In Silico Analysis Predicts Nuclear Factors NR2F6 and YAP1 as Mesenchymal Subtype-Specific Therapeutic Targets for Ovarian Cancer Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
2.1. Gene Expression Profiles and Data Pre-Processing
2.2. Enrichment of Hallmark Gene Signatures
2.3. In Silico Enrichment of Immune and Stromal Cell Type Signatures
2.4. Complementary Single-Cell Analysis of Tumour Composition
2.5. Cell–Cell Communication Analysis Combined with Cytokine Activity Inference
2.6. Correlation of Ligand–Receptor Pair Expression with Cancer Hallmarks
2.7. Inference of Transcription Factor Activity from Gene Expression Data
2.8. Inferred Transcription Factor Activity-Guided Intercellular Communication Network
2.9. Generation of a Prior Knowledge Network
2.10. Contextualization of Signalling Networks via Causal Reasoning
3. Results
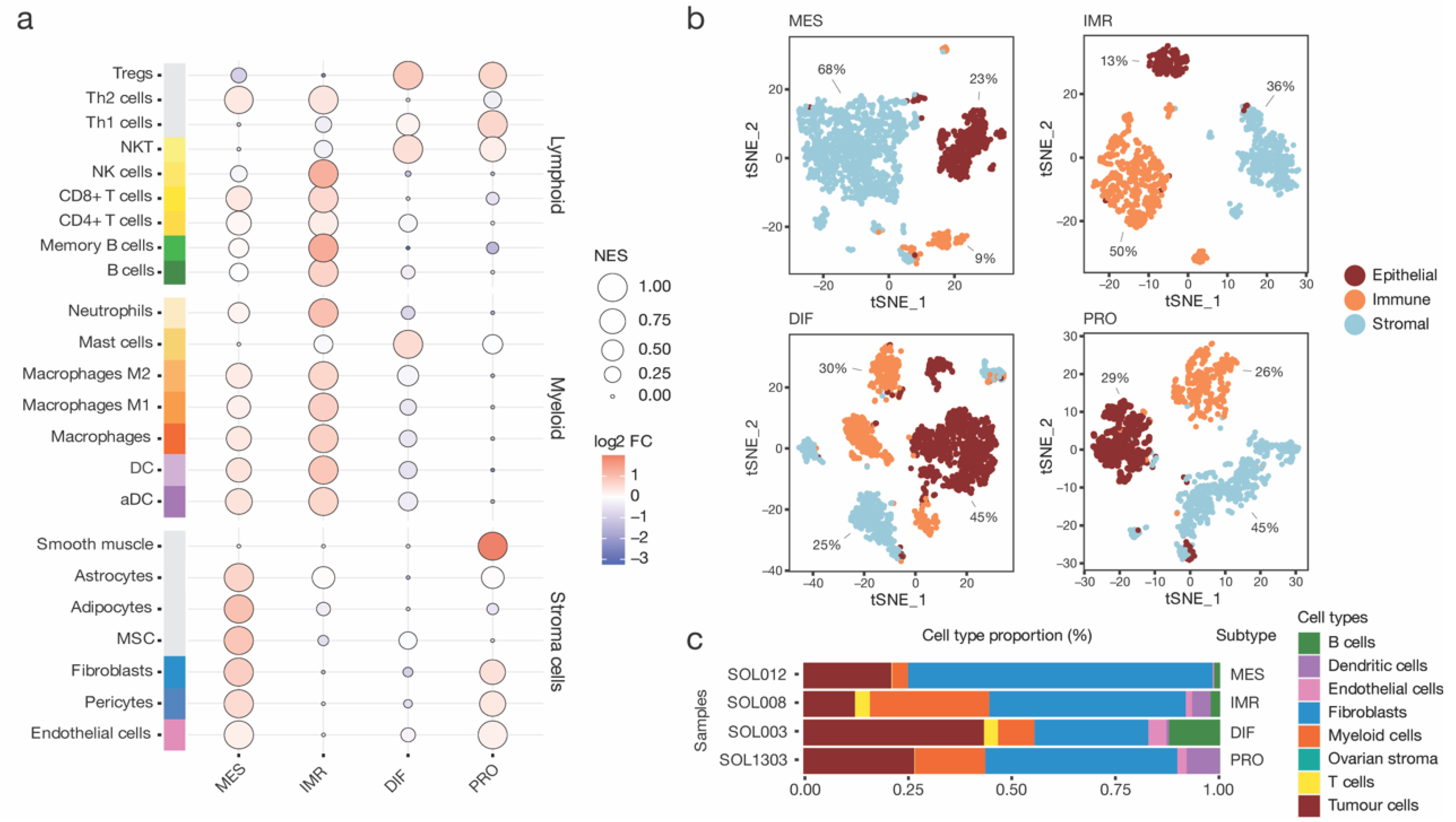
3.1. Classification and Characterization of High-Grade Serous Ovarian Cancer Datasets
3.2. Compositional Analysis of High-Grade Serous Ovarian Cancer Subtypes Reveals Distinct Cell Type Proportions
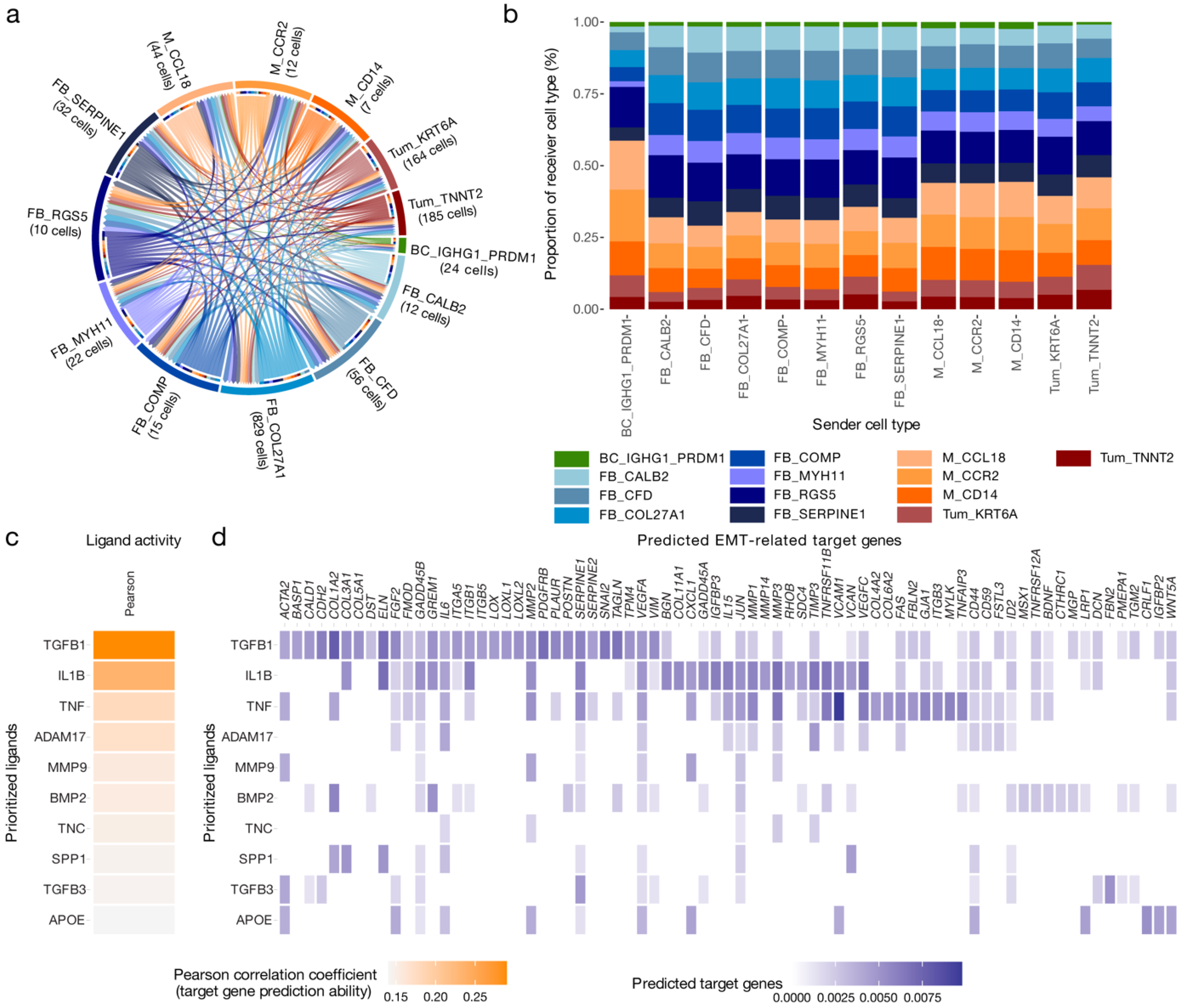
3.3. Cell–Cell Interactions Drive Cancer Hallmarks in High-Grade Serous Ovarian Cancer
3.4. IL1B, TGFB1 and TNF Are Key Drivers of the Cancer Hallmarks in the MES Subtype
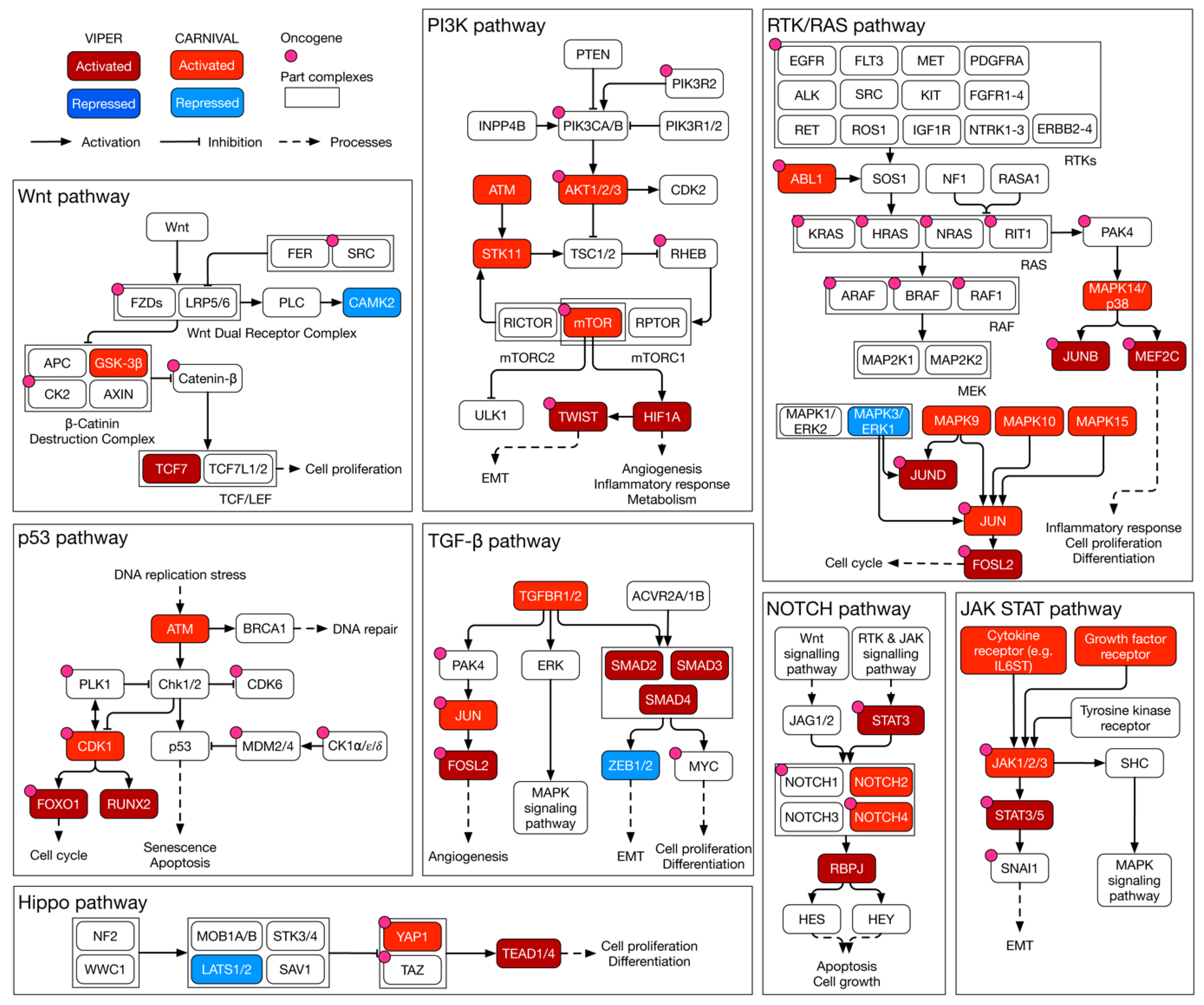
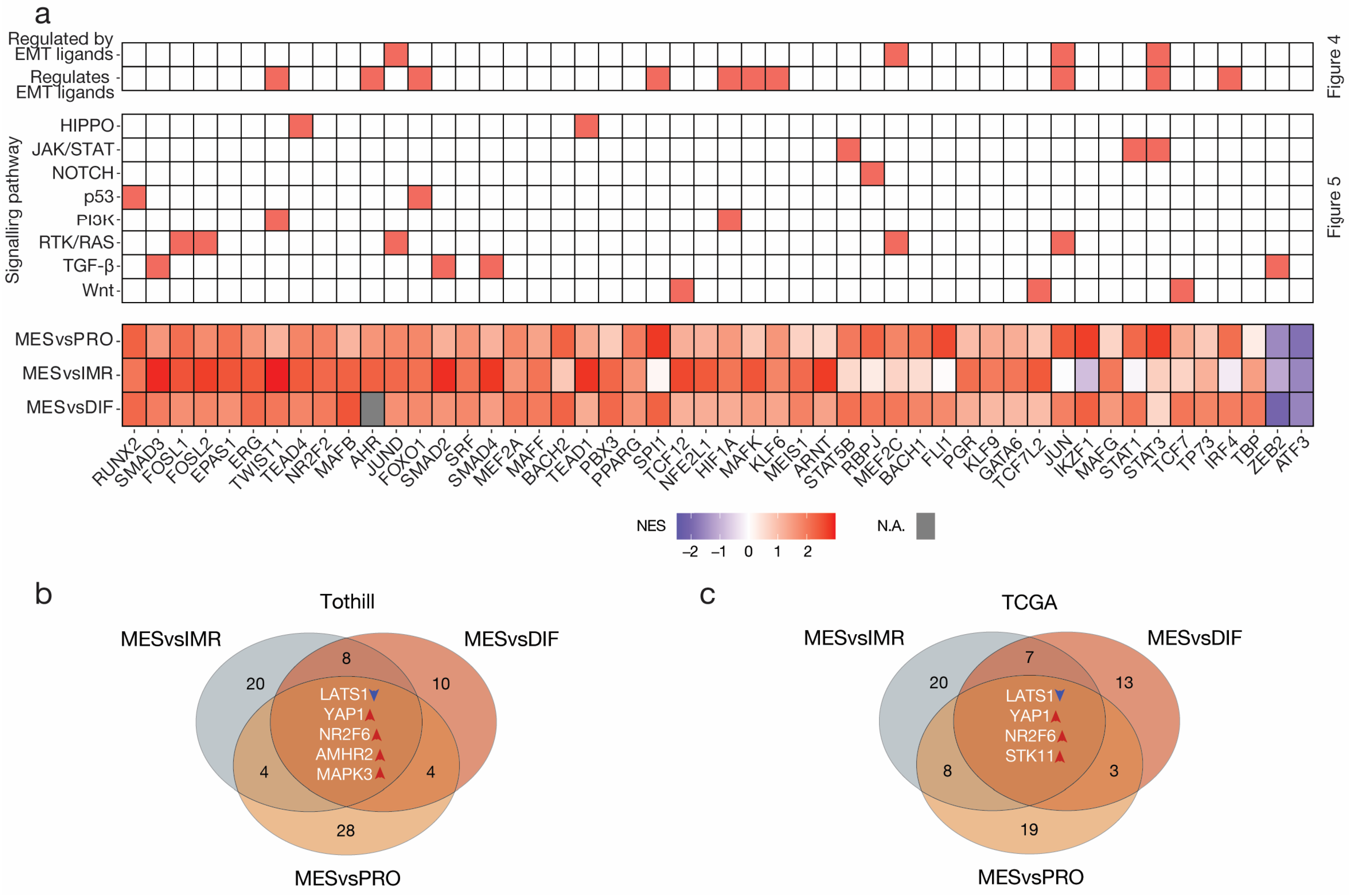
3.5. Intercellular Signalling Mediates Dysregulation of Transcription Factor Activity and Aberrant Cytokine Production in MES
3.6. Causal Inference Analysis Reveals YAP1 and NR2F6 as Novel Therapeutic Targets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bowtell, D.D.; Bohm, S.; Ahmed, A.A.; Aspuria, P.J.; Bast, R.C., Jr.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A.; et al. Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2016. CA A Cancer J. Clin. 2016, 66, 7–30. [Google Scholar] [CrossRef]
- Jiang, X.; Li, W.; Li, X.; Bai, H.; Zhang, Z. Current status and future prospects of PARP inhibitor clinical trials in ovarian cancer. Cancer Manag. Res. 2019, 11, 4371–4390. [Google Scholar] [CrossRef]
- Masoodi, T.; Siraj, S.; Siraj, A.K.; Azam, S.; Qadri, Z.; Parvathareddy, S.K.; Tulbah, A.; Al-Dayel, F.; AlHusaini, H.; AlOmar, O.; et al. Genetic heterogeneity and evolutionary history of high-grade ovarian carcinoma and matched distant metastases. Br. J. Cancer 2020, 122, 1219–1230. [Google Scholar] [CrossRef] [PubMed]
- Tothill, R.W.; Tinker, A.V.; George, J.; Brown, R.; Fox, S.B.; Lade, S.; Johnson, D.S.; Trivett, M.K.; Etemadmoghadam, D.; Locandro, B.; et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin. Cancer Res. 2008, 14, 5198–5208. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Riester, M.; Wei, W.; Waldron, L.; Culhane, A.C.; Trippa, L.; Oliva, E.; Kim, S.H.; Michor, F.; Huttenhower, C.; Parmigiani, G.; et al. Risk prediction for late-stage ovarian cancer by meta-analysis of 1525 patient samples. J. Natl. Cancer Inst. 2014, 106, dju048. [Google Scholar] [CrossRef] [PubMed]
- Tan, T.Z.; Miow, Q.H.; Huang, R.Y.; Wong, M.K.; Ye, J.; Lau, J.A.; Wu, M.C.; Bin Abdul Hadi, L.H.; Soong, R.; Choolani, M.; et al. Functional genomics identifies five distinct molecular subtypes with clinical relevance and pathways for growth control in epithelial ovarian cancer. EMBO Mol. Med. 2013, 5, 1051–1066. [Google Scholar] [CrossRef]
- Olbrecht, S.; Busschaert, P.; Qian, J.; Vanderstichele, A.; Loverix, L.; Van Gorp, T.; Van Nieuwenhuysen, E.; Han, S.; Van den Broeck, A.; Coosemans, A.; et al. High-grade serous tubo-ovarian cancer refined with single-cell RNA sequencing: Specific cell subtypes influence survival and determine molecular subtype classification. Genome Med. 2021, 13, 111. [Google Scholar] [CrossRef] [PubMed]
- Schwede, M.; Waldron, L.; Mok, S.C.; Wei, W.; Basunia, A.; Merritt, M.A.; Mitsiades, C.S.; Parmigiani, G.; Harrington, D.P.; Quackenbush, J.; et al. The Impact of Stroma Admixture on Molecular Subtypes and Prognostic Gene Signatures in Serous Ovarian Cancer. Cancer Epidemiol. Biomark. Prev. 2020, 29, 509–519. [Google Scholar] [CrossRef]
- Kulbe, H.; Chakravarty, P.; Leinster, D.A.; Charles, K.A.; Kwong, J.; Thompson, R.G.; Coward, J.I.; Schioppa, T.; Robinson, S.C.; Gallagher, W.M. A dynamic inflammatory cytokine network in the human ovarian cancer microenvironment. Cancer Res. 2012, 72, 66–75. [Google Scholar] [CrossRef]
- Liubomirski, Y.; Lerrer, S.; Meshel, T.; Rubinstein-Achiasaf, L.; Morein, D.; Wiemann, S.; Korner, C.; Ben-Baruch, A. Tumor-Stroma-Inflammation Networks Promote Pro-metastatic Chemokines and Aggressiveness Characteristics in Triple-Negative Breast Cancer. Front. Immunol. 2019, 10, 757. [Google Scholar] [CrossRef]
- Bhat, A.A.; Nisar, S.; Maacha, S.; Carneiro-Lobo, T.C.; Akhtar, S.; Siveen, K.S.; Wani, N.A.; Rizwan, A.; Bagga, P.; Singh, M.; et al. Cytokine-chemokine network driven metastasis in esophageal cancer; promising avenue for targeted therapy. Mol. Cancer 2021, 20, 2. [Google Scholar] [CrossRef] [PubMed]
- Nisar, S.; Yousuf, P.; Masoodi, T.; Wani, N.A.; Hashem, S.; Singh, M.; Sageena, G.; Mishra, D.; Kumar, R.; Haris, M.; et al. Chemokine-Cytokine Networks in the Head and Neck Tumor Microenvironment. Int. J. Mol. Sci. 2021, 22, 4594. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Ning, B.; Shi, T. Single-Cell RNA-Seq Technologies and Related Computational Data Analysis. Front. Genet. 2019, 10, 317. [Google Scholar] [CrossRef]
- Bao, X.; Shi, R.; Zhao, T.; Wang, Y.; Anastasov, N.; Rosemann, M.; Fang, W. Integrated analysis of single-cell RNA-seq and bulk RNA-seq unravels tumour heterogeneity plus M2-like tumour-associated macrophage infiltration and aggressiveness in TNBC. Cancer Immunol. Immunother. 2021, 70, 189–202. [Google Scholar] [CrossRef]
- Liu, A.; Trairatphisan, P.; Gjerga, E.; Didangelos, A.; Barratt, J.; Saez-Rodriguez, J. From expression footprints to causal pathways: Contextualizing large signaling networks with CARNIVAL. NPJ Syst. Biol. Appl. 2019, 5, 40. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Chen, G.M.; Kannan, L.; Geistlinger, L.; Kofia, V.; Safikhani, Z.; Gendoo, D.M.A.; Parmigiani, G.; Birrer, M.; Haibe-Kains, B.; Waldron, L. Consensus on Molecular Subtypes of High-Grade Serous Ovarian Carcinoma. Clin. Cancer Res. 2018, 24, 5037–5047. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Smillie, C.S.; Biton, M.; Ordovas-Montanes, J.; Sullivan, K.M.; Burgin, G.; Graham, D.B.; Herbst, R.H.; Rogel, N.; Slyper, M.; Waldman, J.; et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell 2019, 178, 714–730.e22. [Google Scholar] [CrossRef]
- Dimitrov, D.; Turei, D.; Garrido-Rodriguez, M.; Burmedi, P.L.; Nagai, J.S.; Boys, C.; Ramirez Flores, R.O.; Kim, H.; Szalai, B.; Costa, I.G.; et al. Comparison of methods and resources for cell-cell communication inference from single-cell RNA-Seq data. Nat. Commun. 2022, 13, 3224. [Google Scholar] [CrossRef]
- Browaeys, R.; Saelens, W.; Saeys, Y. NicheNet: Modeling intercellular communication by linking ligands to target genes. Nat. Methods 2020, 17, 159–162. [Google Scholar] [CrossRef]
- Alvarez, M.J.; Shen, Y.; Giorgi, F.M.; Lachmann, A.; Ding, B.B.; Ye, B.H.; Califano, A. Functional characterization of somatic mutations in cancer using network-based inference of protein activity. Nat. Genet. 2016, 48, 838–847. [Google Scholar] [CrossRef]
- Garcia-Alonso, L.; Holland, C.H.; Ibrahim, M.M.; Turei, D.; Saez-Rodriguez, J. Benchmark and integration of resources for the estimation of human transcription factor activities. Genome Res. 2019, 29, 1363–1375. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Hu, X.; Wang, M.; Wang, J.; Zou, B.; Tan, P.; Cui, T.; Dou, Y.; Ning, L.; et al. CellCall: Integrating paired ligand-receptor and transcription factor activities for cell-cell communication. Nucleic Acids Res. 2021, 49, 8520–8534. [Google Scholar] [CrossRef] [PubMed]
- Turei, D.; Korcsmaros, T.; Saez-Rodriguez, J. OmniPath: Guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods 2016, 13, 966–967. [Google Scholar] [CrossRef] [PubMed]
- Schubert, M.; Klinger, B.; Klunemann, M.; Sieber, A.; Uhlitz, F.; Sauer, S.; Garnett, M.J.; Bluthgen, N.; Saez-Rodriguez, J. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 2018, 9, 20. [Google Scholar] [CrossRef] [PubMed]
- Liao, S.J.; Luo, J.; Li, D.; Zhou, Y.H.; Yan, B.; Wei, J.J.; Tu, J.C.; Li, Y.R.; Zhang, G.M.; Feng, Z.H. TGF-beta1 and TNF-alpha synergistically induce epithelial to mesenchymal transition of breast cancer cells by enhancing TAK1 activation. J. Cell Commun. Signal. 2019, 13, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Hong, I.S. Double-edged sword of mesenchymal stem cells: Cancer-promoting versus therapeutic potential. Cancer Sci. 2017, 108, 1939–1946. [Google Scholar] [CrossRef]
- Zeng, B.; Zhou, M.; Wu, H.; Xiong, Z. SPP1 promotes ovarian cancer progression via Integrin beta1/FAK/AKT signaling pathway. Onco Targets Ther. 2018, 11, 1333–1343. [Google Scholar] [CrossRef]
- An, Q.; Liu, T.; Wang, M.Y.; Yang, Y.J.; Zhang, Z.D.; Liu, Z.J.; Yang, B. KRT7 promotes epithelialmesenchymal transition in ovarian cancer via the TGFbeta/Smad2/3 signaling pathway. Oncol. Rep. 2021, 45, 481–492. [Google Scholar] [CrossRef]
- Ediriweera, M.K.; Tennekoon, K.H.; Samarakoon, S.R. Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer: Biological and therapeutic significance. Semin. Cancer Biol. 2019, 59, 147–160. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhou, L.; Ling, L.; Meng, X.; Chu, F.; Zhang, S.; Zhou, F. The Crosstalk Between Hippo-YAP Pathway and Innate Immunity. Front. Immunol. 2020, 11, 323. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Colaprico, A.; Silva, T.C.; Chen, J.; An, H.; Ban, Y.; Huang, H.; Wang, L.; James, J.L.; Balko, J.M.; et al. Multi-omics analysis identifies therapeutic vulnerabilities in triple-negative breast cancer subtypes. Nat. Commun. 2021, 12, 6276. [Google Scholar] [CrossRef]
- Dasari, S.; Fang, Y.; Mitra, A.K. Cancer Associated Fibroblasts: Naughty Neighbors That Drive Ovarian Cancer Progression. Cancers 2018, 10, 406. [Google Scholar] [CrossRef] [PubMed]
- Sonvilla, G.; Allerstorfer, S.; Stattner, S.; Karner, J.; Klimpfinger, M.; Fischer, H.; Grasl-Kraupp, B.; Holzmann, K.; Berger, W.; Wrba, F.; et al. FGF18 in colorectal tumour cells: Autocrine and paracrine effects. Carcinogenesis 2008, 29, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Nowak, M.; Klink, M. The Role of Tumor-Associated Macrophages in the Progression and Chemoresistance of Ovarian Cancer. Cells 2020, 9, 1299. [Google Scholar] [CrossRef]
- Olivera, I.; Sanz-Pamplona, R.; Bolanos, E.; Rodriguez, I.; Etxeberria, I.; Cirella, A.; Egea, J.; Garasa, S.; Migueliz, I.; Eguren-Santamaria, I.; et al. A therapeutically actionable pro-tumoral axis of cytokines involving interleukin-8, TNFa and IL-1B. Cancer Discov. 2022, 2, 2140–2157. [Google Scholar] [CrossRef]
- Fleming, N.I.; Jorissen, R.N.; Mouradov, D.; Christie, M.; Sakthianandeswaren, A.; Palmieri, M.; Day, F.; Li, S.; Tsui, C.; Lipton, L.; et al. SMAD2, SMAD3 and SMAD4 mutations in colorectal cancer. Cancer Res. 2013, 73, 725–735. [Google Scholar] [CrossRef]
- Zi, Z.; Chapnick, D.A.; Liu, X. Dynamics o.of TGF-beta/Smad signaling. FEBS Lett. 2012, 586, 1921–1928. [Google Scholar] [CrossRef]
- Lee, C.; Jeong, H.; Bae, Y.; Shin, K.; Kang, S.; Kim, H.; Oh, J.; Bae, H. Targeting of M2-like tumor-associated macrophages with a melittin-based pro-apoptotic peptide. J. Immunother. Cancer 2019, 7, 147. [Google Scholar] [CrossRef]
- Lecker, L.S.M.; Berlato, C.; Maniati, E.; Delaine-Smith, R.; Pearce, O.M.T.; Heath, O.; Nichols, S.J.; Trevisan, C.; Novak, M.; McDermott, J.; et al. TGFBI Production by Macrophages Contributes to an Immunosuppressive Microenvironment in Ovarian Cancer. Cancer Res. 2021, 81, 5706–5719. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Hutti, J.E.; Pfefferle, A.D.; Russell, S.C.; Sircar, M.; Perou, C.M.; Baldwin, A.S. Oncogenic PI3K mutations lead to NF-kappaB-dependent cytokine expression following growth factor deprivation. Cancer Res. 2012, 72, 3260–3269. [Google Scholar] [CrossRef]
- De Monte, L.; Reni, M.; Tassi, E.; Clavenna, D.; Papa, I.; Recalde, H.; Braga, M.; Di Carlo, V.; Doglioni, C.; Protti, M.P. Intratumor T helper type 2 cell infiltrate correlates with cancer-associated fibroblast thymic stromal lymphopoietin production and reduced survival in pancreatic cancer. J. Exp. Med. 2011, 208, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Ancrile, B.; Lim, K.H.; Counter, C.M. Oncogenic Ras-induced secretion of IL6 is required for tumorigenesis. Genes Dev. 2007, 21, 1714–1719. [Google Scholar] [CrossRef]
- Lambert, M.; Jambon, S.; Depauw, S.; David-Cordonnier, M.H. Targeting Transcription Factors for Cancer Treatment. Molecules 2018, 23, 1479. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lyu, Z.; Qin, Y.; Wang, X.; Sun, L.; Zhang, Y.; Gong, L.; Wu, S.; Han, S.; Tang, Y.; et al. FOXO1 promotes tumor progression by increased M2 macrophage infiltration in esophageal squamous cell carcinoma. Theranostics 2020, 10, 11535–11548. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, L.; Jiang, H.; Lin, L.; Li, Y. Inhibition of FOSL2 aggravates the apoptosis of ovarian cancer cells by promoting the formation of inflammasomes. Genes Genom. 2022, 44, 29–38. [Google Scholar] [CrossRef] [PubMed]
- Nayak, D.; Kumar, A.; Chakraborty, S.; Rasool, R.U.; Amin, H.; Katoch, A.; Gopinath, V.; Mahajan, V.; Zilla, M.K.; Rah, B.; et al. Inhibition of Twist1-mediated invasion by Chk2 promotes premature senescence in p53-defective cancer cells. Cell Death Differ. 2017, 24, 1275–1287. [Google Scholar] [CrossRef]
- Guo, Q.; Quan, M.; Dong, J.; Bai, J.; Wang, J.; Han, R.; Wang, W.; Cai, Y.; Lv, Y.Q.; Chen, Q.; et al. The WW domains dictate isoform-specific regulation of YAP1 stability and pancreatic cancer cell malignancy. Theranostics 2020, 10, 4422–4436. [Google Scholar] [CrossRef]
- Shen, J.; Huang, Q.; Jia, W.; Feng, S.; Liu, L.; Li, X.; Tao, D.; Xie, D. YAP1 induces invadopodia formation by transcriptionally activating TIAM1 through enhancer in breast cancer. Oncogene 2022, 41, 3830–3845. [Google Scholar] [CrossRef]
- Sanz-Moreno, V. Tumour invasion: A new twist on Rac-driven mesenchymal migration. Curr. Biol. 2012, 22, R449–R451. [Google Scholar] [CrossRef]
- Yang, K.C.; Kalloger, S.E.; Aird, J.J.; Lee, M.K.C.; Rushton, C.; Mungall, K.L.; Mungall, A.J.; Gao, D.; Chow, C.; Xu, J.; et al. Proteotranscriptomic classification and characterization of pancreatic neuroendocrine neoplasms. Cell Rep. 2021, 37, 109817. [Google Scholar] [CrossRef]
- Bhandari, A.; Guan, Y.; Xia, E.; Huang, Q.; Chen, Y. VASN promotes YAP/TAZ and EMT pathway in thyroid carcinogenesis in vitro. Am. J. Transl. Res. 2019, 11, 3589–3599. [Google Scholar] [PubMed]
- Klepsch, V.; Hermann-Kleiter, N.; Do-Dinh, P.; Jakic, B.; Offermann, A.; Efremova, M.; Sopper, S.; Rieder, D.; Krogsdam, A.; Gamerith, G.; et al. Nuclear receptor NR2F6 inhibition potentiates responses to PD-L1/PD-1 cancer immune checkpoint blockade. Nat. Commun. 2018, 9, 1538. [Google Scholar] [CrossRef] [PubMed]
- Santoso, C.S.; Li, Z.; Lal, S.; Yuan, S.; Gan, K.A.; Agosto, L.M.; Liu, X.; Pro, S.C.; Sewell, J.A.; Henderson, A.; et al. Comprehensive mapping of the human cytokine gene regulatory network. Nucleic Acids Res. 2020, 48, 12055–12073. [Google Scholar] [CrossRef]
- Li, H.; Zhang, W.; Niu, C.; Lin, C.; Wu, X.; Jian, Y.; Li, Y.; Ye, L.; Dai, Y.; Ouyang, Y.; et al. Nuclear orphan receptor NR2F6 confers cisplatin resistance in epithelial ovarian cancer cells by activating the Notch3 signaling pathway. Int. J. Cancer 2019, 145, 1921–1934. [Google Scholar] [CrossRef]
- Ajani, J.A.; Xu, Y.; Huo, L.; Wang, R.; Li, Y.; Wang, Y.; Pizzi, M.P.; Scott, A.; Harada, K.; Ma, L.; et al. YAP1 mediates gastric adenocarcinoma peritoneal metastases that are attenuated by YAP1 inhibition. Gut 2021, 70, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.; Wang, F.; Wang, Y.; Li, T.; Xiu, P.; Zhong, J.; Sun, X.; Li, J. Verteporfin suppresses cell survival, angiogenesis and vasculogenic mimicry of pancreatic ductal adenocarcinoma via disrupting the YAP-TEAD complex. Cancer Sci. 2017, 108, 478–487. [Google Scholar] [CrossRef] [PubMed]
- Sajinovic, T.; Baier, G. New Insights into the Diverse Functions of the NR2F Nuclear Orphan Receptor Family. Front. Biosci. (Landmark. Ed.) 2023, 28, 13. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Campbell, S.; Wilson, A.N.; Shumate, J.; Baillargeon, P.; Scampavia, L.; Kamenecka, T.M.; Spicer, T.P.; Solt, L.A. High throughput screening for compounds to the orphan nuclear receptor NR2F6. SLAS Discov. 2022, 27, 242–248. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kassuhn, W.; Cutillas, P.R.; Kessler, M.; Sehouli, J.; Braicu, E.I.; Blüthgen, N.; Kulbe, H. In Silico Analysis Predicts Nuclear Factors NR2F6 and YAP1 as Mesenchymal Subtype-Specific Therapeutic Targets for Ovarian Cancer Patients. Cancers 2023, 15, 3155. https://doi.org/10.3390/cancers15123155
Kassuhn W, Cutillas PR, Kessler M, Sehouli J, Braicu EI, Blüthgen N, Kulbe H. In Silico Analysis Predicts Nuclear Factors NR2F6 and YAP1 as Mesenchymal Subtype-Specific Therapeutic Targets for Ovarian Cancer Patients. Cancers. 2023; 15(12):3155. https://doi.org/10.3390/cancers15123155
Chicago/Turabian StyleKassuhn, Wanja, Pedro R. Cutillas, Mirjana Kessler, Jalid Sehouli, Elena I. Braicu, Nils Blüthgen, and Hagen Kulbe. 2023. "In Silico Analysis Predicts Nuclear Factors NR2F6 and YAP1 as Mesenchymal Subtype-Specific Therapeutic Targets for Ovarian Cancer Patients" Cancers 15, no. 12: 3155. https://doi.org/10.3390/cancers15123155
APA StyleKassuhn, W., Cutillas, P. R., Kessler, M., Sehouli, J., Braicu, E. I., Blüthgen, N., & Kulbe, H. (2023). In Silico Analysis Predicts Nuclear Factors NR2F6 and YAP1 as Mesenchymal Subtype-Specific Therapeutic Targets for Ovarian Cancer Patients. Cancers, 15(12), 3155. https://doi.org/10.3390/cancers15123155






