Spectrum and Frequency of Germline FANCM Protein-Truncating Variants in 44,803 European Female Breast Cancer Cases
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results and Discussion
3.1. Frequency of Germline FANCM PTVs
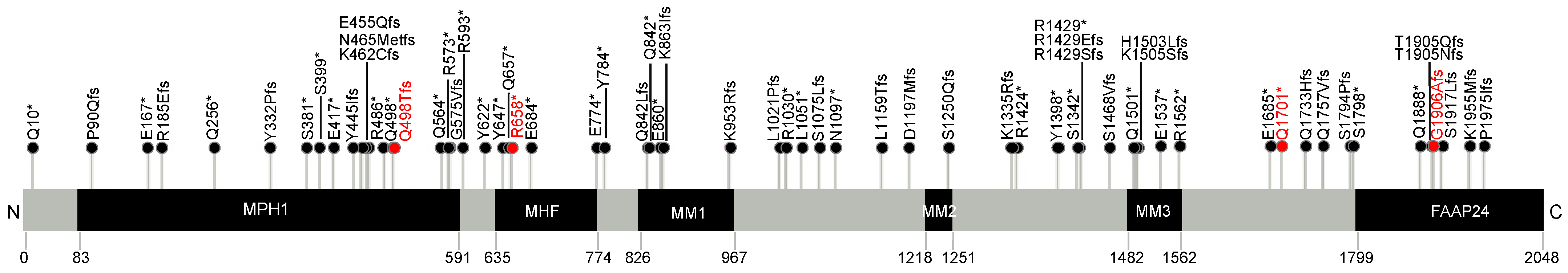
3.2. Spectrum of Common and Rare FANCM PTVs
3.3. Comprehensive Spectrum of FANCM PTVs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lilyquist, J.; Ruddy, K.J.; Vachon, C.M.; Couch, F.J. Common Genetic Variation and Breast Cancer Risk-Past, Present, and Future. Cancer Epidemiol. Biomark. Prev. 2018, 27, 380–394. [Google Scholar] [CrossRef]
- Kuchenbaecker, K.B.; Hopper, J.L.; Barnes, D.R.; Phillips, K.A.; Mooij, T.M.; Roos-Blom, M.J.; Jervis, S.; van Leeuwen, F.E.; Milne, R.L.; Andrieu, N.; et al. Risks of Breast, Ovarian, and Contralateral Breast Cancer for BRCA1 and BRCA2 Mutation Carriers. JAMA 2017, 317, 2402–2416. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Leslie, G.; Doroszuk, A.; Schneider, S.; Allen, J.; Decker, B.; Dunning, A.M.; Redman, J.; Scarth, J.; Plaskocinska, I.; et al. Cancer Risks Associated With Germline PALB2 Pathogenic Variants: An International Study of 524 Families. J. Clin. Oncol. 2020, 38, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Breast Cancer Association Consortium; Dorling, L.; Carvalho, S.; Allen, J.; Gonzalez-Neira, A.; Luccarini, C.; Wahlstrom, C.; Pooley, K.A.; Parsons, M.T.; Fortuno, C.; et al. Breast Cancer Risk Genes—Association Analysis in More than 113,000 Women. N. Engl. J. Med. 2021, 384, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Hart, S.N.; Gnanaolivu, R.; Huang, H.; Lee, K.Y.; Na, J.; Gao, C.; Lilyquist, J.; Yadav, S.; Boddicker, N.J.; et al. A Population-Based Study of Genes Previously Implicated in Breast Cancer. N. Engl. J. Med. 2021, 384, 440–451. [Google Scholar] [CrossRef]
- Laitman, Y.; Friebel, T.M.; Yannoukakos, D.; Fostira, F.; Konstantopoulou, I.; Figlioli, G.; Bonanni, B.; Manoukian, S.; Zuradelli, M.; Tondini, C.; et al. The spectrum of BRCA1 and BRCA2 pathogenic sequence variants in Middle Eastern, North African, and South European countries. Hum. Mutat. 2019, 40, e1–e23. [Google Scholar] [CrossRef]
- Rebbeck, T.R.; Friebel, T.M.; Friedman, E.; Hamann, U.; Huo, D.; Kwong, A.; Olah, E.; Olopade, O.I.; Solano, A.R.; Teo, S.H.; et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum. Mutat. 2018, 39, 593–620. [Google Scholar] [CrossRef]
- Janssen, B.; Bellis, S.; Koller, T.; Tischkowitz, M.; Liau, S.S. A systematic review of predicted pathogenic PALB2 variants: An analysis of mutational overlap between epithelial cancers. J. Hum. Genet. 2020, 65, 199–205. [Google Scholar] [CrossRef]
- Suszynska, M.; Kozlowski, P. Summary of BARD1 Mutations and Precise Estimation of Breast and Ovarian Cancer Risks Associated with the Mutations. Genes 2020, 11, 798. [Google Scholar] [CrossRef]
- Boni, J.; Idani, A.; Roca, C.; Feliubadalo, L.; Tomiak, E.; Weber, E.; Foulkes, W.D.; Orthwein, A.; El Haffaf, Z.; Lazaro, C.; et al. A decade of RAD51C and RAD51D germline variants in cancer. Hum. Mutat. 2022, 43, 285–298. [Google Scholar] [CrossRef]
- Pavlovica, K.; Irmejs, A.; Noukas, M.; Palover, M.; Kals, M.; Tonisson, N.; Metspalu, A.; Gronwald, J.; Lubinski, J.; Murmane, D.; et al. Spectrum and frequency of CHEK2 variants in breast cancer affected and general population in the Baltic states region, initial results and literature review. Eur. J. Med. Genet. 2022, 65, 104477. [Google Scholar] [CrossRef]
- Foulkes, W.D. The ten genes for breast (and ovarian) cancer susceptibility. Nat. Rev. Clin. Oncol. 2021, 18, 259–260. [Google Scholar] [CrossRef]
- Peterlongo, P.; Figlioli, G.; Deans, A.J.; Couch, F.J. Protein truncating variants in FANCM and risk for ER-negative/triple negative breast cancer. NPJ Breast Cancer 2021, 7, 130. [Google Scholar] [CrossRef]
- Figlioli, G.; Bogliolo, M.; Catucci, I.; Caleca, L.; Lasheras, S.V.; Pujol, R.; Kiiski, J.I.; Muranen, T.A.; Barnes, D.R.; Dennis, J.; et al. The FANCM:p.Arg658* truncating variant is associated with risk of triple-negative breast cancer. NPJ Breast Cancer 2019, 5, 38. [Google Scholar] [CrossRef]
- Peterlongo, P.; Catucci, I.; Colombo, M.; Caleca, L.; Mucaki, E.; Bogliolo, M.; Marin, M.; Damiola, F.; Bernard, L.; Pensotti, V.; et al. FANCM c.5791C>T nonsense mutation (rs144567652) induces exon skipping, affects DNA repair activity and is a familial breast cancer risk factor. Hum. Mol. Genet. 2015, 24, 5345–5355. [Google Scholar] [CrossRef]
- Kiiski, J.I.; Pelttari, L.M.; Khan, S.; Freysteinsdottir, E.S.; Reynisdottir, I.; Hart, S.N.; Shimelis, H.; Vilske, S.; Kallioniemi, A.; Schleutker, J.; et al. Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. Proc. Natl. Acad. Sci. USA 2014, 111, 15172–15177. [Google Scholar] [CrossRef]
- Kiiski, J.I.; Tervasmaki, A.; Pelttari, L.M.; Khan, S.; Mantere, T.; Pylkas, K.; Mannermaa, A.; Tengstrom, M.; Kvist, A.; Borg, A.; et al. FANCM mutation c.5791C>T is a risk factor for triple-negative breast cancer in the Finnish population. Breast Cancer Res. Treat. 2017, 166, 217–226. [Google Scholar] [CrossRef]
- Figlioli, G.; Kvist, A.; Tham, E.; Soukupova, J.; Kleiblova, P.; Muranen, T.A.; Andrieu, N.; Azzollini, J.; Balmana, J.; Barroso, A.; et al. The Spectrum of FANCM Protein Truncating Variants in European Breast Cancer Cases. Cancers 2020, 12, 292. [Google Scholar] [CrossRef]
- CHEK2 Breast Cancer Case-Control Consortium. CHEK2*1100delC and susceptibility to breast cancer: A collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am. J. Hum. Genet. 2004, 74, 1175–1182. [Google Scholar] [CrossRef]
- Weischer, M.; Bojesen, S.E.; Ellervik, C.; Tybjaerg-Hansen, A.; Nordestgaard, B.G. CHEK2*1100delC genotyping for clinical assessment of breast cancer risk: Meta-analyses of 26,000 patient cases and 27,000 controls. J. Clin. Oncol. 2008, 26, 542–548. [Google Scholar] [CrossRef]
- Deans, A.J.; West, S.C. FANCM connects the genome instability disorders Bloom’s Syndrome and Fanconi Anemia. Mol. Cell 2009, 36, 943–953. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021, 49, D480–D489. [Google Scholar] [CrossRef]
- Walden, H.; Deans, A.J. The Fanconi anemia DNA repair pathway: Structural and functional insights into a complex disorder. Annu. Rev. Biophys. 2014, 43, 257–278. [Google Scholar] [CrossRef] [PubMed]
- Cavaille, M.; Uhrhammer, N.; Privat, M.; Ponelle-Chachuat, F.; Gay-Bellile, M.; Lepage, M.; Molnar, I.; Viala, S.; Bidet, Y.; Bignon, Y.J. Analysis of 11 candidate genes in 849 adult patients with suspected hereditary cancer predisposition. Genes Chromosomes Cancer 2021, 60, 73–78. [Google Scholar] [CrossRef]
- Del Valle, J.; Rofes, P.; Moreno-Cabrera, J.M.; Lopez-Doriga, A.; Belhadj, S.; Vargas-Parra, G.; Teule, A.; Cuesta, R.; Munoz, X.; Campos, O.; et al. Exploring the Role of Mutations in Fanconi Anemia Genes in Hereditary Cancer Patients. Cancers 2020, 12, 829. [Google Scholar] [CrossRef]
- Helgadottir, H.T.; Thutkawkorapin, J.; Lagerstedt-Robinson, K.; Lindblom, A. Sequencing for germline mutations in Swedish breast cancer families reveals novel breast cancer risk genes. Sci. Rep. 2021, 11, 14737. [Google Scholar] [CrossRef]
- Jarhelle, E.; Riise Stensland, H.M.F.; Hansen, G.A.M.; Skarsfjord, S.; Jonsrud, C.; Ingebrigtsen, M.; Stromsvik, N.; Van Ghelue, M. Identifying sequence variants contributing to hereditary breast and ovarian cancer in BRCA1 and BRCA2 negative breast and ovarian cancer patients. Sci. Rep. 2019, 9, 19986. [Google Scholar] [CrossRef]
- Neidhardt, G.; Hauke, J.; Ramser, J.; Gross, E.; Gehrig, A.; Muller, C.R.; Kahlert, A.K.; Hackmann, K.; Honisch, E.; Niederacher, D.; et al. Association Between Loss-of-Function Mutations Within the FANCM Gene and Early-Onset Familial Breast Cancer. JAMA Oncol. 2017, 3, 1245–1248. [Google Scholar] [CrossRef]
- Nurmi, A.K.; Suvanto, M.; Dennis, J.; Aittomaki, K.; Blomqvist, C.; Nevanlinna, H. Pathogenic Variant Spectrum in Breast Cancer Risk Genes in Finnish Patients. Cancers 2022, 14, 6158. [Google Scholar] [CrossRef]
- Schubert, S.; van Luttikhuizen, J.L.; Auber, B.; Schmidt, G.; Hofmann, W.; Penkert, J.; Davenport, C.F.; Hille-Betz, U.; Wendeburg, L.; Bublitz, J.; et al. The identification of pathogenic variants in BRCA1/2 negative, high risk, hereditary breast and/or ovarian cancer patients: High frequency of FANCM pathogenic variants. Int. J. Cancer 2019, 144, 2683–2694. [Google Scholar] [CrossRef]
- Southey, M.C.; Dowty, J.G.; Riaz, M.; Steen, J.A.; Renault, A.L.; Tucker, K.; Kirk, J.; James, P.; Winship, I.; Pachter, N.; et al. Population-based estimates of breast cancer risk for carriers of pathogenic variants identified by gene-panel testing. NPJ Breast Cancer 2021, 7, 153. [Google Scholar] [CrossRef]
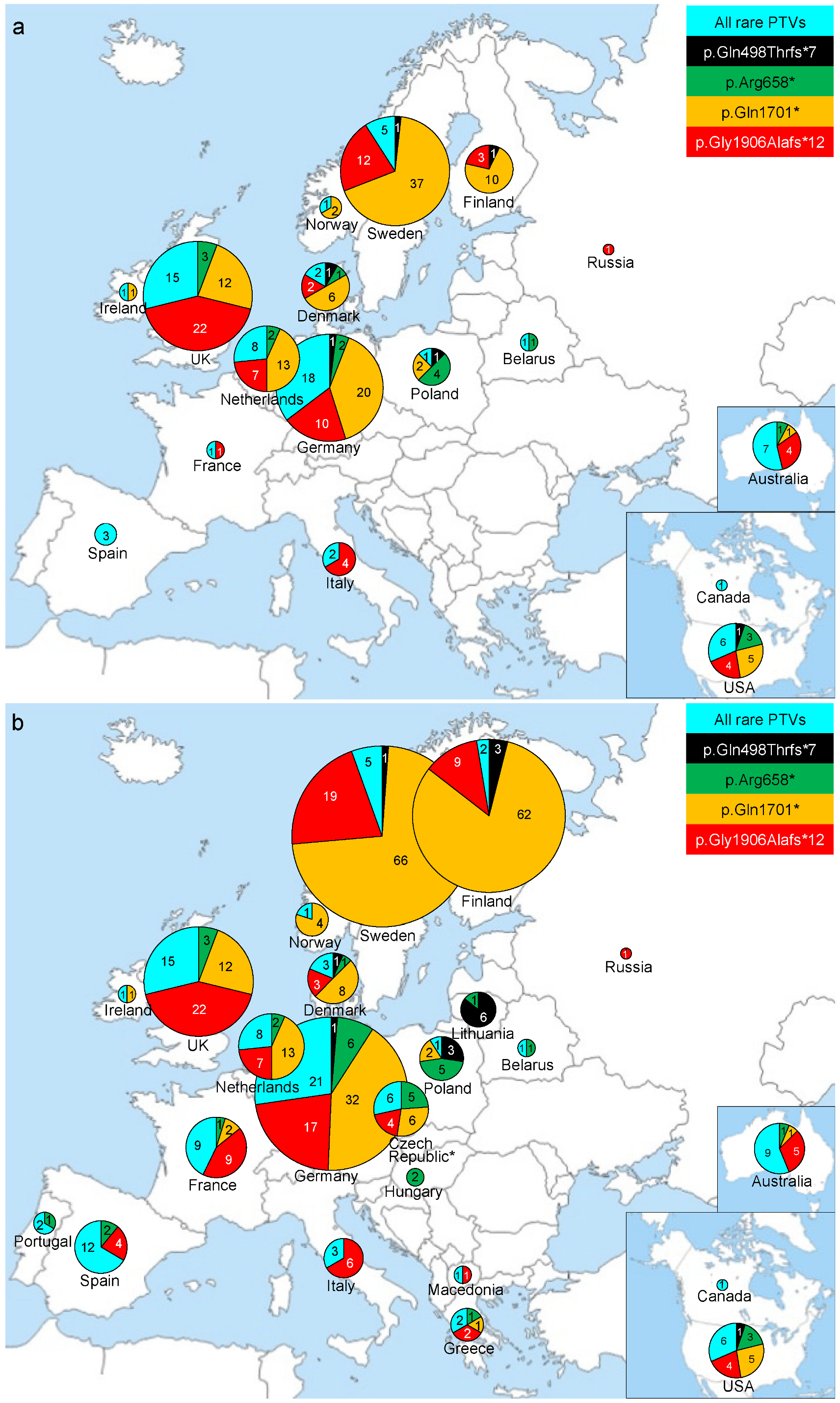
| Country | Breast Cancer Cases | Carriers of All PTV (Freq%) | Carriers of all PTV Excluding p.Gln1701* and p.Gly1906Alafs*12 (Freq%) |
|---|---|---|---|
| UK | 10,683 | 52 (0.9) | 18 (0.17) |
| Germany | 8659 | 51 (0.59) | 21 (0.24) |
| Sweden | 4607 | 55 (1.19) | 6 (0.13) |
| Netherlands | 3705 | 30 (0.81) | 10 (0.27) |
| USA | 2800 | 19 (0.68) | 10 (0.36) |
| Denmark | 2800 | 12 (0.43) | 4 (0.14) |
| Australia | 2460 | 13 (0.53) | 8 (0.32) |
| Poland | 2103 | 8 (0.38) | 6 (0.28) |
| Spain | 1126 | 3 (0.27) | 3 (0.27) |
| Cyprus | 974 | 0 (0) | 0 (0) |
| France | 938 | 2 (0.21) | 1 (0.11) |
| Italy | 933 | 6 (0.64) | 2 (0.21) |
| Norway | 565 | 3 (0.53) | 1 (0.18) |
| Finland | 560 | 14 (2.50) | 1 (0.178) |
| Canada | 491 | 1 (0.20) | 1 (0.20) |
| Greece | 472 | 0 (0) | 0 (0) |
| Ireland | 369 | 2 (0.54) | 1 (0.27) |
| Belarus | 319 | 2 (0.63) | 2 (0.63) |
| Russia | 239 | 1 (0.42) | 0 (0) |
| Total | 44,803 | 274 (0.61) | 95 (0.21) |
| Breast Cancer Cases | Carriers of All PTV (Freq%) | p-Value | Carriers of All PTV Excluding p.Gln1701* and p.Gly1906Alafs*12 (Freq%) | p-Value | |
|---|---|---|---|---|---|
| Group | Number | ||||
| All | 44,803 | 274 (0.61) | - | 95 (0.21) | - |
| Sporadic | 26,539 | 150 (0.56) | 0.032 | 49 (0.18) | 0.021 |
| Familial | 10,680 | 81 (0.76) | 33 (0.31) | ||
| ER-positive | 25,679 | 137 (0.53) | 0.048 | 44 (0.17) | 0.0005 |
| ER-negative | 6572 | 49 (0.74) | 26 (0.39) | ||
| PTV | Number of Carriers | Study, Geographic Origin |
|---|---|---|
| c.551dup; p.Arg185Glufs*13 | 3 | This study, Germany (1) Netherlands (1); Neidhardt et al. [28], Germany (1) |
| c.1196C>G; p.Ser399* | 3 | This study, UK (2), Figlioli et al. [18], Spain (1) |
| c.1492C>T; p.Gln498* | 3 | This study, Germany (1) Netherlands (2) |
| c.2260C>T; p.Arg754* | 3 | Figlioli et al. [18], Czech Republic (2) France (1) |
| c.2320G>T; p.Glu774* | 4 | This study, Spain (1); Figlioli et al. [18], Spain (2) Portugal (1) |
| c.2586_2589del; p.Lys863Ilefs*12 | 3 | This study, Spain (1); Figlioli et al. [18], Spain (1); Cavaillé et al. [24], France (1) |
| c.3088C>T; p.Arg1030* | 3 | This study, Spain (1); Figlioli et al. [18], Czech Republic (1); Neidhardt et al. [28], Germany (1) |
| c.4194T>G; p.Tyr1398* | 3 | This study, UK (3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Figlioli, G.; Billaud, A.; Wang, Q.; Bolla, M.K.; Dennis, J.; Lush, M.; Kvist, A.; Adank, M.A.; Ahearn, T.U.; Antonenkova, N.N.; et al. Spectrum and Frequency of Germline FANCM Protein-Truncating Variants in 44,803 European Female Breast Cancer Cases. Cancers 2023, 15, 3313. https://doi.org/10.3390/cancers15133313
Figlioli G, Billaud A, Wang Q, Bolla MK, Dennis J, Lush M, Kvist A, Adank MA, Ahearn TU, Antonenkova NN, et al. Spectrum and Frequency of Germline FANCM Protein-Truncating Variants in 44,803 European Female Breast Cancer Cases. Cancers. 2023; 15(13):3313. https://doi.org/10.3390/cancers15133313
Chicago/Turabian StyleFiglioli, Gisella, Amandine Billaud, Qin Wang, Manjeet K. Bolla, Joe Dennis, Michael Lush, Anders Kvist, Muriel A. Adank, Thomas U. Ahearn, Natalia N. Antonenkova, and et al. 2023. "Spectrum and Frequency of Germline FANCM Protein-Truncating Variants in 44,803 European Female Breast Cancer Cases" Cancers 15, no. 13: 3313. https://doi.org/10.3390/cancers15133313
APA StyleFiglioli, G., Billaud, A., Wang, Q., Bolla, M. K., Dennis, J., Lush, M., Kvist, A., Adank, M. A., Ahearn, T. U., Antonenkova, N. N., Auvinen, P., Behrens, S., Bermisheva, M., Bogdanova, N. V., Bojesen, S. E., Bonanni, B., Brüning, T., Camp, N. J., Campbell, A., ... Peterlongo, P. (2023). Spectrum and Frequency of Germline FANCM Protein-Truncating Variants in 44,803 European Female Breast Cancer Cases. Cancers, 15(13), 3313. https://doi.org/10.3390/cancers15133313

















