Prognostic and Clinicopathological Significance of the Loss of Expression of Retinoblastoma Protein (pRb) in Oral Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Protocol
2.2. Search Strategy
2.3. Eligibility Criteria
2.4. Study Selection Process
2.5. Data Extraction
2.6. Evaluation of Quality and Risk of Bias
2.7. Statistical Analysis
3. Results
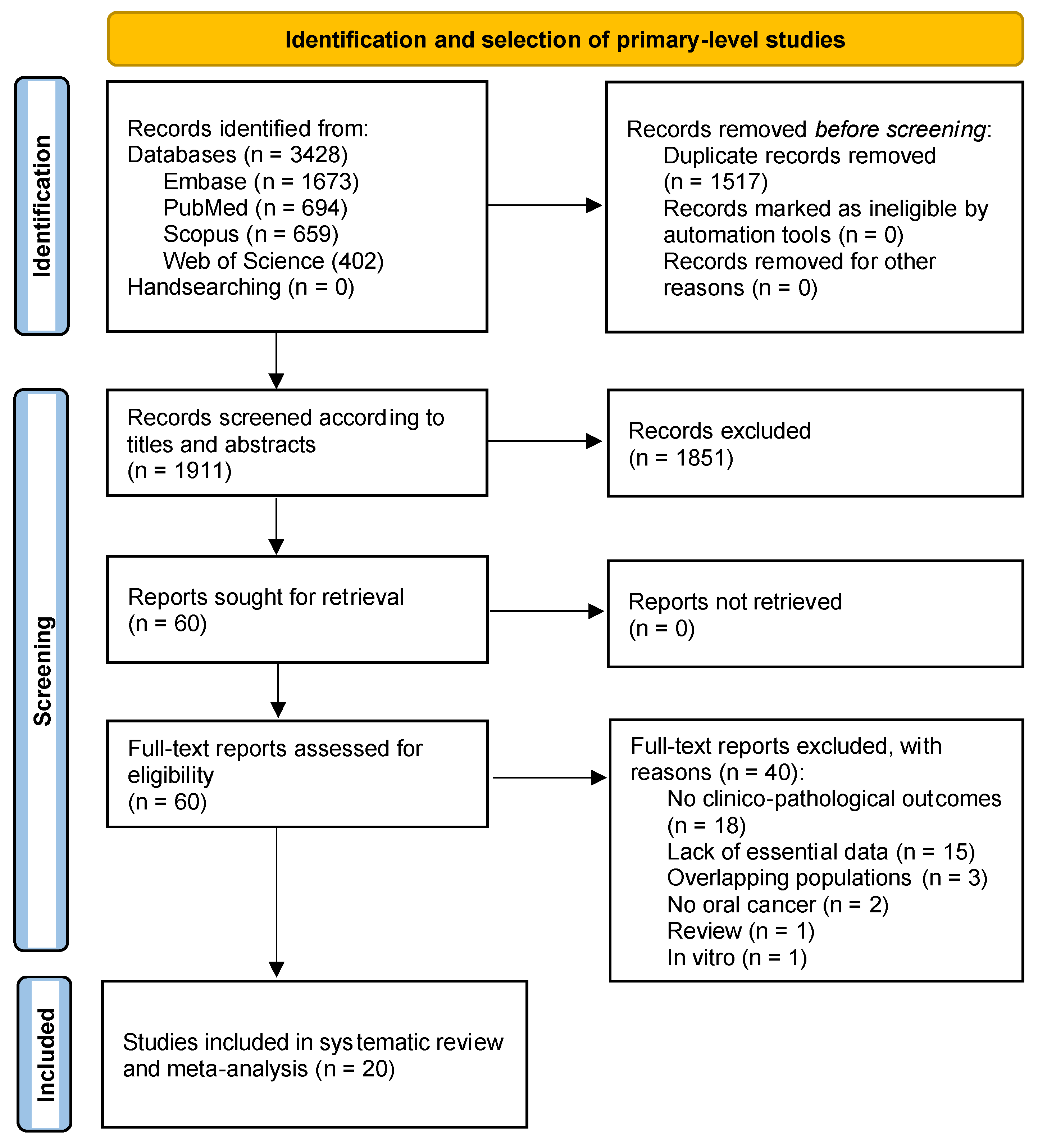
3.1. Results of the Literature Search
3.2. Study Characteristics
3.3. Qualitative Evaluation
3.4. Quantitative Evaluation (Meta-Analysis)
3.4.1. Association between the Loss of pRb Expression and Prognostic Variables
3.4.2. Association between the Loss of pRb Expression and Clinico-Pathological Variables
3.5. Quantitative Evaluation (Secondary Analyses)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. The Hallmarks of Cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- González-Moles, M.Á.; Warnakulasuriya, S.; López-Ansio, M.; Ramos-García, P. Hallmarks of Cancer Applied to Oral and Oropharyngeal Carcinogenesis: A Scoping Review of the Evidence Gaps Found in Published Systematic Reviews. Cancers 2022, 14, 3834. [Google Scholar] [CrossRef] [PubMed]
- Dannenberg, J.-H.; Riele, H.P. The retinoblastoma gene family in cell cycle regulation and suppression of tumorigenesis. Results Probl. Cell Differ. 2006, 42, 183–225. [Google Scholar] [CrossRef]
- Zheng, L.; Lee, W.-H. The retinoblastoma gene: A prototypic and multifunctional tumor suppressor. Exp. Cell Res. 2001, 264, 2–18. [Google Scholar] [CrossRef]
- Dimaras, H.; Khetan, V.; Halliday, W.; Orlic, M.; Prigoda, N.L.; Piovesan, B.; Marrano, P.; Corson, T.W.; Eagle, R.C.; Squire, J.A.; et al. Loss of RB1 induces non-proliferative retinoma: Increasing genomic instability correlates with progression to retinoblastoma. Hum. Mol. Genet. 2008, 17, 1363–1372. [Google Scholar] [CrossRef]
- Corson, T.W.; Gallie, B.L. One hit, two hits, three hits, more? Genomic changes in the development of retinoblastoma. Genes Chromosom. Cancer 2007, 46, 617–634. [Google Scholar] [CrossRef]
- Knudson, A.G. Genetic predisposition to cancer. Cancer Detect. Prev. 1984, 7, 1–8. [Google Scholar]
- Perez-Ordonez, B.; Beauchemin, M.; Jordan, R.C.K. Molecular biology of squamous cell carcinoma of the head and neck. J. Clin. Pathol. 2006, 59, 445–453. [Google Scholar] [CrossRef]
- Doorbar, J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 2006, 110, 525–541. [Google Scholar] [CrossRef]
- Munakata, T.; Liang, Y.; Kim, S.; McGivern, D.R.; Huibregtse, J.; Nomoto, A.; Lemon, S.M. Hepatitis C virus induces E6AP-dependent degradation of the retinoblastoma protein. PLoS Pathog. 2007, 3, 1335–1347. [Google Scholar] [CrossRef] [PubMed]
- González-Moles, M.A.; Scully, C.; Ruiz-Ávila, I.; Plaza-Campillo, J.J. The cancer stem cell hypothesis applied to oral carcinoma. Oral Oncol. 2013, 49, 738–746. [Google Scholar] [CrossRef]
- Slack, R.; El-Bizri, H.; Wong, J.; Belliveau, D.J.; Miller, F.D. A Critical Temporal Requirement for the Retinoblastoma Protein Family During Neuronal Determination. J. Cell Biol. 1998, 140, 1497–1509. [Google Scholar] [CrossRef] [PubMed]
- Kleinerman, R.A.; Tucker, M.A.; Tarone, R.E.; Abramson, D.H.; Seddon, J.M.; Stovall, M.; Li, F.P.; Fraumeni, J.F. Risk of new cancers after radiotherapy in long-term survivors of retinoblastoma: An extended follow-up. J. Clin. Oncol. 2005, 23, 2272–2279. [Google Scholar] [CrossRef] [PubMed]
- MacLeod, K.F.; Hu, Y.; Jacks, T. Loss of Rb activates both p53-dependent and independent cell death pathways in the developing mouse nervous system. EMBO J. 1996, 15, 6178–6188. [Google Scholar] [CrossRef]
- Tsai, K.Y.; Hu, Y.; Macleod, K.F.; Crowley, D.; Yamasaki, L.; Jacks, T. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol. Cell 1998, 2, 293–304. [Google Scholar] [CrossRef]
- Polager, S.; Ofir, M.; Ginsberg, D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 2008, 27, 4860–4864. [Google Scholar] [CrossRef]
- Tracy, K.; Dibling, B.C.; Spike, B.T.; Knabb, J.R.; Schumacker, P.; Macleod, K.F. BNIP3 Is an RB/E2F Target Gene Required for Hypoxia-Induced Autophagy. Mol. Cell. Biol. 2007, 27, 6229–6242. [Google Scholar] [CrossRef]
- Sanseverino, F.; Santopietro, R.; Torricelli, M.; D’Andrilli, G.; Russo, G.; Cevenini, G.; Bovicelli, A.; Leoncini, L.; Scambia, G.; Petraglia, F.; et al. pRb2/p130 and VEGF expression in endometrial carcinoma in relation to angiogenesis and histopathologic tumor grade. Cancer Biol. Ther. 2006, 5, 84–88. [Google Scholar] [CrossRef]
- Riley, R.D.; Ridley, G.; Williams, K.; Altman, D.G.; Hayden, J.; de Vet, H.C. Prognosis research: Toward evidence-based results and a Cochrane methods group. J. Clin. Epidemiology 2007, 60, 863–865. [Google Scholar] [CrossRef]
- Higgins, J.P.; Green, S. Cochrane Handbook for Systematic Reviews of Interventions: Cochrane Book Series. John Wiley & Sons: Hoboken, NJ, USA, 2019. [Google Scholar]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Stroup, D.F.; Berlin, J.A.; Morton, S.C.; Olkin, I.; Williamson, G.D.; Rennie, D.; Moher, D.; Becker, B.J.; Sipe, T.A.; Thacker, S.B. Meta-analysis of observational studies in epidemiology: A proposal for reporting. J. Am. Med. Assoc. 2000, 283, 2008–2012. [Google Scholar] [CrossRef] [PubMed]
- Shamseer, L.; Moher, D.; Clarke, M.; Ghersi, D.; Liberati, A.; Petticrew, M.; Shekelle, P.; Stewart, L.A. PRISMA-P Group Preferred reporting items for systematic review and meta-analysis protocols (PRISMA-P) 2015: Elaboration and explanation. BMJ 2015, 350, g7647. [Google Scholar] [CrossRef] [PubMed]
- Hayden, J.A.; Côté, P.; Bombardier, C. Evaluation of the quality of prognosis studies in systematic reviews. Ann. Intern. Med. 2006, 144, 427–437. [Google Scholar] [CrossRef] [PubMed]
- González-Moles, M.Á.; Ayén, Á.; González-Ruiz, I.; De Porras-Carrique, T.; González-Ruiz, L.; Ruiz-Ávila, I.; Ramos-García, P. Prognostic and Clinicopathological Significance of FADD Upregulation in Head and Neck Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis. Cancers 2020, 12, 2393. [Google Scholar] [CrossRef]
- Ramos-García, P.; González-Moles, M.Á. Prognostic and Clinicopathological Significance of the Aberrant Expression of β-Catenin in Oral Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis. Cancers 2022, 14, 479. [Google Scholar] [CrossRef]
- Ramos-García, P.; González-Moles, M.Á.; Warnakulasuriya, S. Significance of p53 overexpression in the prediction of the malignant transformation risk of oral potentially malignant disorders: A systematic review and meta-analysis. Oral Oncol. 2022, 126, 105734. [Google Scholar] [CrossRef]
- González-Moles, M.Á.; Moya-González, E.; García-Ferrera, A.; Nieto-Casado, P.; Ramos-García, P. Prognostic and Clinicopathological Significance of Telomerase Reverse Transcriptase Upregulation in Oral Cancer: A Systematic Review and Meta-Analysis. Cancers 2022, 14, 3673. [Google Scholar] [CrossRef]
- Tierney, J.F.; Stewart, L.A.; Ghersi, D.; Burdett, S.; Sydes, M.R. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials 2007, 8, 16. [Google Scholar] [CrossRef]
- Parmar, M.K.; Torri, V.; Stewart, L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat. Med. 1998, 17, 2815–2834. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef]
- Thompson, S.G.; Higgins, J.P.T. How should meta-regression analyses be undertaken and interpreted? Stat. Med. 2002, 21, 1559–1573. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G. Controlling the risk of spurious findings from meta-regression. Stat. Med. 2004, 23, 1663–1682. [Google Scholar] [CrossRef]
- Manly, B.F.J. Randomization, Bootstrap and Monte Carlo Methods in Biology; CRC Press: Boca Raton, FL, USA, 2006; Volume 53. [Google Scholar] [CrossRef]
- Egger, M.; Smith, G.D.; Schneider, M.; Minder, C. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997, 315, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Pavelic, Z.P.; Lasmar, M.; Pavelic, L.; Sorensen, C.; Stambrook, P.J.; Zimmermann, N.; Gluckman, J.L. Absence of Retinoblastoma gene product in human primary oral cavity carcinoma. Eur. J. Cancer B. Oral Oncol. 1996, 32, 347–351. [Google Scholar] [CrossRef]
- Pande, P.; Mathur, M.; Shukla, N.K.; Ralhan, R. pRb and p16 protein alterations in human oral tumorigenesis. Oral Oncol. 1998, 34, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Muirhead, D.M.; Hoffman, H.T.; Robinson, R.A. Correlation of clinicopathological features with immunohistochemical expression of cell cycle regulatory proteins p16 and retinoblastoma: Distinct association with keratinisation and differentiation in oral cavity squamous cell carcinoma. J. Clin. Pathol. 2006, 59, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Balan, A.; Balaram, P. The expression of retinoblastoma tumor suppressor protein in oral cancers and precancers: A clinicopathological study. Dent. Res. J. 2015, 12, 307–314. [Google Scholar] [CrossRef]
- Vallonthaiel, A.G.; Singh, M.K.; Dinda, A.K.; Kakkar, A.; Thakar, A.; Das, S.N. Expression of Cell Cycle-associated Proteins p53, pRb, p16, p27, and Correlation With Survival: A Comparative Study on Oral Squamous Cell Carcinoma and Verrucous Carcinoma. Appl. Immunohistochem. Mol. Morphol. AIMM 2016, 24, 193–200. [Google Scholar] [CrossRef]
- Kühn, J.P.; Schmid, W.; Körner, S.; Bochen, F.; Wemmert, S.; Rimbach, H.; Smola, S.; Radosa, J.C.; Wagner, M.; Morris, L.G.T.; et al. HPV Status as Prognostic Biomarker in Head and Neck Cancer—Which Method Fits the Best for Outcome Prediction? Cancers 2021, 13, 4730. [Google Scholar] [CrossRef] [PubMed]
- Chen, B. Expression of human papillomavirus and pRb in head and neck squamous cell carcinoma. Lin Chuang Er Bi Yan Hou Ke Za Zhi 2004, 18, 154–156. [Google Scholar]
- Mena, M.; Taberna, M.; Tous, S.; Marquez, S.; Clavero, O.; Quiros, B.; Lloveras, B.; Alejo, M.; Leon, X.; Quer, M.; et al. Double positivity for HPV-DNA/p16ink4a is the biomarker with strongest diagnostic accuracy and prognostic value for human papillomavirus related oropharyngeal cancer patients. Oral Oncol. 2018, 78, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.G.; Trivedi, T.I.; Tankshali, R.A.; Goswami, J.V.; Jetly, D.H.; Shukla, S.N.; Shah, P.M.; Verma, R.J. Prognostic significance of molecular markers in oral squamous cell carcinoma: A multivariate analysis. Head Neck 2009, 31, 1544–1556. [Google Scholar] [CrossRef] [PubMed]
- Soni, S.; Kaur, J.; Kumar, A.; Chakravarti, N.; Mathur, M.; Bahadur, S.; Shukla, N.K.; Deo, S.V.; Ralhan, R. Alterations of Rb pathway components are frequent events in patients with oral epithelial dysplasia and predict clinical outcome in patients with squamous cell carcinoma. Oncology 2005, 68, 314–325. [Google Scholar] [CrossRef]
- Liu, C.-J.; Chang, K.-W.; Chao, S.-Y.; Kwan, P.-C.; Chang, S.-M.; Yen, R.-Y.; Wang, C.-Y.; Wong, Y.-K. The molecular markers for prognostic evaluation of areca-associated buccal squamous cell carcinoma. J. Oral Pathol. Med. 2004, 33, 327–334. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Gimenez-Conti, I.B.; Cunningham, J.E.; Collet, A.M.; Luna, M.A.; Lanfranchi, H.E.; Spitz, M.R.; Conti, C.J. Alterations of p53, cyclin D1, rb, and H-ras in human oral carcinomas related to tobacco use. Cancer 1998, 83, 204–212. [Google Scholar] [CrossRef]
- Tanaka, N.; Odajima, T.; Nakano, T.; Kimijima, Y.; Yamada, S.; Ogi, K.; Kohama, G. Immunohistochemical investigation of new suppressor oncogene p130 in oral squamous cell carcinoma. Oral Oncol. 1999, 35, 321–325. [Google Scholar] [CrossRef]
- Partridge, M.; Emilion, G.; Falworth, M.; A’Hern, R.; Phillips, E.; Pateromichelakis, S.; Langdon, J. Patient-specific mutation databases for oral cancer. Int. J. Cancer 1999, 84, 284–292. [Google Scholar] [CrossRef]
- Sano, T.; Hikino, T.; Xue, Q.; Saito, T.; Kashiwabara, K.; Oyama, T.; Nakajima, T. Immunohistochemical inactivation of p14ARF concomitant with MDM2 overexpression inversely correlates with p53 overexpression in oral squamous cell carcinoma. Pathol. Int. 2000, 50, 709–716. [Google Scholar] [CrossRef]
- Nakahara, Y.; Shintani, S.; Mihara, M.; Kiyota, A.; Ueyama, Y.; Matsumura, T. Alterations of Rb, p16INK4A and cyclin D1 in the tumorigenesis of oral squamous cell carcinomas. Cancer Lett. 2000, 160, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Takes, R.P.; De Jong, R.J.B.; Alles, M.J.R.C.; Meeuwis, C.A.; Marres, H.A.M.; Knegt, P.P.M.; De La Riviere, G.B.; De Wilde, P.C.M.; Mooi, W.J.; Hermans, J.; et al. Markers for nodal metastasis in head and neck squamous cell cancer. Arch. Otolaryngol. Head. Neck Surg. 2002, 128, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Van Heerden, W.F.P.; Dreyer, L.; Swart, T.J.P.; Van Heerden, M.B.; Boy, S.C. The suitability of paraffin-embedded material to predict metastatic potential of oral squamous cell carcinoma. Anticancer Res. 2002, 22, 4147–4150. [Google Scholar] [PubMed]
- Li, W.; Thompson, C.H.; O’Brien, C.J.; McNeil, E.B.; Scolyer, R.A.; Cossart, Y.E.; Veness, M.J.; Walker, D.M.; Morgan, G.J.; Rose, B.R. Human papillomavirus positivity predicts favourable outcome for squamous carcinoma of the tonsil. Int. J. Cancer 2003, 106, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Jayasurya, R.; Sathyan, K.M.; Lakshminarayanan, K.; Abraham, T.; Nalinakumari, K.R.; Abraham, E.K.; Nair, M.K.; Kannan, S. Phenotypic alterations in Rb pathway have more prognostic influence than p53 pathway proteins in oral carcinoma. Mod. Pathol. 2005, 18, 1056–1066. [Google Scholar] [CrossRef]
- Lim, K.P.; Hamid, S.; Lau, S.-H.; Teo, S.-H.; Cheong, S.C. HPV infection and the alterations of the pRB pathway in oral carcinogenesis. Oncol. Rep. 2007, 17, 1321–1326. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef]
- Mulder, F.J.; Klufah, F.; Janssen, F.M.E.; Farshadpour, F.; Willems, S.M.; de Bree, R.; Hausen, A.Z.; Hout, M.F.C.M.V.D.; Kremer, B.; Speel, E.-J.M. Presence of Human Papillomavirus and Epstein–Barr Virus, but Absence of Merkel Cell Polyomavirus, in Head and Neck Cancer of Non-Smokers and Non-Drinkers. Front. Oncol. 2021, 10, 560434. [Google Scholar] [CrossRef]
- Soares, R.C.; Oliveira, M.C.; De Souza, L.B.; Costa, A.D.L.L.; Pinto, L.P. Detection of HPV DNA and immunohistochemical expression of cell cycle proteins in oral carcinoma in a population of brazilian patients. J. Appl. Oral Sci. 2008, 16, 340–344. [Google Scholar] [CrossRef]
- Sannigrahi, M.; Singh, V.; Sharma, R.; Panda, N.; Radotra, B.; Khullar, M. Detection of active human papilloma virus-16 in head and neck cancers of Asian North Indian patients. Oral Dis. 2016, 22, 62–68. [Google Scholar] [CrossRef]
- Nemes, J.A.; Deli, L.; Nemes, Z.; Márton, I.J. Expression of p16INK4A, p53, and Rb proteins are independent from the presence of human papillomavirus genes in oral squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endodontology 2006, 102, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Reyes, M.; Rojas-Alcayaga, G.; Pennacchiotti, G.; Carrillo, D.; Muñoz, J.P.; Peña, N.; Montes, R.; Lobos, N.; Aguayo, F. Human papillomavirus infection in oral squamous cell carcinomas from Chilean patients. Exp. Mol. Pathol. 2015, 99, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Emmett, S.; Jenkins, G.; Boros, S.; Whiteman, D.C.; Panizza, B.; Antonsson, A. Low prevalence of human papillomavirus in oral cavity squamous cell carcinoma in Queensland, Australia. ANZ J. Surg. 2017, 87, 714–719. [Google Scholar] [CrossRef]
- Castellsagué, X.; Alemany, L.; Quer, M.; Halec, G.; Quirós, B.; Tous, S.; Clavero, O.; Alòs, L.; Biegner, T.; Szafarowski, T.; et al. HPV Involvement in Head and Neck Cancers: Comprehensive Assessment of Biomarkers in 3680 Patients. J. Natl. Cancer Inst. 2016, 108, djv403. [Google Scholar] [CrossRef] [PubMed]


| Total | 20 Studies |
|---|---|
| Year of publication | 1996–2021 |
| Total patients (range) | 2451 (9–784) |
| Study design | |
| Retrospective cohort | 20 studies |
| Experimental methods for pRb expression determination | |
| Immunohistochemistry | 20 studies |
| Anti-pRb antibody | |
| Clone IF8 | 4 studies |
| Clone G3-245 | 2 studies |
| Other | 5 studies |
| Not reported | 9 studies |
| Anti-pRb antibody dilution | |
| 1:5–1:50 | 6 studies |
| 1:100 | 5 studies |
| 1:150–1:700 | 4 studies |
| Not reported | 5 studies |
| Anti-pRb antibody incubation time | |
| Overnight | 8 studies |
| 1 h or less | 2 studies |
| Not reported | 10 studies |
| Anti-pRb antibody incubation temperature | |
| 4 °C | 7 studies |
| Room temperature | 2 studies |
| Not reported | 11 studies |
| Geographical region | |
| Asian countries | 11 studies |
| Non-Asian countries | 9 studies |
| Pooled Data | Heterogeneity | |||||||
|---|---|---|---|---|---|---|---|---|
| Meta-Analyses | No. of Studies | No. of Patients | Stat. Model | Wt | ES (95% CI) | p-Value | Phet | I2 (%) |
| SURVIVAL PARAMETERS | ||||||||
| Overall survival | ||||||||
| Loss of pRb expression (all) a | 10 | 1636 | REM | D–L | HR = 0.79 (0.64–0.98) | 0.03 | 0.35 | 10.5 |
| Subgroup analysis by geographical area b | 0.98 c | |||||||
| Asian | 5 | 599 | REM | D–L | HR = 0.77 (0.55–1.07) | 0.12 | 0.47 | 0.0 |
| Non-Asian | 5 | 1037 | REM | D–L | HR = 0.77 (0.49–1.19) | 0.23 | 0.17 | 37.6 |
| Subgroup analysis by anti-pRb antibody b | 0.68 c | |||||||
| Clone IF8 | 1 | 340 | - | - | HR = 0.66 (0.42–1.02) | 0.07 | - | - |
| Clone G3-245 | 1 | 55 | - | - | HR = 1.05 (0.45–2.44) | 0.91 | - | - |
| Other | 2 | 122 | REM | D–L | HR = 1.36 (0.28–6.52) | 0.70 | 0.09 | 65.8 |
| Not reported | 6 | 1119 | REM | D–L | HR = 0.78 (0.58–1.05) | 0.10 | 0.33 | 12.7 |
| Subgroup analysis by anti-pRb antibody dilution b | 0.76 c | |||||||
| 1:0–50 | 3 | 224 | REM | D–L | HR = 1.07 (0.51–2.25) | 0.85 | 0.27 | 24.4 |
| 1:100 | 1 | 60 | - | - | HR = 0.96 (0.31–2.97) | 0.94 | - | - |
| 1:150–700 | 3 | 142 | REM | D–L | HR = 0.78 (0.38–1.59) | 0.49 | 0.27 | 24.6 |
| Not reported | 3 | 1210 | REM | D–L | HR = 0.70 (0.49–1.01) | 0.06 | 0.14 | 49.6 |
| Subgroup analysis by anti-pRb antibody incubation time b | 0.97 c | |||||||
| 1 h or less | 2 | 123 | REM | D–L | HR = 0.71 (0.36–1.38) | 0.31 | 0.94 | 0.0 |
| Overnight | 3 | 535 | REM | D–L | HR = 0.75 (0.52–1.07) | 0.11 | 0.65 | 0.0 |
| Not reported | 5 | 978 | REM | D–L | HR = 0.79 (0.43–1.47) | 0.47 | 0.07 | 54.2 |
| Subgroup analysis by anti-pRb antibody incubation temperature b | 0.97 c | |||||||
| 4 °C | 3 | 535 | REM | D–L | HR = 0.75 (0.52–1.07) | 0.11 | 0.65 | 0.0 |
| Room temperature | 2 | 123 | REM | D–L | HR = 0.71 (0.36–1.38) | 0.31 | 0.94 | 0.0 |
| Not reported | 5 | 978 | REM | D–L | HR = 0.79 (0.43–1.47) | 0.47 | 0.07 | 54.2 |
| Subgroup analysis by cut-off point b | 0.74 c | |||||||
| <10 | 2 | 123 | REM | D–L | HR = 0.71 (0.36–1.38) | 0.31 | 0.94 | 0.0 |
| 10 | 2 | 190 | REM | D–L | HR = 0.98 (0.57–1.68) | 0.94 | 0.83 | 0.0 |
| >10 | 5 | 983 | REM | D–L | HR = 0.77 (0.39–1.50) | 0.44 | 0.08 | 53.0 |
| Intensity-based | 1 | 340 | - | - | HR = 0.66 (0.42–1.03) | 0.07 | - | - |
| Subgroup analysis by overall risk of bias in primary-level studies b | 0.14 c | |||||||
| Low RoB | 1 | 340 | - | - | HR = 0.66 (0.42–1.03) | 0.07 | - | - |
| Moderate RoB | 5 | 1078 | REM | D–L | HR = 0.88 (0.73–1.06) | 0.19 | 0.55 | 0.0 |
| High RoB | 4 | 218 | REM | D–L | HR = 0.53 (0.31–0.92) | 0.03 | 0.38 | 1.8 |
| Univariable meta-regressions by study design and patients’ characteristics d | ||||||||
| Follow up (months, mean) | 10 | 1636 | random-effects meta-regression | Coef = −0.005 (−0.052 to 0.042) | 0.69 ± 0.005 e | hetexplained = −1320% f | ||
| Sex (proportion of males, %) | 7 | 1417 | random-effects meta-regression | Coef = 0.005 (−0.021 to 0.030) | 0.94 ± 0.002 e | hetexplained = 0.00% f | ||
| Age (years, mean) | 8 | 1472 | random-effects meta-regression | Coef = −0.010 (−0.058 to 0.039) | 0.53 ± 0.005 e | hetexplained= 0.00% f | ||
| Clinical stage (proportion of stage-III/IV patients, %) | 4 | 233 | - | - | - | - | ||
| Tobacco consumption (proportion of smokers, %) | 4 | 1303 | - | - | - | - | ||
| Areca nut/Betel quid consumption (proportion of chewers, %) | 1 | 55 | - | - | - | - | ||
| Alcohol consumption (% of patients with positive habit) | 2 | 828 | - | - | - | - | ||
| Disease-free survival | ||||||||
| Loss of pRb expression (all) a | 5 | 799 | REM | D–L | HR = 1.09 (0.59–2.02) | 0.79 | 0.02 | 67.5 |
| CLINICO-PATHOLOGICAL CHARACTERISTICS | ||||||||
| T status | ||||||||
| Loss of pRb expression (all) a | 8 | 756 | REM | D–L | OR = 1.89 (0.97–3.69) | 0.06 | 0.003 | 67.9 |
| N status | ||||||||
| Loss of pRb expression (all) a | 11 | 786 | REM | D–L | OR = 1.25 (0.76–2.10) | 0.40 | 0.06 | 43.4 |
| Clinical Stage | ||||||||
| Loss of pRb expression (all) a | 5 | 453 | REM | D–L | OR = 1.25 (0.65–2.39) | 0.50 | 0.18 | 36.6 |
| Histological grade | ||||||||
| Loss of pRb expression (all) a | 11 | 812 | REM | D–L | OR = 0.95 (0.67–1.34) | 0.77 | 0.42 | 2.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López-Ansio, M.; Ramos-García, P.; González-Moles, M.Á. Prognostic and Clinicopathological Significance of the Loss of Expression of Retinoblastoma Protein (pRb) in Oral Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis. Cancers 2023, 15, 3132. https://doi.org/10.3390/cancers15123132
López-Ansio M, Ramos-García P, González-Moles MÁ. Prognostic and Clinicopathological Significance of the Loss of Expression of Retinoblastoma Protein (pRb) in Oral Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis. Cancers. 2023; 15(12):3132. https://doi.org/10.3390/cancers15123132
Chicago/Turabian StyleLópez-Ansio, María, Pablo Ramos-García, and Miguel Ángel González-Moles. 2023. "Prognostic and Clinicopathological Significance of the Loss of Expression of Retinoblastoma Protein (pRb) in Oral Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis" Cancers 15, no. 12: 3132. https://doi.org/10.3390/cancers15123132
APA StyleLópez-Ansio, M., Ramos-García, P., & González-Moles, M. Á. (2023). Prognostic and Clinicopathological Significance of the Loss of Expression of Retinoblastoma Protein (pRb) in Oral Squamous Cell Carcinoma: A Systematic Review and Meta-Analysis. Cancers, 15(12), 3132. https://doi.org/10.3390/cancers15123132









