The RHOA Mutation G17V Does Not Lead to Increased Migration of Human Malignant T Cells but Is Associated with Matrix Remodelling
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Cell Culture, Cloning, and Transfection
2.2. Microchannel Experiments and 3D Collagen Gels
2.3. Proteomics, Cleaved Collagen, and Matrix Metalloproteinase Assay
2.4. Live Cell Imaging and Analysis of Collagen Fibrosis
3. Results
3.1. The Growth Behavior of the T-Cell Lymphoma Cell Lines HuT78 and HH Is Not Significantly Influenced by the Overexpression of RHOA-G17V
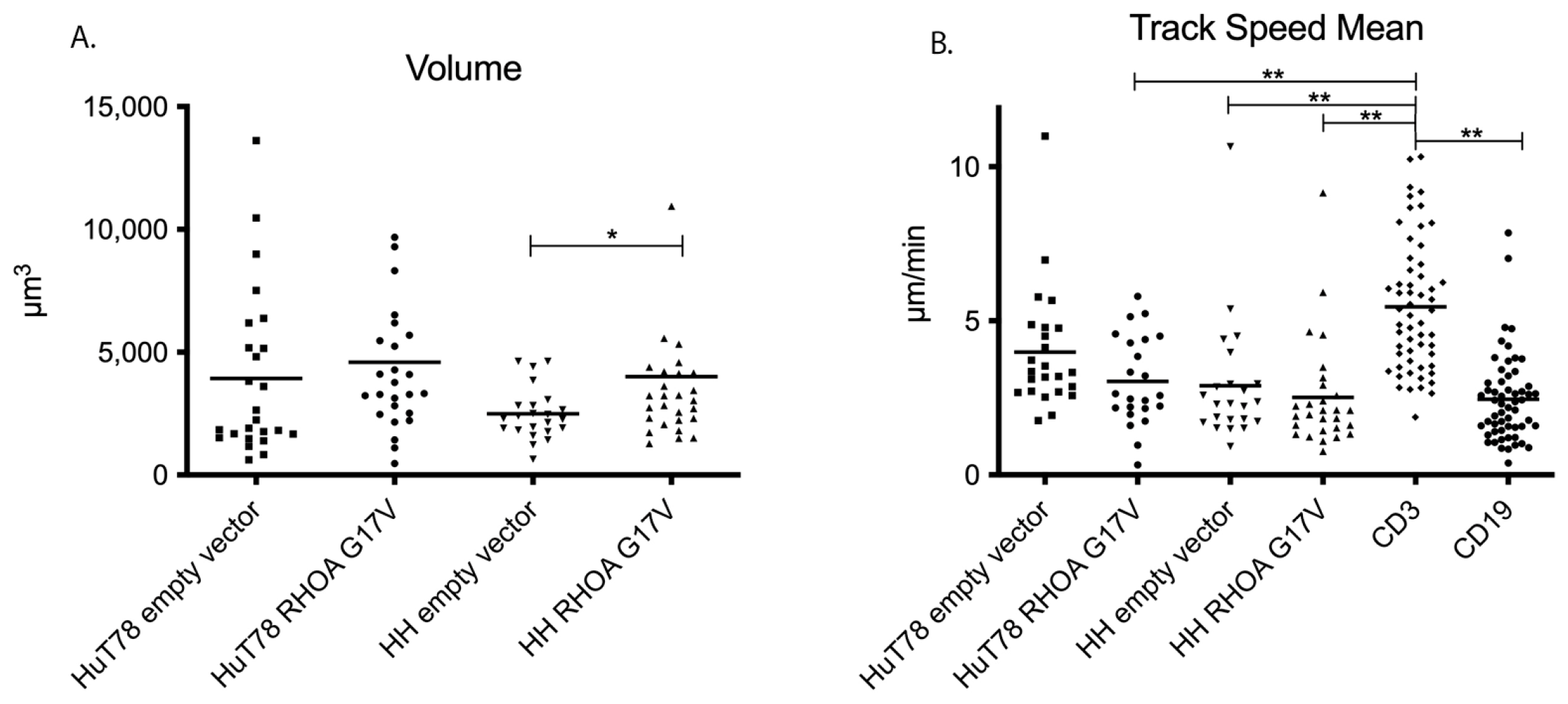
3.2. HH and Hut78 Cells Expressing RHOA-G17V Exhibited Impaired Motility in Straight Microchannels
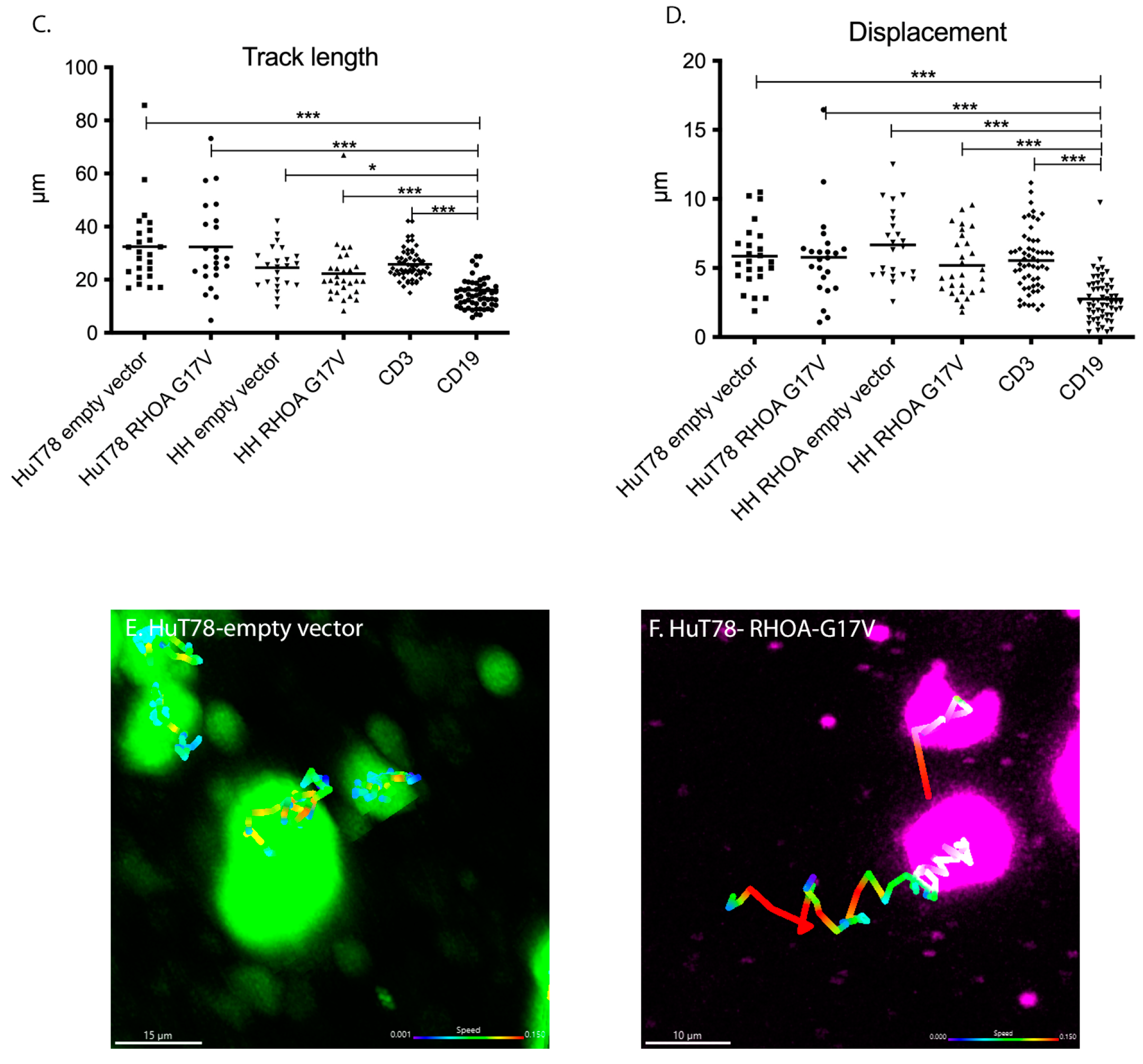
3.3. HuT78 and HH Cells Expressing RHOA-G17V Do Not Differ in Their Movement Patterns in 3D Collagen Gels and Human Lymphoid Tissue
3.4. Global Proteomics Screening Reveals Decreased Expression of RHOG and RHOF in HuT78 and HH Cells Expressing RHOA-G17V
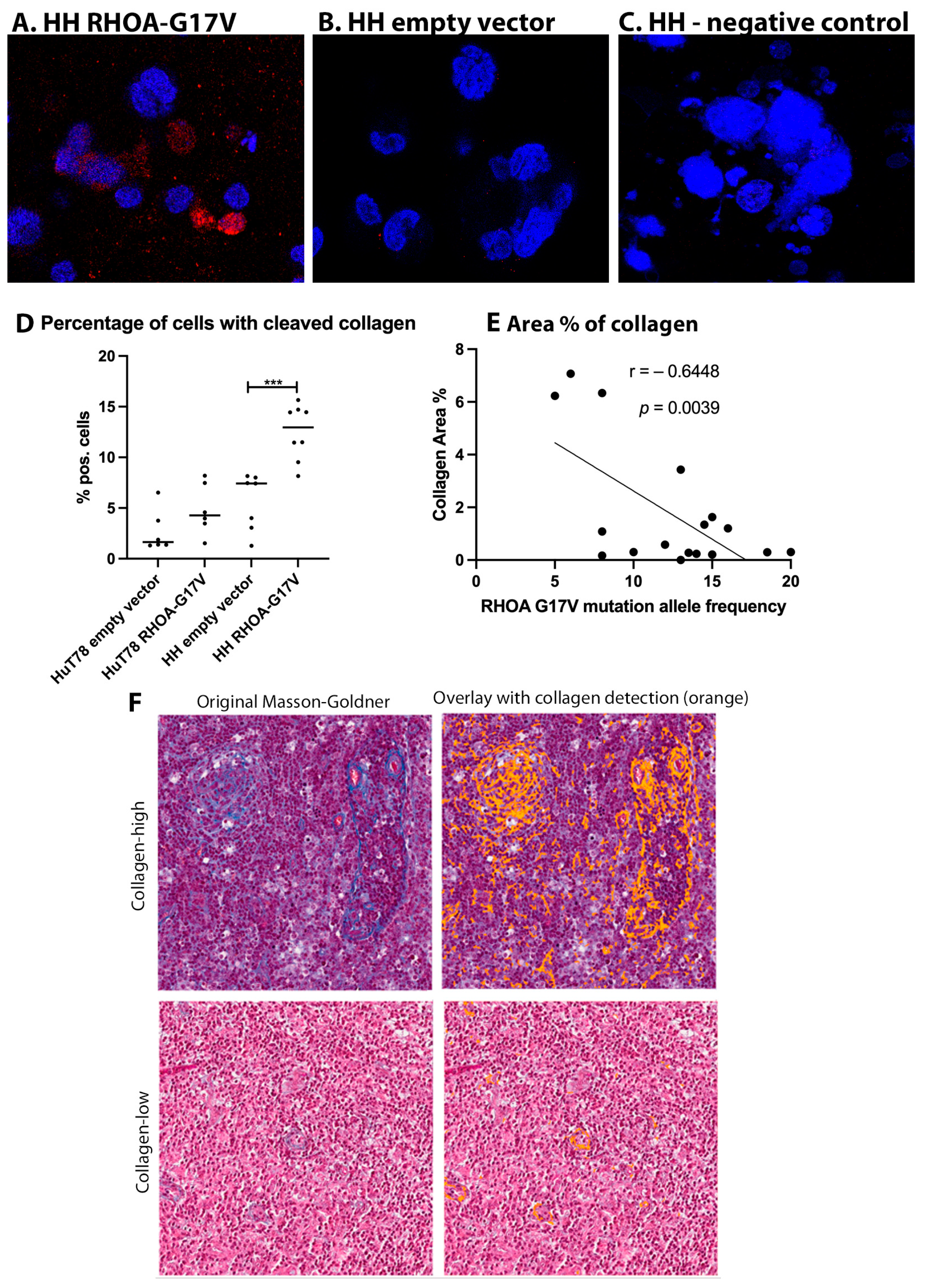
3.5. Collagen Cleavage Is Observed in HH Cells Expressing RHOA-G17V and Reduced Collagen Content Is Found in AITL Patients with High RHOA-G17V Allele Frequencies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- de Leval, L.; Parrens, M.; Le Bras, F.; Jais, J.P.; Fataccioli, V.; Martin, A.; Lamant, L.; Delarue, R.; Berger, F.; Arbion, F.; et al. Angioimmunoblastic T-cell lymphoma is the most common T-cell lymphoma in two distinct French information data sets. Haematologica 2015, 100, e361–e364. [Google Scholar] [CrossRef] [PubMed]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef] [PubMed]
- Campo, E.; Jaffe, E.S.; Cook, J.R.; Quintanilla-Martinez, L.; Swerdlow, S.H.; Anderson, K.C.; Brousset, P.; Cerroni, L.; de Leval, L.; Dirnhofer, S.; et al. The International Consensus Classification of Mature Lymphoid Neoplasms: A report from the Clinical Advisory Committee. Blood 2022, 140, 1229–1253. [Google Scholar] [CrossRef]
- Frizzera, G.; Moran, E.M.; Rappaport, H. Angio-immunoblastic lymphadenopathy with dysproteinaemia. Lancet 1974, 1, 1070–1073. [Google Scholar] [CrossRef]
- International Agency for Research on Cancer. WHO Classification of Haematolymphoid Tumours; International Agency for Research on Cancer: Lyon, France, 2022. [Google Scholar]
- Mourad, N.; Mounier, N.; Briere, J.; Raffoux, E.; Delmer, A.; Feller, A.; Meijer, C.J.; Emile, J.F.; Bouabdallah, R.; Bosly, A.; et al. Clinical, biologic, and pathologic features in 157 patients with angioimmunoblastic T-cell lymphoma treated within the Groupe d’Etude des Lymphomes de l’Adulte (GELA) trials. Blood 2008, 111, 4463–4470. [Google Scholar] [CrossRef] [PubMed]
- Advani, R.H.; Skrypets, T.; Civallero, M.; Spinner, M.A.; Manni, M.; Kim, W.S.; Shustov, A.R.; Horwitz, S.M.; Hitz, F.; Cabrera, M.E.; et al. Outcomes and prognostic factors in angioimmunoblastic T-cell lymphoma: Final report from the international T-cell Project. Blood 2021, 138, 213–220. [Google Scholar] [CrossRef]
- Attygalle, A.D.; Kyriakou, C.; Dupuis, J.; Grogg, K.L.; Diss, T.C.; Wotherspoon, A.C.; Chuang, S.S.; Cabecadas, J.; Isaacson, P.G.; Du, M.Q.; et al. Histologic evolution of angioimmunoblastic T-cell lymphoma in consecutive biopsies: Clinical correlation and insights into natural history and disease progression. Am. J. Surg. Pathol. 2007, 31, 1077–1088. [Google Scholar] [CrossRef]
- de Leval, L.; Rickman, D.S.; Thielen, C.; Reynies, A.; Huang, Y.L.; Delsol, G.; Lamant, L.; Leroy, K.; Briere, J.; Molina, T.; et al. The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood 2007, 109, 4952–4963. [Google Scholar] [CrossRef]
- Piccaluga, P.P.; Agostinelli, C.; Califano, A.; Carbone, A.; Fantoni, L.; Ferrari, S.; Gazzola, A.; Gloghini, A.; Righi, S.; Rossi, M.; et al. Gene expression analysis of angioimmunoblastic lymphoma indicates derivation from T follicular helper cells and vascular endothelial growth factor deregulation. Cancer Res. 2007, 67, 10703–10710. [Google Scholar] [CrossRef] [PubMed]
- Sakata-Yanagimoto, M.; Enami, T.; Yoshida, K.; Shiraishi, Y.; Ishii, R.; Miyake, Y.; Muto, H.; Tsuyama, N.; Sato-Otsubo, A.; Okuno, Y.; et al. Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat. Genet. 2014, 46, 171–175. [Google Scholar] [CrossRef] [PubMed]
- Palomero, T.; Couronne, L.; Khiabanian, H.; Kim, M.Y.; Ambesi-Impiombato, A.; Perez-Garcia, A.; Carpenter, Z.; Abate, F.; Allegretta, M.; Haydu, J.E.; et al. Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat. Genet. 2014, 46, 166–170. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.Y.; Sung, M.K.; Lee, S.H.; Kim, S.; Lee, H.; Park, S.; Kim, S.C.; Lee, B.; Rho, K.; Lee, J.E.; et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat. Genet. 2014, 46, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Manso, R.; Sanchez-Beato, M.; Monsalvo, S.; Gomez, S.; Cereceda, L.; Llamas, P.; Rojo, F.; Mollejo, M.; Menarguez, J.; Alves, J.; et al. The RHOA G17V gene mutation occurs frequently in peripheral T-cell lymphoma and is associated with a characteristic molecular signature. Blood 2014, 123, 2893–2894. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ning, J.; Liu, X.; Chan, W.C.J. Mutations Affecting Genes in the Proximal T-Cell Receptor Signaling Pathway in Peripheral T-Cell Lymphoma. Cancers 2022, 14, 3716. [Google Scholar] [CrossRef]
- Sahai, E.; Marshall, C.J. RHO-GTPases and cancer. Nat. Rev. Cancer 2002, 2, 133–142. [Google Scholar] [CrossRef]
- O’Connor, K.; Chen, M. Dynamic functions of RhoA in tumor cell migration and invasion. Small GTPases 2013, 4, 141–147. [Google Scholar] [CrossRef]
- Fukumoto, K.; Nguyen, T.B.; Chiba, S.; Sakata-Yanagimoto, M. Review of the biologic and clinical significance of genetic mutations in angioimmunoblastic T-cell lymphoma. Cancer Sci. 2018, 109, 490–496. [Google Scholar] [CrossRef]
- Cortes, J.R.; Ambesi-Impiombato, A.; Couronne, L.; Quinn, S.A.; Kim, C.S.; da Silva Almeida, A.C.; West, Z.; Belver, L.; Martin, M.S.; Scourzic, L.; et al. RHOA G17V Induces T Follicular Helper Cell Specification and Promotes Lymphomagenesis. Cancer Cell 2018, 33, 259–273.e257. [Google Scholar] [CrossRef]
- Kataoka, K.; Nagata, Y.; Kitanaka, A.; Shiraishi, Y.; Shimamura, T.; Yasunaga, J.; Totoki, Y.; Chiba, K.; Sato-Otsubo, A.; Nagae, G.; et al. Integrated molecular analysis of adult T cell leukemia/lymphoma. Nat. Genet. 2015, 47, 1304–1315. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Yang, J.; Wenzel, A.T.; Ramachandran, A.; Lee, W.J.; Daniels, J.C.; Kim, J.; Martinez-Escala, E.; Amankulor, N.; Pro, B.; et al. Genomic analysis of 220 CTCLs identifies a novel recurrent gain-of-function alteration in RLTPR (p.Q575E). Blood 2017, 130, 1430–1440. [Google Scholar] [CrossRef] [PubMed]
- Rohde, M.; Richter, J.; Schlesner, M.; Betts, M.J.; Claviez, A.; Bonn, B.R.; Zimmermann, M.; Damm-Welk, C.; Russell, R.B.; Borkhardt, A.; et al. Recurrent RHOA mutations in pediatric Burkitt lymphoma treated according to the NHL-BFM protocols. Genes Chromosom. Cancer 2014, 53, 911–916. [Google Scholar] [CrossRef] [PubMed]
- O’Hayre, M.; Inoue, A.; Kufareva, I.; Wang, Z.; Mikelis, C.M.; Drummond, R.A.; Avino, S.; Finkel, K.; Kalim, K.W.; DiPasquale, G.; et al. Inactivating mutations in GNA13 and RHOA in Burkitt’s lymphoma and diffuse large B-cell lymphoma: A tumor suppressor function for the Galpha13/RhoA axis in B cells. Oncogene 2016, 35, 3771–3780. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research, N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Kowarz, E.; Loscher, D.; Marschalek, R. Optimized Sleeping Beauty transposons rapidly generate stable transgenic cell lines. Biotechnol. J. 2015, 10, 647–653. [Google Scholar] [CrossRef]
- Bein, J.; Flinner, N.; Häupl, B.; Mathur, A.; Schneider, O.; Abu-Ayyad, M.; Hansmann, M.L.; Piel, M.; Oellerich, T.; Hartmann, S. T-cell-derived Hodgkin lymphoma has motility characteristics intermediate between Hodgkin and anaplastic large cell lymphoma. J. Cell. Mol. Med. 2022, 26, 3495–3505. [Google Scholar] [CrossRef]
- Friedl, P.; Brocker, E.B. TCR triggering on the move: Diversity of T-cell interactions with antigen-presenting cells. Immunol. Rev. 2002, 186, 83–89. [Google Scholar] [CrossRef]
- Goncharova, O.; Flinner, N.; Bein, J.; Doring, C.; Donnadieu, E.; Rikirsch, S.; Herling, M.; Kuppers, R.; Hansmann, M.L.; Hartmann, S. Migration Properties Distinguish Tumor Cells of Classical Hodgkin Lymphoma from Anaplastic Large Cell Lymphoma Cells. Cancers 2019, 11, 1484. [Google Scholar] [CrossRef]
- Katoh, H.; Hiramoto, K.; Negishi, M. Activation of Rac1 by RhoG regulates cell migration. J. Cell Sci. 2006, 119, 56–65. [Google Scholar] [CrossRef]
- Haga, R.B.; Ridley, A.J. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 2016, 7, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Sadjadi, Z.; Zhao, R.; Hoth, M.; Qu, B.; Rieger, H. Migration of Cytotoxic T Lymphocytes in 3D Collagen Matrices. Biophys. J. 2020, 119, 2141–2152. [Google Scholar] [CrossRef]
- Thorseth, M.L.; Carretta, M.; Jensen, C.; Molgaard, K.; Jurgensen, H.J.; Engelholm, L.H.; Behrendt, N.; Willumsen, N.; Madsen, D.H. Uncovering mediators of collagen degradation in the tumor microenvironment. Matrix Biol. Plus 2022, 13, 100101. [Google Scholar] [CrossRef] [PubMed]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef]
- Sorokin, L. The impact of the extracellular matrix on inflammation. Nat. Rev. Immunol. 2010, 10, 712–723. [Google Scholar] [CrossRef]
- Cheng, J.; Haas, M. Frequent mutations in the p53 tumor suppressor gene in human leukemia T-cell lines. Mol. Cell Biol. 1990, 10, 5502–5509. [Google Scholar] [CrossRef] [PubMed]
- Davies, H.; Bignell, G.R.; Cox, C.; Stephens, P.; Edkins, S.; Clegg, S.; Teague, J.; Woffendin, H.; Garnett, M.J.; Bottomley, W.; et al. Mutations of the BRAF gene in human cancer. Nature 2002, 417, 949–954. [Google Scholar] [CrossRef] [PubMed]
- Lemonnier, F.; Couronne, L.; Parrens, M.; Jais, J.P.; Travert, M.; Lamant, L.; Tournillac, O.; Rousset, T.; Fabiani, B.; Cairns, R.A.; et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 2012, 120, 1466–1469. [Google Scholar] [CrossRef]

| Gene Symbol | HH-RHOA G17V/Empty Vector Ratio | HuT78-RHOA G17V/Empty Vector Ratio | Mean Ratio | Name |
|---|---|---|---|---|
| ANXA4 | 1.27 | 1.13 | 1.20 | Annexin A4 |
| ASS1 | 1.14 | 1.16 | 1.15 | Argininosuccinate Synthase 1 |
| CDC26 | 1.13 | 1.13 | 1.13 | Cell Division Cycle 26 |
| RFFL | 1.19 | 1.39 | 1.29 | Ring Finger And FYVE-Like Domain Containing E3 Ubiquitin Protein Ligase |
| Gene Symbol | HH-RHOA G17V/Empty Vector Ratio | HuT78-RHOA G17V/Empty Vector Ratio | Mean Ratio | Name |
|---|---|---|---|---|
| ZNF770 | 0.65 | 0.80 | 0.72 | Zinc Finger Protein 770 |
| RHOF | 0.81 | 0.68 | 0.75 | Ras Homolog Family Member F, Filopodia Associated |
| BRPF1 | 0.84 | 0.69 | 0.77 | Bromodomain and PHD Finger Containing 1 |
| PECR | 0.73 | 0.83 | 0.78 | Peroxisomal Trans-2-Enoyl-CoA Reductase |
| KAT5 | 0.72 | 0.86 | 0.79 | Lysine Acetyltransferase 5 |
| SLC43A3 | 0.72 | 0.88 | 0.80 | Solute Carrier Family 43 Member 3 |
| RHOG | 0.88 | 0.74 | 0.81 | Ras Homolog Family Member G |
| MED19 | 0.83 | 0.82 | 0.82 | Mediator Complex Subunit 19 |
| RELCH | 0.85 | 0.83 | 0.84 | RAB11 Binding and LisH Domain, Coiled-Coil and HEAT Repeat Containing |
| RETSAT | 0.88 | 0.87 | 0.88 | Retinol Saturase |
| CAV1 | 0.88 | 0.88 | 0.88 | Caveolin 1 |
| PRNP | 0.88 | 0.89 | 0.88 | Prion Protein |
| TYMS | 0.89 | 0.88 | 0.89 | Thymidylate Synthetase |
| DDX10 | 0.90 | 0.88 | 0.89 | DEAD-Box Helicase 10 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merk-Ahmad, K.; Bein, J.; Scharf, S.; Schäfer, H.; Bexte, T.; Ullrich, E.; Loth, A.G.; Flinner, N.; Senff, T.; Schneider, O.; et al. The RHOA Mutation G17V Does Not Lead to Increased Migration of Human Malignant T Cells but Is Associated with Matrix Remodelling. Cancers 2023, 15, 3226. https://doi.org/10.3390/cancers15123226
Merk-Ahmad K, Bein J, Scharf S, Schäfer H, Bexte T, Ullrich E, Loth AG, Flinner N, Senff T, Schneider O, et al. The RHOA Mutation G17V Does Not Lead to Increased Migration of Human Malignant T Cells but Is Associated with Matrix Remodelling. Cancers. 2023; 15(12):3226. https://doi.org/10.3390/cancers15123226
Chicago/Turabian StyleMerk-Ahmad, Katrin, Julia Bein, Sonja Scharf, Hendrik Schäfer, Tobias Bexte, Evelyn Ullrich, Andreas G. Loth, Nadine Flinner, Tina Senff, Olga Schneider, and et al. 2023. "The RHOA Mutation G17V Does Not Lead to Increased Migration of Human Malignant T Cells but Is Associated with Matrix Remodelling" Cancers 15, no. 12: 3226. https://doi.org/10.3390/cancers15123226
APA StyleMerk-Ahmad, K., Bein, J., Scharf, S., Schäfer, H., Bexte, T., Ullrich, E., Loth, A. G., Flinner, N., Senff, T., Schneider, O., Hansmann, M.-L., Piel, M., Häupl, B., Oellerich, T., Donnadieu, E., & Hartmann, S. (2023). The RHOA Mutation G17V Does Not Lead to Increased Migration of Human Malignant T Cells but Is Associated with Matrix Remodelling. Cancers, 15(12), 3226. https://doi.org/10.3390/cancers15123226





