Neuroblastoma. Theranostics 2017, 7, 4099–4117. [Google Scholar] [CrossRef]
- Wan, X.; Wang, W.; Liang, Z. Epigallocatechin-3-Gallate Inhibits the Growth of Three-Dimensional in Vitro Models of Neuroblastoma Cell SH-SY5Y. Mol. Cell Biochem. 2021, 476, 3141–3148. [Google Scholar] [CrossRef] [PubMed]
- Ning, L.; Shim, J.; Tomov, M.L.; Liu, R.; Mehta, R.; Mingee, A.; Hwang, B.; Jin, L.; Mantalaris, A.; Xu, C.; et al. A 3D Bioprinted in Vitro Model of Neuroblastoma Recapitulates Dynamic Tumor-Endothelial Cell Interactions Contributing to Solid Tumor Aggressive Behavior. Adv. Sci. 2022, 9, 2200244. [Google Scholar] [CrossRef]
- Barberio, C.; Withers, A.; Mishra, Y.; Couraud, P.O.; Romero, I.A.; Weksler, B.; Owens, R.M. A Human-Derived Neurovascular Unit In Vitro Model to Study the Effects of Cellular Cross-Talk and Soluble Factors on Barrier Integrity. Front. Cell Neurosci. 2022, 16, 1065193. [Google Scholar] [CrossRef] [PubMed]
- Weng, Z.; Lin, J.; He, J.; Gao, L.; Lin, S.; Tsang, L.L.; Zhang, H.; He, X.; Wang, G.; Yang, X.; et al. Human Embryonic Stem Cell-Derived Neural Crest Model Unveils CD55 as a Cancer Stem Cell Regulator for Therapeutic Targeting in MYCN-Amplified Neuroblastoma. Neuro Oncol. 2022, 24, 872–885. [Google Scholar] [CrossRef] [PubMed]
- Saxena, S.; Wahl, J.; Huber-Lang, M.S.; Stadel, D.; Braubach, P.; Debatin, K.M.; Beltinger, C. Generation of Murine Sympathoadrenergic Progenitor-Like Cells from Embryonic Stem Cells and Postnatal Adrenal Glands. PLoS ONE 2013, 8, e64454. [Google Scholar] [CrossRef]
- Schloo, H.; Kutscher, L.M. Modeling Brain and Neural Crest Neoplasms with Human Pluripotent Stem Cells. Neuro Oncol. 2023. online ahead of print. [Google Scholar] [CrossRef]
- Carr-Wilkinson, J.; Prathalingam, N.; Pal, D.; Moad, M.; Lee, N.; Sundaresh, A.; Forgham, H.; James, P.; Herbert, M.; Lako, M.; et al. Differentiation of Human Embryonic Stem Cells to Sympathetic Neurons: A Potential Model for Understanding Neuroblastoma Pathogenesis. Stem Cells Int. 2018, 2018, 4391641. [Google Scholar] [CrossRef]
- Kramer, M.; Ribeiro, D.; Arsenian-Henriksson, M.; Deller, T.; Rohrer, H. Proliferation and Survival of Embryonic Sympathetic Neuroblasts by MYCN and Activated ALK Signaling. J. Neurosci. 2016, 36, 10425–10439. [Google Scholar] [CrossRef]
- Huang, M.; Weiss, W.A. Neuroblastoma and MYCN. Cold Spring Harb. Perspect. Med. 2013, 3, a014415. [Google Scholar] [CrossRef]
- Weiss, W.A.; Aldape, K.; Mohapatra, G.; Feuerstein, B.G.; Michael Bishop, J. Targeted Expression of MYCN Causes Neuroblastoma in Transgenic Mice. EMBO J. 1997, 16, 2985–2995. [Google Scholar] [CrossRef]
- Teitz, T.; Stanke, J.J.; Federico, S.; Bradley, C.L.; Brennan, R.; Zhang, J.; Johnson, M.D.; Sedlacik, J.; Inoue, M.; Zhang, Z.M.; et al. Preclinical Models for Neuroblastoma: Establishing a Baseline for Treatment. PLoS ONE 2011, 6, e19133. [Google Scholar] [CrossRef]
- Webb, E.R.; Lanati, S.; Wareham, C.; Easton, A.; Dunn, S.N.; Inzhelevskaya, T.; Sadler, F.M.; James, S.; Ashton-Key, M.; Cragg, M.S.; et al. Immune Characterization of Pre-Clinical Murine Models of Neuroblastoma. Sci. Rep. 2020, 10, 16695. [Google Scholar] [CrossRef]
- Hong, S.H.; Wietlisbach, L.; Galli, S.; Mahajan, A.; Zhu, S.; Tilan, J.; Lee, Y.; Rodriguez, O.; Albanese, C.; Kitlinska, J. Abstract 1940: Prenatal Stress Increases Malignancy of Neuroblastoma Tumors in TH-MYCN Animal Model. Cancer Res. 2017, 77, 1940. [Google Scholar] [CrossRef]
- Faisal, A.; Vaughan, L.; Bavetsias, V.; Sun, C.; Atrash, B.; Avery, S.; Jamin, Y.; Robinson, S.P.; Workman, P.; Blagg, J.; et al. The Aurora Kinase Inhibitor CCT137690 Downregulates MYCN and Sensitizes MYCN-Amplified Neuroblastoma In Vivo. Mol. Cancer Ther. 2011, 10, 2115–2123. [Google Scholar] [CrossRef]
- Morowitz, M.J.; Barr, R.; Wang, Q.; King, R.; Rhodin, N.; Pawel, B.; Zhao, H.; Erickson, S.A.; Sheppard, G.S.; Wang, J.; et al. Methionine Aminopeptidase 2 Inhibition Is an EffectiveTreatment Strategy for Neuroblastoma in Preclinical Models. Clin. Cancer Res. 2005, 11, 2680–2685. [Google Scholar] [CrossRef]
- Yogev, O.; Almeida, G.S.; Barker, K.T.; George, S.L.; Kwok, C.; Campbell, J.; Zarowiecki, M.; Kleftogiannis, D.; Smith, L.M.; Hallsworth, A.; et al. In Vivo Modeling of Chemoresistant Neuroblastoma Provides New Insights into Chemorefractory Disease and Metastasis. Cancer Res. 2019, 79, 5382–5393. [Google Scholar] [CrossRef] [PubMed]
- Chesler, L.; Weiss, W.A. Genetically Engineered Murine Models-Contribution to Our Understanding of the Genetics, Molecular Pathology and Therapeutic Targeting of Neuroblastoma. Semin. Cancer Biol. 2011, 21, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Nijhuis, A.; Sikka, A.; Yogev, O.; Herendi, L.; Balcells, C.; Ma, Y.; Poon, E.; Eckold, C.; Valbuena, G.N.; Xu, Y.; et al. Indisulam Targets RNA Splicing and Metabolism to Serve as a Therapeutic Strategy for High-Risk Neuroblastoma. Nat. Commun. 2022, 13, 1380. [Google Scholar] [CrossRef] [PubMed]
- Kamili, A.; Atkinson, C.; Trahair, T.N.; Fletcher, J.I. Mouse Models of High-Risk Neuroblastoma. Cancer Metastasis Rev. 2020, 39, 261–274. [Google Scholar] [CrossRef]
- Yogev, O.; Barker, K.; Sikka, A.; Almeida, G.S.; Hallsworth, A.; Smith, L.M.; Jamin, Y.; Ruddle, R.; Koers, A.; Twebber, H.; et al. P53 Loss in MYC-Driven Neuroblastoma Leads to Metabolic Adaptations Supporting Radioresistance. Cancer Res. 2016, 76, 3025–3035. [Google Scholar] [CrossRef] [PubMed]
- Kim, P.Y.; Tan, O.; Diakiw, S.M.; Carter, D.; Sekerye, E.O.; Wasinger, V.C.; Liu, T.; Kavallaris, M.; Norris, M.D.; Haber, M.; et al. Identification of Plasma Complement C3 as a Potential Biomarker for Neuroblastoma Using a Quantitative Proteomic Approach. J. Proteom. 2014, 96, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shawraba, F.; Hammoud, H.; Mrad, Y.; Saker, Z.; Fares, Y.; Harati, H.; Bahmad, H.F.; Nabha, S. Biomarkers in Neuroblastoma: An Insight into Their Potential Diagnostic and Prognostic Utilities. Curr. Treat Options Oncol. 2021, 22, 102. [Google Scholar] [CrossRef]
- Teitz, T.; Inoue, M.; Valentine, M.B.; Zhu, K.; Rehg, J.E.; Zhao, W.; Finkelstein, D.; Wang, Y.D.; Johnson, M.D.; Calabrese, C.; et al. Th-MYCN Mice with Caspase-8 Deficiency Develop Advanced Neuroblastoma with Bone Marrow Metastasis. Cancer Res. 2013, 73, 4086–4097. [Google Scholar] [CrossRef] [PubMed]
- Althoff, K.; Schramm, A. MYCN-Mediated Murine Cancer Models. Aging 2017, 9, 1084–1085. [Google Scholar] [CrossRef]
- Althoff, K.; Beckers, A.; Bell, E.; Nortmeyer, M.; Thor, T.; Sprüssel, A.; Lindner, S.; De Preter, K.; Florin, A.; Heukamp, L.C.; et al. A Cre-Conditional MYCN-Driven Neuroblastoma Mouse Model as an Improved Tool for Preclinical Studies. Oncogene 2015, 34, 3357–3368. [Google Scholar] [CrossRef]
- Chen, Z.; Lin, Y.; Barbieri, E.; Burlingame, S.; Hicks, J.; Ludwig, A.; Shohet, J.M. Mdm2 Deficiency Suppresses MYCN-Driven Neuroblastoma Tumorigenesis In Vivo. Neoplasia 2009, 11, 753–762. [Google Scholar] [CrossRef]
- Galli, S.; Naranjo, A.; Van Ryn, C.; Tilan, J.U.; Trinh, E.; Yang, C.; Tsuei, J.; Hong, S.H.; Wang, H.; Izycka-Swieszewska, E.; et al. Neuropeptide Y as a Biomarker and Therapeutic Target for Neuroblastoma. Am. J. Pathol. 2016, 186, 3040–3053. [Google Scholar] [CrossRef] [PubMed]
- Schleiermacher, G.; Janoueix-Lerosey, I.; Delattre, O. Recent Insights into the Biology of Neuroblastoma. Int. J. Cancer 2014, 135, 2249–2261. [Google Scholar] [CrossRef]
- Kiyonari, S.; Kadomatsu, K. Neuroblastoma Models for Insights into Tumorigenesis and New Therapies. Expert Opin. Drug Discov. 2015, 10, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Heukamp, L.C.; Thor, T.; Schramm, A.; De Preter, K.; Kumps, C.; De Wilde, B.; Odersky, A.; Peifer, M.; Lindner, S.; Spruessel, A.; et al. Targeted Expression of Mutated ALK Induces Neuroblastoma in Transgenic Mice. Sci. Transl. Med. 2012, 4, 141ra91. [Google Scholar] [CrossRef] [PubMed]
- Molenaar, J.J.; Domingo-Fernández, R.; Ebus, M.E.; Lindner, S.; Koster, J.; Drabek, K.; Mestdagh, P.; Van Sluis, P.; Valentijn, L.J.; Van Nes, J.; et al. LIN28B Induces Neuroblastoma and Enhances MYCN Levels via Let-7 Suppression. Nat. Genet. 2012, 44, 1199–1206. [Google Scholar] [CrossRef]
- Berry, T.; Luther, W.; Bhatnagar, N.; Jamin, Y.; Poon, E.; Sanda, T.; Pei, D.; Sharma, B.; Vetharoy, W.R.; Hallsworth, A.; et al. The ALKF1174L Mutation Potentiates the Oncogenic Activity of MYCN in Neuroblastoma. Cancer Cell 2012, 22, 117–130. [Google Scholar] [CrossRef] [PubMed]
- Ono, S.; Saito, T.; Terui, K.; Yoshida, H.; Enomoto, H. Generation of Conditional ALK F1174L Mutant Mouse Models for the Study of Neuroblastoma Pathogenesis. Genesis 2019, 57, e23323. [Google Scholar] [CrossRef]
- Tao, T.; Shi, H.; Mariani, L.; Abraham, B.J.; Durbin, A.D.; Zimmerman, M.W.; Powers, J.T.; Missios, P.; Ross, K.N.; Perez-Atayde, A.R.; et al. LIN28B Regulates Transcription and Potentiates MYCN-Induced Neuroblastoma through Binding to ZNF143 at Target Gene Promotors. Proc. Natl. Acad. Sci. USA 2020, 117, 16516–16526. [Google Scholar] [CrossRef]
- De Wilde, B.; Beckers, A.; Lindner, S.; Kristina, A.; De Preter, K.; Depuydt, P.; Mestdagh, P.; Sante, T.; Lefever, S.; Hertwig, F.; et al. The Mutational Landscape of MYCN, Lin28b and ALK F1174L Driven Murine Neuroblastoma Mimics Human Disease. Oncotarget 2018, 9, 8334. [Google Scholar] [CrossRef]
- Singh, S.; Quarni, W.; Goralski, M.; Wan, S.; Jin, H.; Van de Velde, L.-A.; Fang, J.; Wu, Q.; Abu-Zaid, A.; Wang, T.; et al. Targeting the Spliceosome through RBM39 Degradation Results in Exceptional Responses in High-Risk Neuroblastoma Models. Sci. Adv. 2021, 7, eabj5405. [Google Scholar] [CrossRef]
- Iwakura, H.; Akamizu, T. Neuroblastoma Mouse Model; Hayat, M.A., Ed.; Pediatric Cancer; Springer: Berlin/Heidelberg, Germany, 2012. [Google Scholar]
- Koike, K.; Jay, G.; Hartley, J.W.; Schrenzel, M.D.; Higgins, R.J.; Hinrichs, S.H. Activation of Retrovirus in Transgenic Mice: Association with Development of Olfactory Neuroblastoma. J. Virol. 1990, 64, 3988–3991. [Google Scholar] [CrossRef]
- Small, J.A.; Khoury, G.; Jayt, G.; Howley, P.M.; Scangos, G.A. Early Regions of JC Virus and BK Virus Induce Distinct and Tissue-Specific Tumors in Transgenic Mice. Proc. Natl. Acad. Sci. USA 1986, 83, 8288–8292. [Google Scholar] [CrossRef]
- Iwamoto, T.; Taniguchi, M.; Wajjwalku, W.; Nakashimal, I.; Takahashi, M. Neuroblastoma in a Transgenic Mouse Carrying a Metallothionein/Ret Fusion Gene. Br. J. Cancer 1993, 67, 504. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, C.; Sakata, S.; Kakinoki, S.; Takeyama, Y.; Ohyanagi, H.; Shiozaki, H. In Vivo Evaluation of Microspheres Containing the Angiogenesis Inhibitor, TNP-470, and the Metastasis Suppression with Liver Metastatic Model Implanted Neuroblastoma. Pathophysiology 2010, 17, 149–155. [Google Scholar] [CrossRef]
- Beltinger, C.; Debatin, K.M. Murine Models for Experimental Therapy of Pediatric Solid Tumors with Poor Prognosis. Int. J. Cancer 2001, 92, 313–318. [Google Scholar] [CrossRef]
- Khanna, C.; Jaboin, J.J.; Drakos, E.; Tsokos, M.; Thiele, C.J. Biologically Relevant Orthotopic Neuroblastoma Xenograft Models: Primary Adrenal Tumor Growth and Spontaneous Distant Metastasis. In Vivo 2002, 16, 77–86. [Google Scholar]
- Shultz, L.D.; Goodwin, N.; Ishikawa, F.; Hosur, V.; Lyons, B.L.; Greiner, D.L. Human Cancer Growth and Therapy in Immunodeficient Mouse Models. Cold Spring Harb. Protoc. 2014, 2014, 694–708. [Google Scholar] [CrossRef]
- Kaneko, Y.; Tsuchida, Y.; Maseki, N.; Takasaki, N.; Sakurai, M.; Saito, S. Chromosome findings in human neuroblastomas xenografted in nude mice. J. Cancer Res. GANN 1985, 76, 359–364. [Google Scholar]
- Hidalgo, M.; Bruckheimer, E.; Rajeshkumar, N.V.; Garrido-Laguna, I.; De Oliveira, E.; Rubio-Viqueira, B.; Strawn, S.; Wick, M.J.; Martell, J.; Sidransky, D. A Pilot Clinical Study of Treatment Guided by Personalized Tumorgrafts in Patients with Advanced Cancer. Mol. Cancer Ther. 2011, 10, 1311–1316. [Google Scholar] [CrossRef]
- Gao, Q.; Chen, C.F.; Dong, Q.; Hou, L.; Chen, X.; Zhi, Y.L.; Li, X.; Lu, H.T.; Zhang, H.Y. Establishment of a Neuroblastoma Mouse Model by Subcutaneous Xenograft Transplantation and Its Use to Study Metastatic Neuroblastoma. Genet. Mol. Res. 2015, 14, 16297–16307. [Google Scholar] [CrossRef]
- Braekeveldt, N.; Wigerup, C.; Gisselsson, D.; Mohlin, S.; Merselius, M.; Beckman, S.; Jonson, T.; Börjesson, A.; Backman, T.; Tadeo, I.; et al. Neuroblastoma Patient-Derived Orthotopic Xenografts Retain Metastatic Patterns and Geno-and Phenotypes of Patient Tumours. Int. J. Cancer 2015, 136, E252–E261. [Google Scholar] [CrossRef]
- Braekeveldt, N.; Bexell, D. Patient-Derived Xenografts as Preclinical Neuroblastoma Models. Cell Tissue Res. 2018, 372, 233–243. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.N.; Wills, C.A.; Liu, X.; Spiegelman, V.S.; Wang, H.G. Thoracic Neuroblastoma: A Novel Surgical Model for the Study of Extra-Adrenal Neuroblastoma. In Vivo 2022, 36, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.; Shelat, A.; Bradley, C.; Chen, X.; Federico, S.; Thiagarajan, S.; Shirinifard, A.; Bahrami, A.; Pappo, A.; Qu, C.; et al. Development and Characterization of a Human Orthotopic Neuroblastoma Xenograft. Dev. Biol. 2015, 407, 344–355. [Google Scholar] [CrossRef]
- Braekeveldt, N.; Wigerup, C.; Tadeo, I.; Beckman, S.; Sandén, C.; Jönsson, J.; Erjefält, J.S.; Berbegall, A.P.; Börjesson, A.; Backman, T.; et al. Neuroblastoma Patient-Derived Orthotopic Xenografts Reflect the Microenvironmental Hallmarks of Aggressive Patient Tumours. Cancer Lett. 2016, 375, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Cao, Y.; Okada, R.; Reyes-González, J.M.; Stack, H.G.; Qin, H.; Li, N.; Seibert, C.; Kelly, M.C.; Ruppin, E.; et al. Preclinical Optimization of a GPC2-Targeting CAR T-Cell Therapy for Neuroblastoma. J. Immunother. Cancer 2023, 11, e005881. [Google Scholar] [CrossRef]
- Quinn, C.H.; Beierle, A.M.; Hutchins, S.C.; Marayati, R.; Bownes, L.V.; Stewart, J.E.; Markert, H.R.; Erwin, M.H.; Aye, J.M.; Yoon, K.J.; et al. Targeting High-Risk Neuroblastoma Patient-Derived Xenografts with Oncolytic Virotherapy. Cancers 2022, 14, 762. [Google Scholar] [CrossRef]
- Labitzky, V.; Baranowsky, A.; Maar, H.; Hanika, S.; Starzonek, S.; Ahlers, A.K.; Stübke, K.; Koziolek, E.J.; Heine, M.; Schäfer, P.; et al. Modeling Spontaneous Bone Metastasis Formation of Solid Human Tumor Xenografts in Mice. Cancers 2020, 12, 385. [Google Scholar] [CrossRef]
- Rokita, J.L.; Rathi, K.S.; Cardenas, M.F.; Upton, K.A.; Jayaseelan, J.; Cross, K.L.; Pfeil, J.; Egolf, L.E.; Way, G.P.; Farrel, A.; et al. Genomic Profiling of Childhood Tumor Patient-Derived Xenograft Models to Enable Rational Clinical Trial Design. Cell Rep. 2019, 29, 1675–1689.e9. [Google Scholar] [CrossRef] [PubMed]
- Morton, J.J.; Bird, G.; Keysar, S.B.; Astling, D.P.; Lyons, T.R.; Anderson, R.T.; Glogowska, M.J.; Estes, P.; Eagles, J.R.; Le, P.N.; et al. XactMice: Humanizing Mouse Bone Marrow Enables Microenvironment Reconstitution in a Patient-Derived Xenograft Model of Head and Neck Cancer. Oncogene 2016, 35, 290–300. [Google Scholar] [CrossRef]
- Cohen, M.A.; Zhang, S.; Sengupta, S.; Ma, H.; Bell, G.W.; Horton, B.; Sharma, B.; George, R.E.; Spranger, S.; Jaenisch, R. Formation of Human Neuroblastoma in Mouse-Human Neural Crest Chimeras. Cell Stem Cell 2020, 26, 579–592.e6. [Google Scholar] [CrossRef]
- Thole, T.M.; Toedling, J.; Sprüssel, A.; Pfeil, S.; Savelyeva, L.; Capper, D.; Messerschmidt, C.; Beule, D.; Groeneveld-Krentz, S.; Eckert, C.; et al. Reflection of Neuroblastoma Intratumor Heterogeneity in the New OHC-NB1 Disease Model. Int. J. Cancer 2020, 146, 1031–1041. [Google Scholar] [CrossRef]
- Li, S.; Yeo, K.S.; Levee, T.M.; Howe, C.J.; Her, Z.P.; Zhu, S. Zebrafish as a Neuroblastoma Model: Progress Made, Promise for the Future. Cells 2021, 10, 580. [Google Scholar] [CrossRef]
- Martinez, D.A.; Kahwash, S.; O’Dorisio, M.S.; Lloyd, T.V.; McGhee, R.B.; Qualman, S.J. Disseminated Neuroblastoma in the Nude Rat: A Xenograft Model of Human Malignancy. Cancer 1996, 77, 409–419. [Google Scholar] [CrossRef]
- Medina-Cuadra, L.; Monsoro-Burq, A.H. Xenopus, an Emerging Model for Studying Pathologies of the Neural Crest. In Current Topics in Developmental Biology; Academic Press Inc.: Cambridge, MA, USA, 2021; Volume 145, pp. 313–348. [Google Scholar]
- Gonzalez Malagon, S.G.; Lopez Muñoz, A.M.; Doro, D.; Bolger, T.G.; Poon, E.; Tucker, E.R.; Adel Al-Lami, H.; Krause, M.; Phiel, C.J.; Chesler, L.; et al. Glycogen Synthase Kinase 3 Controls Migration of the Neural Crest Lineage in Mouse and Xenopus. Nat. Commun. 2018, 9, 1126. [Google Scholar] [CrossRef]
- Corallo, D.; Donadon, M.; Pantile, M.; Sidarovich, V.; Cocchi, S.; Ori, M.; De Sarlo, M.; Candiani, S.; Frasson, C.; Distel, M.; et al. LIN28B Increases Neural Crest Cell Migration and Leads to Transformation of Trunk Sympathoadrenal Precursors. Cell Death Differ. 2020, 27, 1225–1242. [Google Scholar] [CrossRef]
- Zhu, S.; Thomas Look, A. Neuroblastoma and Its Zebrafish Model. In Advances in Experimental Medicine and Biology; Springer: New York, NY, USA, 2016; Volume 916, pp. 451–478. [Google Scholar]
- Her, Z.P.; Yeo, K.S.; Howe, C.; Levee, T.; Zhu, S. Zebrafish Model of Neuroblastoma Metastasis. J. Vis. Exp. 2021, 2021, e62416. [Google Scholar] [CrossRef]
- Sarmiento, B.E.; Callegari, S.; Ghotme, K.A.; Akle, V. Patient-Derived Xenotransplant of CNS Neoplasms in Zebrafish: A Systematic Review. Cells 2022, 11, 1204. [Google Scholar] [CrossRef]
- Delloye-Bourgeois, C.; Bertin, L.; Thoinet, K.; Jarrosson, L.; Kindbeiter, K.; Buffet, T.; Tauszig-Delamasure, S.; Bozon, M.; Marabelle, A.; Combaret, V.; et al. Microenvironment-Driven Shift of Cohesion/Detachment Balance within Tumors Induces a Switch toward Metastasis in Neuroblastoma. Cancer Cell 2017, 32, 427–443.e8. [Google Scholar] [CrossRef]
- Ben Amar, D.; Thoinet, K.; Villalard, B.; Imbaud, O.; Costechareyre, C.; Jarrosson, L.; Reynaud, F.; Novion Ducassou, J.; Couté, Y.; Brunet, J.F.; et al. Environmental Cues from Neural Crest Derivatives Act as Metastatic Triggers in an Embryonic Neuroblastoma Model. Nat. Commun. 2022, 13, e16629. [Google Scholar] [CrossRef] [PubMed]
- Swadi, R.; Mather, G.; Pizer, B.L.; Losty, P.D.; See, V.; Moss, D. Optimising the Chick Chorioallantoic Membrane Xenograft Model of Neuroblastoma for Drug Delivery. BMC Cancer 2018, 18, 28. [Google Scholar] [CrossRef] [PubMed]
- Ribatti, D.; Tamma, R. The Chick Embryo Chorioallantoic Membrane as an In Vivo Experimental Model to Study Human Neuroblastoma. J. Cell Physiol. 2018, 234, 152–157. [Google Scholar] [CrossRef]
- Herrmann, A.; Rice, M.; Lévy, R.; Pizer, B.L.; Losty, P.D.; Moss, D.; Sée, V. Cellular Memory of Hypoxia Elicits Neuroblastoma Metastasis and Enables Invasion by Non-Aggressive Neighbouring Cells. Oncogenesis 2015, 4, e138. [Google Scholar] [CrossRef] [PubMed]
- Bogenmann, E. A Metastatic Neuroblastoma Model in SCID Mice. Int. J. Cancer 1996, 67, 379–385. [Google Scholar] [CrossRef]
- Chen, D.; Cox, J.; Annam, J.; Weingart, M.; Essien, G.; Rathi, K.S.; Rokita, J.L.; Khurana, P.; Cuya, S.M.; Bosse, K.R.; et al. LIN28B Promotes Neuroblastoma Metastasis and Regulates PDZ Binding Kinase. Neoplasia 2020, 22, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Seong, B.K.A.; Fathers, K.E.; Hallett, R.; Yung, C.K.; Stein, L.D.; Mouaaz, S.; Kee, L.; Hawkins, C.E.; Irwin, M.S.; Kaplan, D.R. A Metastatic Mouse Model Identifies Genes That Regulate Neuroblastoma Metastasis. Cancer Res. 2017, 77, 696–706. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.L.; Chan, S.; Fung, M.K.L.; Chan, G.C.F. Mesenchymal Stem Cells Accelerated Growth and Metastasis of Neuroblastoma and Preferentially Homed towards Both Primary and Metastatic Loci in Orthotopic Neuroblastoma Model. BMC Cancer 2021, 21, 393. [Google Scholar] [CrossRef] [PubMed]
- Segura, M.F.; Soriano, A.; Roma, J.; Piskareva, O.; Jiménez, C.; Boloix, A.; Fletcher, J.I.; Haber, M.; Gray, J.C.; Cerdá-Alberich, L.; et al. Methodological Advances in the Discovery of Novel Neuroblastoma Therapeutics. Expert Opin. Drug Discov. 2022, 17, 167–179. [Google Scholar] [CrossRef]
- Chilamakuri, R.; Rouse, D.C.; Agarwal, S. Inhibition of Polo-like Kinase 1 by HMN-214 Blocks Cell Cycle Progression and Inhibits Neuroblastoma Growth. Pharmaceuticals 2022, 15, 523. [Google Scholar] [CrossRef]
- Marzagalli, M.; Pelizzoni, G.; Fedi, A.; Vitale, C.; Fontana, F.; Bruno, S.; Poggi, A.; Dondero, A.; Aiello, M.; Castriconi, R.; et al. A Multi-Organ-on-Chip to Recapitulate the Infiltration and the Cytotoxic Activity of Circulating NK Cells in 3D Matrix-Based Tumor Model. Front. Bioeng. Biotechnol. 2022, 10, 945149. [Google Scholar] [CrossRef]
- Heinze, A.; Grebe, B.; Bremm, M.; Huenecke, S.; Munir, T.A.; Graafen, L.; Frueh, J.T.; Merker, M.; Rettinger, E.; Soerensen, J.; et al. The Synergistic Use of IL-15 and IL-21 for the Generation of NK Cells From CD3/CD19-Depleted Grafts Improves Their Ex Vivo Expansion and Cytotoxic Potential Against Neuroblastoma: Perspective for Optimized Immunotherapy Post Haploidentical Stem Cell Transplantation. Front. Immunol. 2019, 10, 2816. [Google Scholar] [CrossRef]
- Logan, J.A.; Kelly, M.E.; Ayers, D.; Shipillis, N.; Baier, G.; Day, P.J.R. Systems Biology and Modeling in Neuroblastoma: Practicalities and Perspectives. Expert Rev. Mol. Diagn. 2010, 10, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Liu, Q.; Cao, Z.; Wang, J.; Zhang, H.; Liu, J.; Zou, L. Multi-Omics Integration Reveals a Six-Malignant Cell Maker Gene Signature for Predicting Prognosis in High-Risk Neuroblastoma. Front. Neuroinform. 2022, 16, 1034793. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).