Innate Immune Cells in the Tumor Microenvironment of Liver Metastasis from Colorectal Cancer: Contribution to a Comprehensive Therapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants and Sample Collection
2.2. Histological Analysis
2.3. Flow Cytometry Characterization of Innate Immune Cells
2.3.1. Staining Protocol
2.3.2. Sample Acquisition and Analysis
3. Results
3.1. Innate Immune Cells in Liver Colorectal Cancer Metastasis Microenvironment
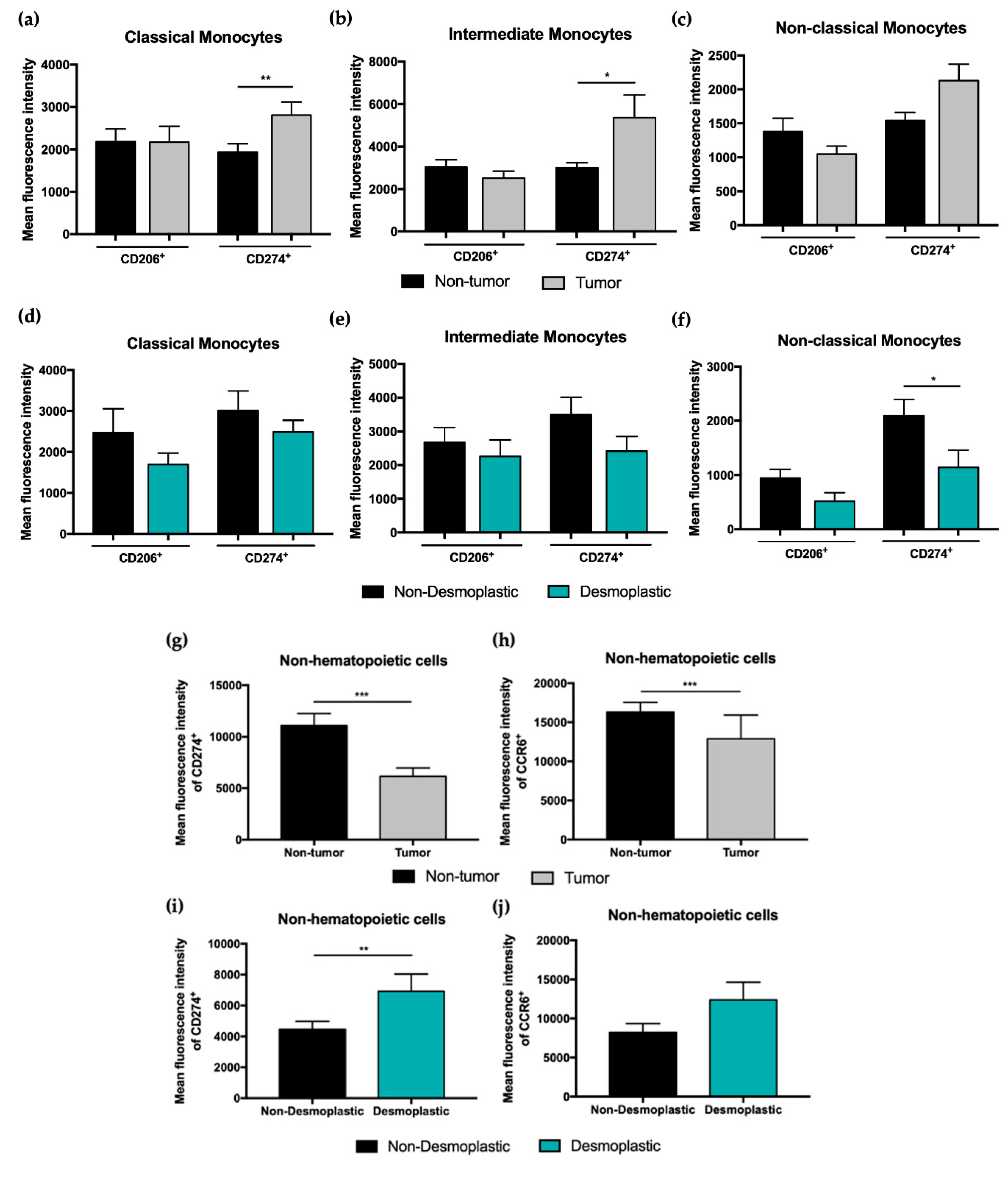
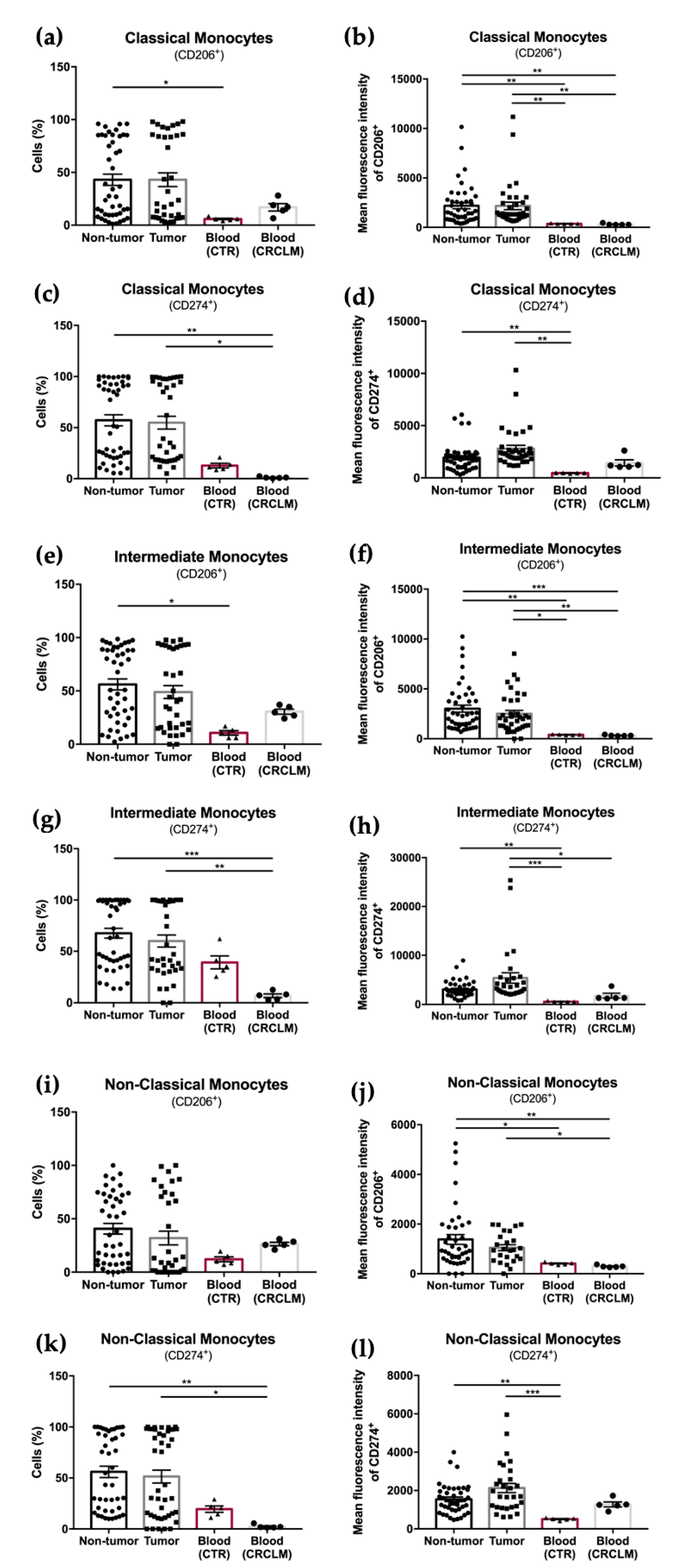
3.2. Mean Fluorescence Intensity (MFI) Analysis of CD206 and CD274 Reveals Differential Expression on Cells from CRC Liver Metastasis
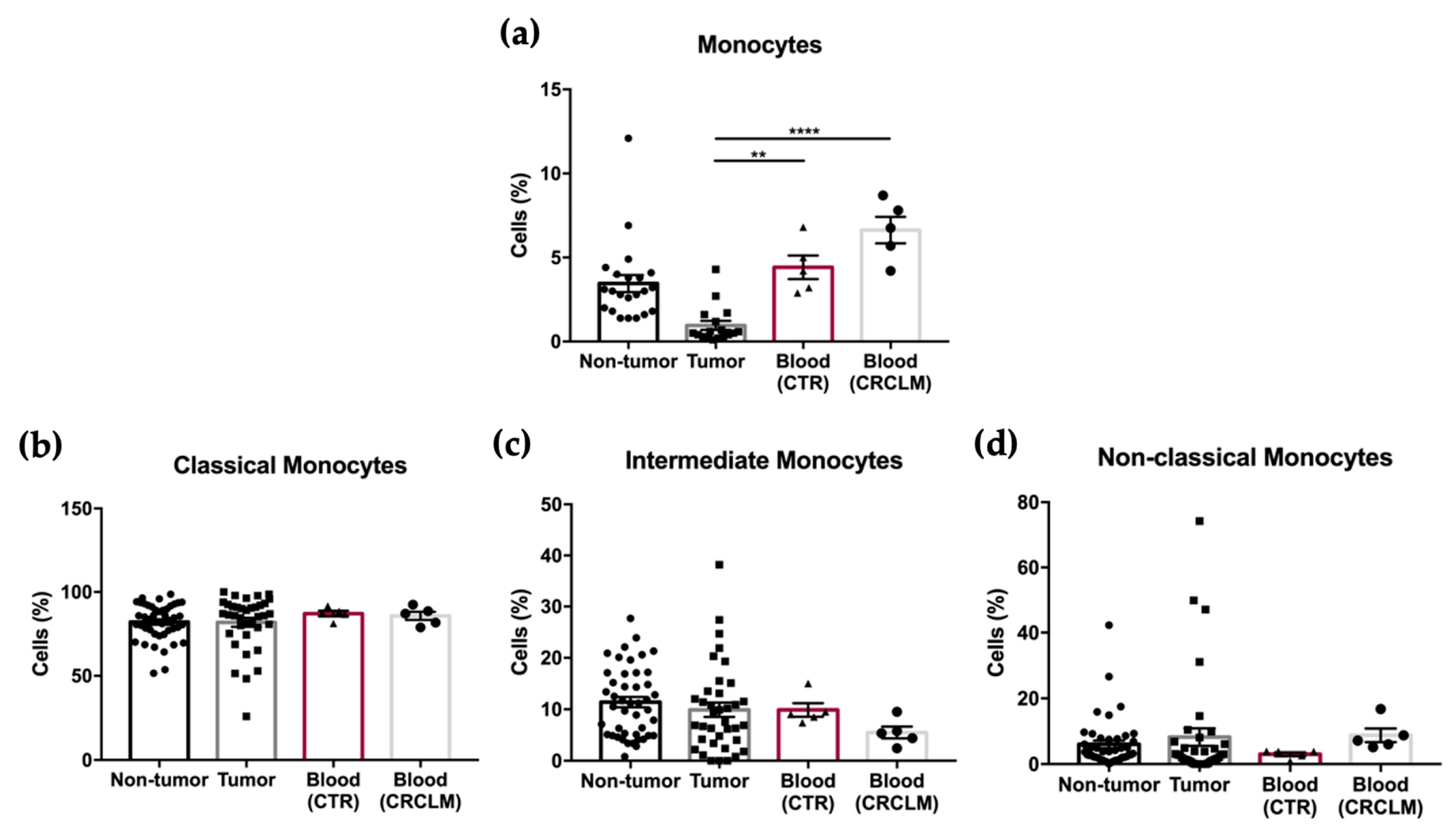
3.3. Blood Sample Analysis of CRCLM Patients
3.4. The Percentage of CD206+ and CD274+ Monocytes Influences the Disease-Free Survival Time of CRCLM Patients
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shihab, I.; Khalil, B.A.; Elemam, N.M.; Hachim, I.Y.; Hachim, M.Y.; Hamoudi, R.A.; Maghazachi, A.A. Understanding the Role of Innate Immune Cells and Identifying Genes in Breast Cancer Microenvironment. Cancers 2020, 12, 2226. [Google Scholar] [CrossRef]
- Vaillant, A.A.J.; Sabir, S.; Jan, A. Physiology, Immune Response; StatPearls: Tampa, FL, USA, 2022. [Google Scholar]
- Zhou, H.; Liu, Z.; Wang, Y.; Wen, X.; Amador, E.H.; Yuan, L.; Ran, X.; Xiong, L.; Ran, Y.; Chen, W.; et al. Colorectal liver metastasis: Molecular mechanism and interventional therapy. Signal Transduct. Target. Ther. 2022, 7, 70. [Google Scholar] [CrossRef] [PubMed]
- Baghdadi, A.; Mirpour, S.; Ghadimi, M.; Motaghi, M.; Hazhirkarzar, B.; Pawlik, T.M.; Kamel, I.R. Imaging of Colorectal Liver Metastasis. J. Gastrointest. Surg. 2021, 26, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.A.; Nisar, S.; Singh, M.; Ashraf, B.; Masoodi, T.; Prasad, C.P.; Sharma, A.; Maacha, S.; Karedath, T.; Hashem, S.; et al. Cytokine- and chemokine-induced inflammatory colorectal tumor microenvironment: Emerging avenue for targeted therapy. Cancer Commun. 2022, 42, 689–715. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Q.; Xing, B.; Luo, N.; Gao, R.; Yu, K.; Hu, X.; Bu, Z.; Peng, J.; Ren, X.; et al. Immune phenotypic linkage between colorectal cancer and liver metastasis. Cancer Cell 2022, 40, 424–437. [Google Scholar] [CrossRef]
- Saraiva, A.L.; Carneiro, F. New Insights into the Role of Tissue Eosinophils in the Progression of Colorectal Cancer: A Literature Review. Acta Med. Port. 2018, 31, 329–337. [Google Scholar] [CrossRef]
- Peña-Romero, A.C.; Orenes-Piñero, E. Dual Effect of Immune Cells within Tumour Microenvironment: Pro- and Anti-Tumour Effects and Their Triggers. Cancers 2022, 14, 1681. [Google Scholar] [CrossRef]
- Benito-Martin, A.; Di Giannatale, A.; Ceder, S.; Peinado, H. The New Deal: A Potential Role for Secreted Vesicles in Innate Immunity and Tumor Progression. Front. Immunol. 2015, 6, 66. [Google Scholar] [CrossRef]
- Colangelo, T.; Polcaro, G.; Muccillo, L.; D’Agostino, G.; Rosato, V.; Ziccardi, P.; Lupo, A.; Mazzoccoli, G.; Sabatino, L.; Colantuoni, V. Friend or foe? Biochim. Biophys. Acta Rev. Cancer 2017, 1867, 1–18. [Google Scholar] [CrossRef]
- Chandra, R.; Karalis, J.D.; Liu, C.; Murimwa, G.Z.; Park, J.V.; Heid, C.A.; Reznik, S.I.; Huang, E.; Minna, J.D.; Brekken, R.A. The Colorectal Cancer Tumor Microenvironment and Its Impact on Liver and Lung Metastasis. Cancers 2021, 13, 6206. [Google Scholar] [CrossRef]
- Yang, P.; Li, Y.; Xie, Y.; Liu, Y. Different Faces for Different Places: Heterogeneity of Neutrophil Phenotype and Function. J. Immunol. Res. 2019, 2019, 8016254. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hou, S.; Liang, Q.; He, W.; Li, R.; Wang, H.; Zhu, Y.; Zhang, B.; Chen, L.; Dai, X.; et al. Localized Degradation of Neutrophil Extracellular Traps by Photoregulated Enzyme Delivery for Cancer Immunotherapy and Metastasis Suppression. ACS Nano 2022, 16, 2585–2597. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, R.; Kawada, K.; Itatani, Y.; Ogawa, R.; Kiyasu, Y.; Sakai, Y. The Role of Tumor-Associated Neutrophils in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 529. [Google Scholar] [CrossRef] [PubMed]
- Inaguma, S.; Lasota, J.; Felisiak-Golabek, A.; Kowalik, A.; Wang, Z.; Zieba, S.; Kalisz, J.; Ikeda, H.; Miettinen, M. Histopathological and genotypic characterization of metastatic colorectal carcinoma with PD-L1 (CD274)-expression: Possible roles of tumour micro environmental factors for CD274 expression. J. Pathol. Clin. Res. 2017, 3, 268–278. [Google Scholar] [CrossRef] [PubMed]
- He, P.-X.; Ma, Z.-L.; Han, H.; Zhang, X.-Y.; Niu, S.-H.; Du, L.-N.; Zheng, Y.-C.; Liu, H.-M. Expression of programmed death ligand 1 (PD-L1) is associated with metastasis and differentiation in gastric cancer. Life Sci. 2020, 242, 117247. [Google Scholar] [CrossRef] [PubMed]
- Engerud, H.; Berg, H.F.; Myrvold, M.; Halle, M.K.; Bjorge, L.; Haldorsen, I.S.; Hoivik, E.A.; Trovik, J.; Krakstad, C. High degree of heterogeneity of PD-L1 and PD-1 from primary to metastatic endometrial cancer. Gynecol. Oncol. 2020, 157, 260–267. [Google Scholar] [CrossRef]
- Yaghoubi, N.; Soltani, A.; Ghazvini, K.; Hassanian, S.M.; Hashemy, S.I. PD-1/ PD-L1 blockade as a novel treatment for colorectal cancer. Biomed. Pharmacother. 2018, 110, 312–318. [Google Scholar] [CrossRef]
- Scodeller, P.; Simón-Gracia, L.; Kopanchuk, S.; Tobi, A.; Kilk, K.; Säälik, P.; Kurm, K.; Squadrito, M.L.; Kotamraju, V.R.; Rinken, A.; et al. Precision Targeting of Tumor Macrophages with a CD206 Binding Peptide. Sci. Rep. 2017, 7, 14655. [Google Scholar] [CrossRef]
- Lin, Y.; Xu, J.; Lan, H. Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. J. Hematol. Oncol. 2019, 12, 76. [Google Scholar] [CrossRef]
- Qi, L.; Chen, J.; Yang, Y.; Hu, W. Hypoxia Correlates with Poor Survival and M2 Macrophage Infiltration in Colorectal Cancer. Front. Oncol. 2020, 10, 566430. [Google Scholar] [CrossRef]
- Wang, X.; Yuwen, T.-J.; Zhong, Y.; Li, Z.-G.; Wang, X.-Y. A new method for predicting the prognosis of colorectal cancer patients through a combination of multiple tumor-associated macrophage markers at the invasive front. Heliyon 2023, 9, e13211. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Mi, Y.; Guan, B.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Cai, S.; Xu, Y.; Li, X.; et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J. Hematol. Oncol. 2020, 13, 156. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Li, T.; Shi, H.; Zhou, Z.; Huang, Z.; Lei, X. The cellular and molecular components involved in pre-metastatic niche formation in colorectal cancer liver metastasis. Expert Rev. Gastroenterol. Hepatol. 2020, 15, 389–399. [Google Scholar] [CrossRef]
- Sawa-Wejksza, K.; Dudek, A.; Lemieszek, M.; Kaławaj, K.; Kandefer-Szerszeń, M. Colon cancer–derived conditioned medium induces differentiation of THP-1 monocytes into a mixed population of M1/M2 cells. Tumor Biol. 2018, 40, 1010428318797880. [Google Scholar] [CrossRef] [PubMed]
- Scheurlen, K.M.; Billeter, A.T.; O’Brien, S.J.; Galandiuk, S. Metabolic dysfunction and early-onset colorectal cancer—How macrophages build the bridge. Cancer Med. 2020, 9, 6679–6693. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.-C.; Chen, C.; Xu, Z.-Q.; Zhao, J.-K.; Ou, B.-C.; Sun, J.; Zheng, M.-H.; Zong, Y.-P.; Lu, A.-G. CCR6 promotes tumor angiogenesis via the AKT/NF-κB/VEGF pathway in colorectal cancer. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 387–397. [Google Scholar] [CrossRef] [PubMed]
- Korbecki, J.; Grochans, S.; Gutowska, I.; Barczak, K.; Baranowska-Bosiacka, I. CC Chemokines in a Tumor: A Review of Pro-Cancer and Anti-Cancer Properties of Receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 Ligands. Int. J. Mol. Sci. 2020, 21, 7619. [Google Scholar] [CrossRef]
- Sampaio-Ribeiro, G.; Ruivo, A.; Silva, A.; Santos, A.L.; Oliveira, R.C.; Laranjeira, P.; Gama, J.; Cipriano, M.A.; Tralhão, J.G.; Paiva, A. Extensive Phenotypic Characterization of T Cells Infiltrating Liver Metastasis from Colorectal Cancer: A Potential Role in Precision Medicine. Cancers 2022, 14, 6069. [Google Scholar] [CrossRef]
- Höppener, D.J.; Nierop, P.M.H.; Hof, J.; Sideras, K.; Zhou, G.; Visser, L.; Gouw, A.S.H.; de Jong, K.P.; Sprengers, D.; Kwekkeboom, J.; et al. Enrichment of the tumour immune microenvironment in patients with desmoplastic colorectal liver metastasis. Br. J. Cancer 2020, 123, 196–206. [Google Scholar] [CrossRef]
- Van Den Eynden, G.G.; Bird, N.C.; Majeed, A.W.; Van Laere, S.; Dirix, L.Y.; Vermeulen, P.B. The histological growth pattern of colorectal cancer liver metastases has prognostic value. Clin. Exp. Metastasis 2012, 29, 541–549. [Google Scholar] [CrossRef]
- Mármol, I.; Sánchez-De-Diego, C.; Pradilla Dieste, A.; Cerrada, E.; Rodriguez Yoldi, M. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int. J. Mol. Sci. 2017, 18, 197. [Google Scholar] [CrossRef] [PubMed]
- Zarour, L.R.; Anand, S.; Billingsley, K.G.; Bisson, W.H.; Cercek, A.; Clarke, M.F.; Coussens, L.M.; Gast, C.E.; Geltzeiler, C.B.; Hansen, L.; et al. Colorectal Cancer Liver Metastasis: Evolving Paradigms and Future Directions. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Zafari, N.; Khosravi, F.; Rezaee, Z.; Esfandyari, S.; Bahiraei, M.; Bahramy, A.; Ferns, G.A.; Avan, A. The role of the tumor microenvironment in colorectal cancer and the potential therapeutic approaches. J. Clin. Lab. Anal. 2022, 36, e24585. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-Q.; Li, W.-L.; Ma, S.-M.; Liang, L.; Kou, Z.-Y.; Yang, J. Discovery of core gene families associated with liver metastasis in colorectal cancer and regulatory roles in tumor cell immune infiltration. Transl. Oncol. 2021, 14, 101011. [Google Scholar] [CrossRef] [PubMed]
- Gatault, S.; Legrand, F.; Delbeke, M.; Loiseau, S.; Capron, M. Involvement of eosinophils in the anti-tumor response. Cancer Immunol. Immunother. 2012, 61, 1527–1534. [Google Scholar] [CrossRef] [PubMed]
- Haram, A.; Boland, M.R.; Kelly, M.E.; Bolger, J.C.; Waldron, R.M.; Kerin, M.J. The prognostic value of neutrophil-to-lymphocyte ratio in colorectal cancer: A systematic review. J. Surg. Oncol. 2017, 115, 470–479. [Google Scholar] [CrossRef] [PubMed]
- Shaul, M.E.; Fridlender, Z.G. Tumour-associated neutrophils in patients with cancer. Nat. Rev. Clin. Oncol. 2019, 16, 601–620. [Google Scholar] [CrossRef]
- Xiao, B.; Peng, J.; Wang, Y.; Deng, Y.; Ou, Q.; Wu, X.; Lin, J.; Pan, Z.; Zhang, L. Prognostic value of tumor infiltrating lymphocytes combined with PD-L1 expression for patients with solitary colorectal cancer liver metastasis. Ann. Transl. Med. 2020, 8, 1221. [Google Scholar] [CrossRef]
- Wang, E.; Shibutani, M.; Nagahara, H.; Fukuoka, T.; Iseki, Y.; Okazaki, Y.; Kashiwagi, S.; Tanaka, H.; Maeda, K. Prognostic value of the density of tumor-infiltrating lymphocytes in colorectal cancer liver metastases. Oncol. Lett. 2021, 22, 837. [Google Scholar] [CrossRef]
- Zhou, Y.; Cheng, L.; Liu, L.; Li, X. NK cells are never alone: Crosstalk and communication in tumour microenvironments. Mol. Cancer 2023, 22, 34. [Google Scholar] [CrossRef]
- Sun, Y.; Hu, H.; Liu, Z.; Xu, J.; Gao, Y.; Zhan, X.; Zhou, S.; Zhong, W.; Wu, D.; Wang, P.; et al. Macrophage STING signaling promotes NK cell to suppress colorectal cancer liver metastasis via 4-1BBL/4-1BB co-stimulation. J. Immunother. Cancer 2023, 11, e006481. [Google Scholar] [CrossRef] [PubMed]
- Cózar, B.; Greppi, M.; Carpentier, S.; Narni-Mancinelli, E.; Chiossone, L.; Vivier, E. Tumor-Infiltrating Natural Killer Cells. Cancer Discov. 2021, 11, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Dai, S.; Yue, L.; Xu, F.; Gu, J.; Dai, X.; Qian, X. Emerging mechanisms progress of colorectal cancer liver metastasis. Front. Endocrinol. 2022, 13, 1081585. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Zou, Z.; Li, H.; Zou, G.; Xu, J.; Wang, L.; Du, X. The Preoperative Peripheral Blood Monocyte Count Is Associated with Liver Metastasis and Overall Survival in Colorectal Cancer Patients. PLoS ONE 2016, 11, e0157486. [Google Scholar] [CrossRef]
- Cortese, N.; Soldani, C.; Franceschini, B.; Barbagallo, M.; Marchesi, F.; Torzilli, G.; Donadon, M. Macrophages in Colorectal Cancer Liver Metastases. Cancers 2019, 11, 633. [Google Scholar] [CrossRef]
- Kasprzak, A. The Role of Tumor Microenvironment Cells in Colorectal Cancer (CRC) Cachexia. Int. J. Mol. Sci. 2021, 22, 1565. [Google Scholar] [CrossRef]
- Gao, Y.; Rosen, J.M.; Zhang, X.H.-F. The tumor-immune ecosystem in shaping metastasis. Am. J. Physiol. Physiol. 2023, 324, C707–C717. [Google Scholar] [CrossRef]
- Valls, A.F.; Knipper, K.; Giannakouri, E.; Sarachaga, V.; Hinterkopf, S.; Wuehrl, M.; Shen, Y.; Radhakrishnan, P.; Klose, J.; Ulrich, A.; et al. VEGFR1+ Metastasis–Associated Macrophages Contribute to Metastatic Angiogenesis and Influence Colorectal Cancer Patient Outcome. Clin. Cancer Res. 2019, 25, 5674–5685. [Google Scholar] [CrossRef]
- Malekghasemi, S.; Majidi, J.; Baghbanzadeh, A.; Abdolalizadeh, J.; Baradaran, B.; Aghebati-Maleki, L. Tumor-Associated Macrophages: Protumoral Macrophages in Inflammatory Tumor Microenvironment. Adv. Pharm. Bull. 2020, 10, 556–565. [Google Scholar] [CrossRef]
- Gallo, G.; Vescio, G.; De Paola, G.; Sammarco, G. Therapeutic Targets and Tumor Microenvironment in Colorectal Cancer. J. Clin. Med. 2021, 10, 2295. [Google Scholar] [CrossRef]
- Ferrucci, V.; Asadzadeh, F.; Collina, F.; Siciliano, R.; Boccia, A.; Marrone, L.; Spano, D.; Carotenuto, M.; Chiarolla, C.M.; De Martino, D.; et al. Prune-1 drives polarization of tumor-associated macrophages (TAMs) within the lung metastatic niche in triple-negative breast cancer. iScience 2020, 24, 101938. [Google Scholar] [CrossRef]
- Han, Y.; Guo, W.; Ren, T.; Huang, Y.; Wang, S.; Liu, K.; Zheng, B.; Yang, K.; Zhang, H.; Liang, X. Tumor-associated macrophages promote lung metastasis and induce epithelial-mesenchymal transition in osteosarcoma by activating the COX-2/STAT3 axis. Cancer Lett. 2018, 440–441, 116–125. [Google Scholar] [CrossRef]
- Cantero-Cid, R.; Casas-Martin, J.; Hernández-Jiménez, E.; Cubillos-Zapata, C.; Varela-Serrano, A.; Avendaño-Ortiz, J.; Casarrubios, M.; Montalbán-Hernández, K.; Villacañas-Gil, I.; Guerra-Pastrián, L.; et al. PD-L1/PD-1 crosstalk in colorectal cancer: Are we targeting the right cells? BMC Cancer 2018, 18, 945. [Google Scholar] [CrossRef]
- Xie, F.; Xu, M.; Lu, J.; Mao, L.; Wang, S. The role of exosomal PD-L1 in tumor progression and immunotherapy. Mol. Cancer 2019, 18, 146. [Google Scholar] [CrossRef]
- Yamaguchi, H.; Hsu, J.-M.; Yang, W.-H.; Hung, M.-C. Mechanisms regulating PD-L1 expression in cancers and associated opportunities for novel small-molecule therapeutics. Nat. Rev. Clin. Oncol. 2022, 19, 287–305. [Google Scholar] [CrossRef]
- Alexander, P.G.; McMillan, D.C.; Park, J.H. A meta-analysis of CD274 (PD-L1) assessment and prognosis in colorectal cancer and its role in predicting response to anti-PD-1 therapy. Crit. Rev. Oncol. 2020, 157, 103147. [Google Scholar] [CrossRef]
- Lee, K.S.; Kwak, Y.; Ahn, S.; Shin, E.; Oh, H.-K.; Kim, D.-W.; Kang, S.-B.; Choe, G.; Kim, W.H.; Lee, H.S. Prognostic implication of CD274 (PD-L1) protein expression in tumor-infiltrating immune cells for microsatellite unstable and stable colorectal cancer. Cancer Immunol. Immunother. 2017, 66, 927–939. [Google Scholar] [CrossRef]
- Ahn, S.B.; Sharma, S.; Mohamedali, A.; Mahboob, S.; Redmond, W.J.; Pascovici, D.; Wu, J.X.; Zaw, T.; Adhikari, S.; Vaibhav, V.; et al. Potential early clinical stage colorectal cancer diagnosis using a proteomics blood test panel. Clin. Proteom. 2019, 16, 34. [Google Scholar] [CrossRef]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Väyrynen, J.P.; Haruki, K.; Lau, M.C.; Väyrynen, S.A.; Zhong, R.; Dias Costa, A.; Borowsky, J.; Zhao, M.; Fujiyoshi, K.; Arima, K.; et al. The Prognostic Role of Macrophage Polarization in the Colorectal Cancer Microenvironment. Cancer Immunol. Res. 2021, 9, 8–19. [Google Scholar] [CrossRef]
- Ugai, T.; Väyrynen, J.P.; Lau, M.C.; Borowsky, J.; Akimoto, N.; Väyrynen, S.A.; Zhao, M.; Zhong, R.; Haruki, K.; Costa, A.D.; et al. Immune cell profiles in the tumor microenvironment of early-onset, intermediate-onset, and later-onset colorectal cancer. Cancer Immunol. Immunother. 2021, 71, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Liu, B.; Cao, Y.; Yao, S.; Liu, Y.; Jin, G.; Qin, Y.; Chen, Y.; Cui, K.; Zhou, L.; et al. Colorectal Cancer-Derived Small Extracellular Vesicles Promote Tumor Immune Evasion by Upregulating PD-L1 Expression in Tumor-Associated Macrophages. Adv. Sci. 2022, 9, 2102620. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wu, F.; Zhao, S.; Shi, P.; Wang, S.; Cui, D. Correlation between PD-1/PD-L1 expression and polarization in tumor-associated macrophages: A key player in tumor immunotherapy. Cytokine Growth Factor Rev. 2022, 67, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, Z.; Li, Y.; Zhao, W.; Wu, J.; Zhang, Z. PD-1/PD-L1 Checkpoint Inhibitors in Tumor Immunotherapy. Front. Pharmacol. 2021, 12, 731798. [Google Scholar] [CrossRef]
- Yi, M.; Zheng, X.; Niu, M.; Zhu, S.; Ge, H.; Wu, K. Combination strategies with PD-1/PD-L1 blockade: Current advances and future directions. Mol. Cancer 2022, 21, 28. [Google Scholar] [CrossRef]
- Du, F.; Liu, Y. Predictive molecular markers for the treatment with immune checkpoint inhibitors in colorectal cancer. J. Clin. Lab. Anal. 2021, 36, e24141. [Google Scholar] [CrossRef]
- Payandeh, Z.; Khalili, S.; Somi, M.H.; Mard-Soltani, M.; Baghbanzadeh, A.; Hajiasgharzadeh, K.; Samadi, N.; Baradaran, B. PD-1/PD-L1-dependent immune response in colorectal cancer. J. Cell. Physiol. 2020, 235, 5461–5475. [Google Scholar] [CrossRef]
- Wang, Y.; Gu, T.; Tian, X.; Li, W.; Zhao, R.; Yang, W.; Gao, Q.; Li, T.; Shim, J.-H.; Zhang, C.; et al. A Small Molecule Antagonist of PD-1/PD-L1 Interactions Acts as an Immune Checkpoint Inhibitor for NSCLC and Melanoma Immunotherapy. Front. Immunol. 2021, 12, 654463. [Google Scholar] [CrossRef]
- Vandeveer, A.J.; Fallon, J.K.; Tighe, R.; Sabzevari, H.; Schlom, J.; Greiner, J.W. Systemic Immunotherapy of Non-Muscle Invasive Mouse Bladder Cancer with Avelumab, an Anti–PD-L1 Immune Checkpoint Inhibitor. Cancer Immunol. Res. 2016, 4, 452–462. [Google Scholar] [CrossRef]
- Tu, W.; Gong, J.; Zhou, Z.; Tian, D.; Wang, Z. TCF4 enhances hepatic metastasis of colorectal cancer by regulating tumor-associated macrophage via CCL2/CCR2 signaling. Cell Death Dis. 2021, 12, 882. [Google Scholar] [CrossRef]
- Zhang, R.; Qi, F.; Zhao, F.; Li, G.; Shao, S.; Zhang, X.; Yuan, L.; Feng, Y. Cancer-associated fibroblasts enhance tumor-associated macrophages enrichment and suppress NK cells function in colorectal cancer. Cell Death Dis. 2019, 10, 273. [Google Scholar] [CrossRef] [PubMed]
- Pei, X.-B.; Wu, X.-Z.; Yi, F.-S.; Zhai, K.; Shi, H.-Z. Diagnostic value of CD206+CD14+ macrophages in diagnosis of lung cancer originated malignant pleural effusion. J. Thorac. Dis. 2019, 11, 2730–2736. [Google Scholar] [CrossRef] [PubMed]
- Larionova, I.; Tuguzbaeva, G.; Ponomaryova, A.; Stakheyeva, M.; Cherdyntseva, N.; Pavlov, V.; Choinzonov, E.; Kzhyshkowska, J. Tumor-Associated Macrophages in Human Breast, Colorectal, Lung, Ovarian and Prostate Cancers. Front. Oncol. 2020, 10, 566511. [Google Scholar] [CrossRef]
- Zhang, C.; Yu, X.; Gao, L.; Zhaofei, L.; Lai, J.; Lu, D.; Bao, R.; Jianhao, L.; Zhong, L.; Wang, F.; et al. Noninvasive Imaging of CD206-Positive M2 Macrophages as an Early Biomarker for Post-Chemotherapy Tumor Relapse and Lymph Node Metastasis. Theranostics 2017, 7, 4276–4288. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Feng, J.; Huang, H.; Wang, Y.; Yi, X.; Wei, S.; Zhang, M.; Li, Z.; Wang, W.; Hu, W. Single-cell transcriptome analysis of tumor immune microenvironment characteristics in colorectal cancer liver metastasis. Ann. Transl. Med. 2022, 10, 1170. [Google Scholar] [CrossRef] [PubMed]
- Teng, S.; Li, Y.E.; Yang, M.; Qi, R.; Huang, Y.; Wang, Q.; Zhang, Y.; Chen, S.; Li, S.; Lin, K.; et al. Tissue-specific transcription reprogramming promotes liver metastasis of colorectal cancer. Cell Res. 2019, 30, 34–49. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Li, P.; Zhang, B.; Feng, L.; Cheng, S. Decoding the colorectal cancer ecosystem emphasizes the cooperative role of cancer cells, TAMs and CAFsin tumor progression. J. Transl. Med. 2022, 20, 462. [Google Scholar] [CrossRef] [PubMed]
- Gerovska, D.; Larrinaga, G.; Solano-Iturri, J.D.; Márquez, J.; Gallastegi, P.G.; Khatib, A.-M.; Poschmann, G.; Stühler, K.; Armesto, M.; Lawrie, C.H.; et al. An Integrative Omics Approach Reveals Involvement of BRCA1 in Hepatic Metastatic Progression of Colorectal Cancer. Cancers 2020, 12, 2380. [Google Scholar] [CrossRef]
- Pan, J.; Weng, Z.; Xue, C.; Lin, B.; Lin, M. The Bioinformatics-Based Analysis Identifies 7 Immune-Related Genes as Prognostic Biomarkers for Colon Cancer. Front. Oncol. 2021, 11, 726701. [Google Scholar] [CrossRef]
- Kudelova, E.; Holubekova, V.; Grendar, M.; Kolkova, Z.; Samec, M.; Vanova, B.; Mikolajcik, P.; Smolar, M.; Kudela, E.; Laca, L.; et al. Circulating miRNA expression over the course of colorectal cancer treatment. Oncol. Lett. 2021, 23, 18. [Google Scholar] [CrossRef]
- Ghadjar, P.; Coupland, S.; Na, I.-K.; Noutsias, M.; Letsch, A.; Stroux, A.; Bauer, S.; Buhr, H.J.; Thiel, E.; Scheibenbogen, C.; et al. Chemokine Receptor CCR6 Expression Level and Liver Metastases in Colorectal Cancer. J. Clin. Oncol. 2006, 24, 1910–1916. [Google Scholar] [CrossRef] [PubMed]
- Rubie, C.; Oliveira, V.; Kempf, K.; Wagner, M.; Tilton, B.; Rau, B.; Kruse, B.; König, J.; Schilling, M. Involvement of Chemokine Receptor CCR6 in Colorectal Cancer Metastasis. Tumor Biol. 2006, 27, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.-N.; Han, Y.-B.; Yang, R.; Yang, Z.-C. Chemokines in colon cancer progression. Semin. Cancer Biol. 2022, 86, 400–407. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Mi, Y.; Zheng, B.; Wei, P.; Gu, Y.; Zhang, Z.; Xu, Y.; Cai, S.; Li, X.; Li, D. Highly-metastatic colorectal cancer cell released miR-181a-5p-rich extracellular vesicles promote liver metastasis by activating hepatic stellate cells and remodelling the tumour microenvironment. J. Extracell. Vesicles 2022, 11, e12186. [Google Scholar] [CrossRef] [PubMed]
- Heidrich, I.; Ačkar, L.; Mossahebi Mohammadi, P.; Pantel, K. Liquid biopsies: Potential and challenges. Int. J. Cancer 2021, 148, 528–545. [Google Scholar] [CrossRef] [PubMed]
- Raza, A.; Khan, A.Q.; Inchakalody, V.P.; Mestiri, S.; Yoosuf, Z.S.K.M.; Bedhiafi, T.; El-Ella, D.M.A.; Taib, N.; Hydrose, S.; Akbar, S.; et al. Dynamic liquid biopsy components as predictive and prognostic biomarkers in colorectal cancer. J. Exp. Clin. Cancer Res. 2022, 41, 99. [Google Scholar] [CrossRef]
- Nakashima-Nakasuga, C.; Hazama, S.; Suzuki, N.; Nakagami, Y.; Xu, M.; Yoshida, S.; Tomochika, S.; Fujiwara, N.; Matsukuma, S.; Matsui, H.; et al. Serum LOX-1 is a novel prognostic biomarker of colorectal cancer. Int. J. Clin. Oncol. 2020, 25, 1308–1317. [Google Scholar] [CrossRef]
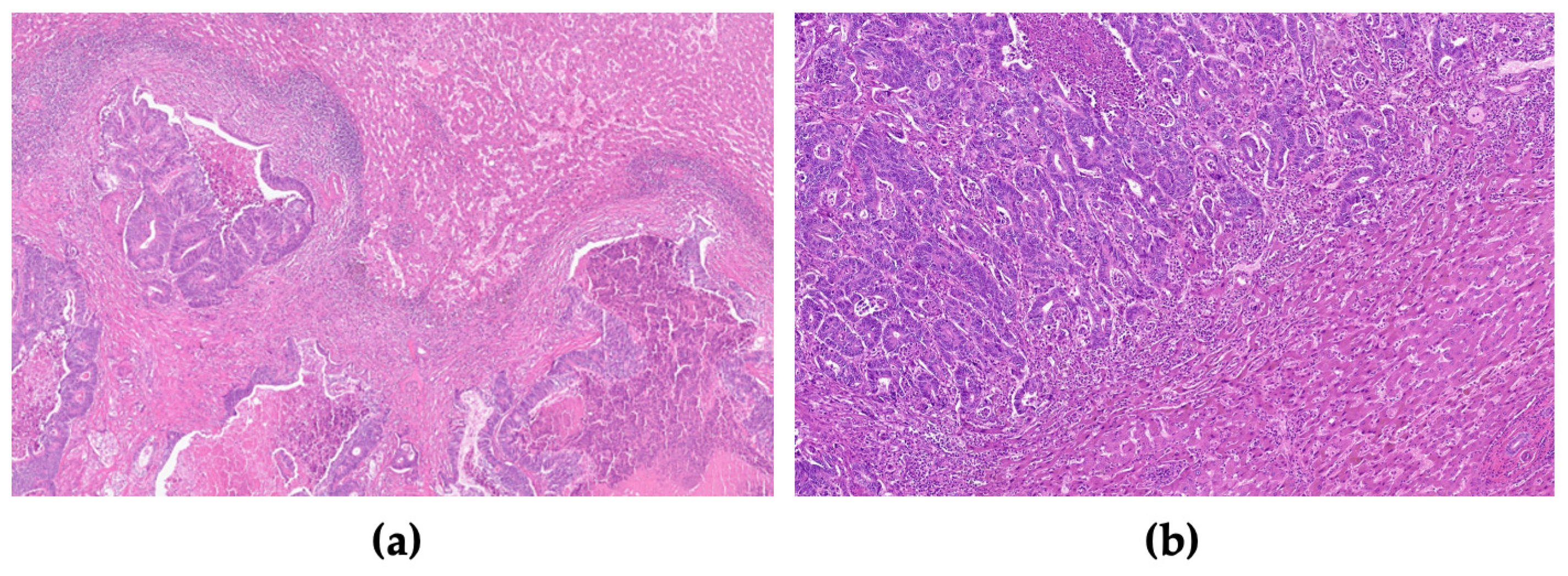


| Variable | (n = 47) |
|---|---|
| Age at surgical resection | |
| Mean ± SD; range | 59.1 ± 12.8; 36–90 |
| Variable | Number (%) |
| Gender | |
| Male | 31 (66) |
| Female | 16 (34) |
| Presentation | |
| Synchronous | 26 (55) |
| Metachronous | 21 (45) |
| Primary tumor location | |
| Sigmoid | 16 (34) |
| Ascending | 10 (21) |
| Descending | 5 (11) |
| Splenic Flexure | 4 (9) |
| Hepatic Flexure | 1 (2) |
| Transverse | 2 (4) |
| Rectal | 9 (19) |
| T stage of the primary colorectal tumor | |
| 3 | 33 (70) |
| 4a | 12 (26) |
| 4b | 2 (4) |
| First approach | |
| Liver First | 6 (13) |
| Colectomy | 34 (72) |
| Synchronous resection | 7 (15) |
| Preoperative systemic chemotherapy | 27 (57) |
| Variable | (n = 47) |
| Number of colorectal liver metastases | |
| Mean ± SD; range | 2.3 ± 1.8; 1–10 |
| Size of the largest colorectal liver metastases (cm) | |
| Mean ± SD; range | 3.8 ± 2.4; 0.4–12 |
| Variable | Number (%) |
| Histologic growth pattern | |
| Desmoplastic | 17 (36) |
| Non-desmoplastic | 30 (64) |
| Status | |
| Dead | 3 (6) |
| Alive | 44 (94) |
| Fluorochromes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Tube | PB | PO | FITC | PE | PerCP-Cy5.5 | PE-Cy7 | APC | APC-H7 |
| 1 | CD274 Biolegend (29E.2A3) (5 µL) | CD45 BD Biosciences (2D1) (2.5 µL) | Cito18 Cytognos (Ks18.04) (2.5 µL) | EpCAM BD Biosciences (EBA-1) (10 µL) | CD206 Biolegend (15–2) (5 µL) | CD16 BD Pharmingen (3G8) (2 µL) | CD33 BD Biosciences (P67.6) (2.5 µL) | CD14 BD Biosciences (mφp9) (2.5 µL) |
| Cell Types | Non-Tumor (%) ± SEM | Tumor (%) ± SEM | p Value | Tumor | p Value | |
|---|---|---|---|---|---|---|
| Non-Desmoplastic (%) ± SEM | Desmoplastic (%) ± SEM | |||||
| Eosinophils | 0.53 ± 0.17 | 0.01 ± 0.01 | 0.0001 a | 0.002 ± 0.002 | 0.02 ± 0.01 | 0.38 |
| Eosinophils (after removing non-hematopoietic cells) | 0.81 ± 0.26 | 0.04 ± 0.03 | 0.0001 a | 0.01 ± 0.01 | 0.11 ± 0.08 | 0.38 |
| Neutrophils | 22.50 ± 2.50 | 6.87 ± 1.56 | <0.0001 b | 5.63 ± 1.57 | 4.20 ± 1.59 | 0.38 |
| Neutrophils (after removing non-hematopoietic cells) | 34.88 ± 3.88 | 32.66 ± 7.43 | 0.50 | 27.65 ± 7.71 | 24.03 ± 9.05 | 0.64 |
| Lymphocytes | 31.36 ± 2.93 | 11.87 ± 1.99 | <0.0001 b | 12.17 ± 1.62 | 11.96 ± 4.30 | 0.44 |
| Lymphocytes (after removing non-hematopoietic cells) | 48.61 ± 4.53 | 56.44 ± 9.44 | 0.79 | 59.75 ± 7.94 | 68.44 ± 24.62 | 0.67 |
| Monocytes | 3.45 ± 0.50 | 0.96 ± 0.27 | <0.0001 b | 0.88 ± 0.29 | 0.55 ± 0.16 | 0.77 |
| Monocytes (after removing non-hematopoietic cells) | 5.35 ± 0.78 | 4.56 ± 1.27 | 0.07 | 4.33 ± 1.44 | 3.17 ± 0.93 | 0.93 |
| Classical | 82.08 ± 1.63 | 81.87 ± 2.68 | 0.43 | 80.37 ± 3.97 | 84.07 ± 3.19 | 0.96 |
| CD206+ | 43.04 ± 5.33 | 43.04 ± 6.46 | 0.93 | 49.70 ± 7.99 | 22.16 ± 9.36 | 0.02 * |
| CD274+ | 57.03 ± 5.50 | 54.86 ± 6.26 | 0.82 | 62.40 ± 7.52 | 43.01 ± 10.49 | 0.03 * |
| Intermediate | 11.38 ± 1.00 | 9.91 ± 1.40 | 0.17 | 10.85 ± 2.18 | 8.54 ± 1.33 | 0.98 |
| CD206+ | 56.06 ± 5.08 | 48.95 ± 6.01 | 0.50 | 58.94 ± 7.19 | 28.29 ± 8.57 | 0.02 * |
| CD274+ | 67.53 ± 4.80 | 59.93 ± 5.90 | 0.25 | 66.14 ± 7.47 | 41.96 ± 8.80 | 0.04 * |
| Non-Classical | 6.01 ± 1.16 | 8.22 ± 2.66 | 0.17 | 8.81 ± 3.84 | 7.35 ± 3.50 | 0.38 |
| CD206+ | 40.71 ± 5.00 | 31.97 ± 6.44 | 0.09 | 34.37 ± 7.91 | 22.86 ± 10.50 | 0.23 |
| CD274+ | 56.04 ± 5.54 | 51.42 ± 6.32 | 0.46 | 54.60 ± 9.09 | 29.72 ± 10.73 | 0.03 * |
| NK cells | 4.25 ± 0.46 | 0.37 ± 0.06 | <0.0001 b | 0.36 ± 0.08 | 0.29 ± 0.07 | >0.99 |
| NK cells (after removing non-hematopoietic cells) | 6.60 ± 0.71 | 1.74 ± 0.29 | <0.0001 b | 1.75 ± 0.39 | 1.65 ± 0.38 | 0.44 |
| Non-Hematopoietic cells | 35.49 ± 3.63 | 78.97 ± 2.37 | <0.0001 b | 79.64 ± 2.85 | 82.52 ± 4.19 | 0.72 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sampaio-Ribeiro, G.; Ruivo, A.; Silva, A.; Santos, A.L.; Oliveira, R.C.; Gama, J.; Cipriano, M.A.; Tralhão, J.G.; Paiva, A. Innate Immune Cells in the Tumor Microenvironment of Liver Metastasis from Colorectal Cancer: Contribution to a Comprehensive Therapy. Cancers 2023, 15, 3222. https://doi.org/10.3390/cancers15123222
Sampaio-Ribeiro G, Ruivo A, Silva A, Santos AL, Oliveira RC, Gama J, Cipriano MA, Tralhão JG, Paiva A. Innate Immune Cells in the Tumor Microenvironment of Liver Metastasis from Colorectal Cancer: Contribution to a Comprehensive Therapy. Cancers. 2023; 15(12):3222. https://doi.org/10.3390/cancers15123222
Chicago/Turabian StyleSampaio-Ribeiro, Gabriela, Ana Ruivo, Ana Silva, Ana Lúcia Santos, Rui Caetano Oliveira, João Gama, Maria Augusta Cipriano, José Guilherme Tralhão, and Artur Paiva. 2023. "Innate Immune Cells in the Tumor Microenvironment of Liver Metastasis from Colorectal Cancer: Contribution to a Comprehensive Therapy" Cancers 15, no. 12: 3222. https://doi.org/10.3390/cancers15123222
APA StyleSampaio-Ribeiro, G., Ruivo, A., Silva, A., Santos, A. L., Oliveira, R. C., Gama, J., Cipriano, M. A., Tralhão, J. G., & Paiva, A. (2023). Innate Immune Cells in the Tumor Microenvironment of Liver Metastasis from Colorectal Cancer: Contribution to a Comprehensive Therapy. Cancers, 15(12), 3222. https://doi.org/10.3390/cancers15123222








