Investigating the Role of Heparanase in Breast Cancer Development Utilising the MMTV-PyMT Murine Model of Mammary Carcinoma
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of MMTV-PyMTxHPSE−/− Mice
2.2. Genotyping Strategy
2.3. HPSE Enzymatic Activity Assay
2.4. Measurement of HPSE Activity of Mouse Splenic Lysate
2.5. Purification of Human HPSE
2.6. Validation of Purified HPSE by Western Blot
2.7. Mammary Tumour Measurements
2.8. Dissection of Mammary Glands and Mammary Tumours
2.9. Whole Mounting of Mouse Mammary Glands
2.10. H&E Staining
2.11. Immunohistochemistry (IHC)
2.12. Pathological Grading of MMTV-PyMT and MMTV-PyMTxHPSE−/− Mammary Tumour Development
2.13. Microvessel Density Quantification following Anti-CD31 IHC
2.14. H-Scoring of Mammary Tumours
2.15. Extraction of Total Lung RNA and Relative Tumour Burden (RTB) Determination by qPCR
3. Results
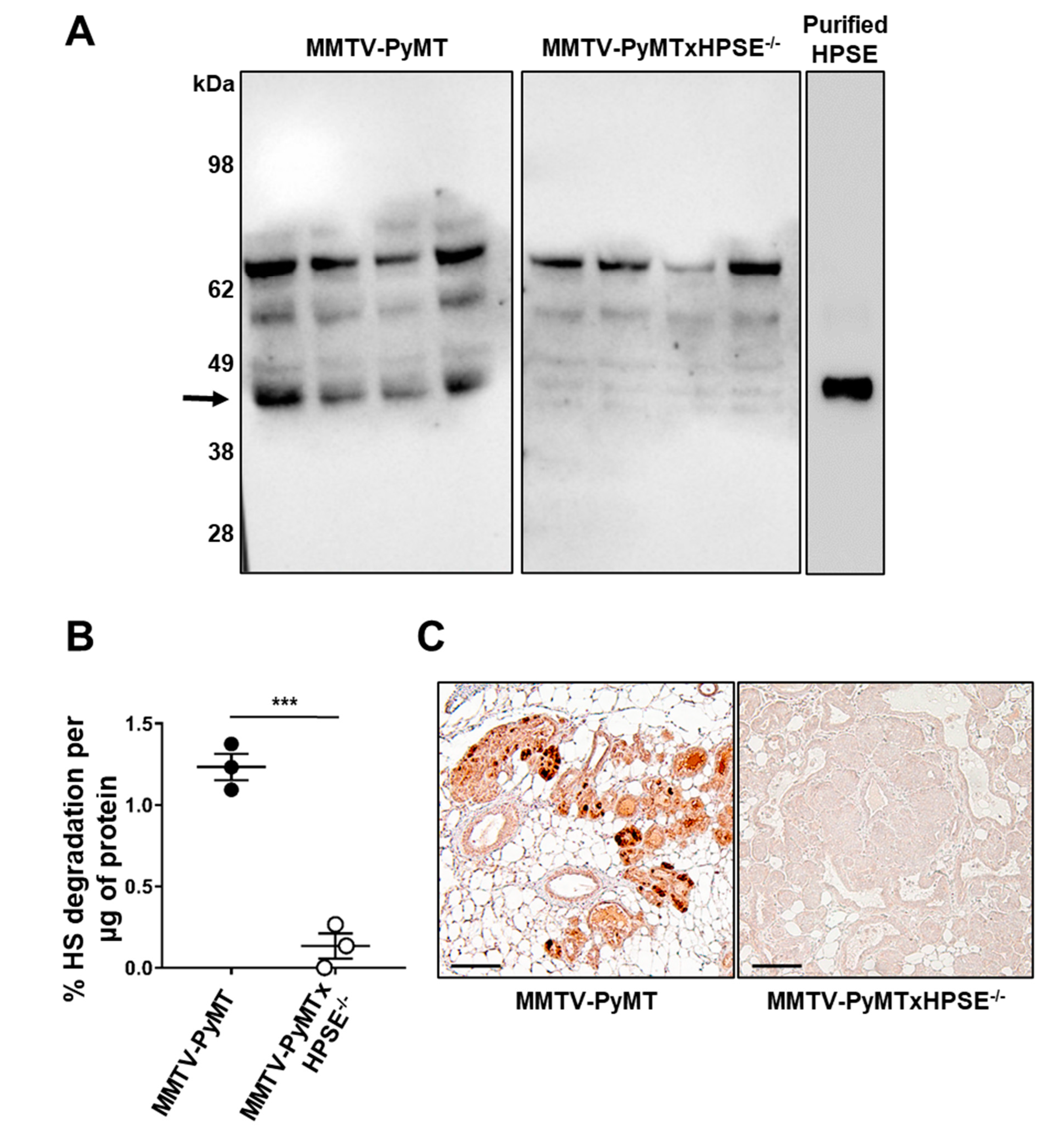
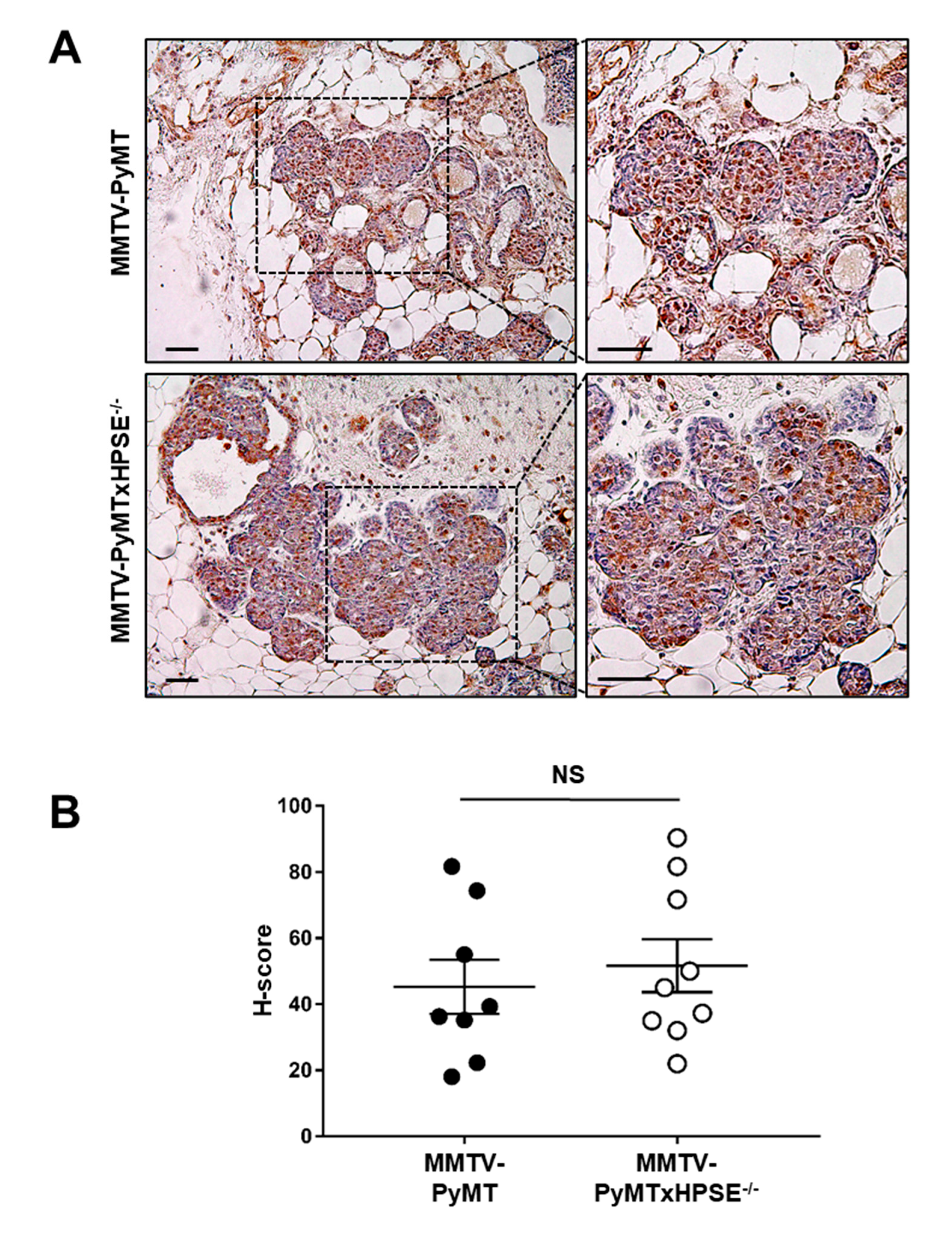
3.1. Characterisation of HPSE Expression and Activity Status of MMTV-PyMT and MMTV-PyMTxHPSE−/− Mice
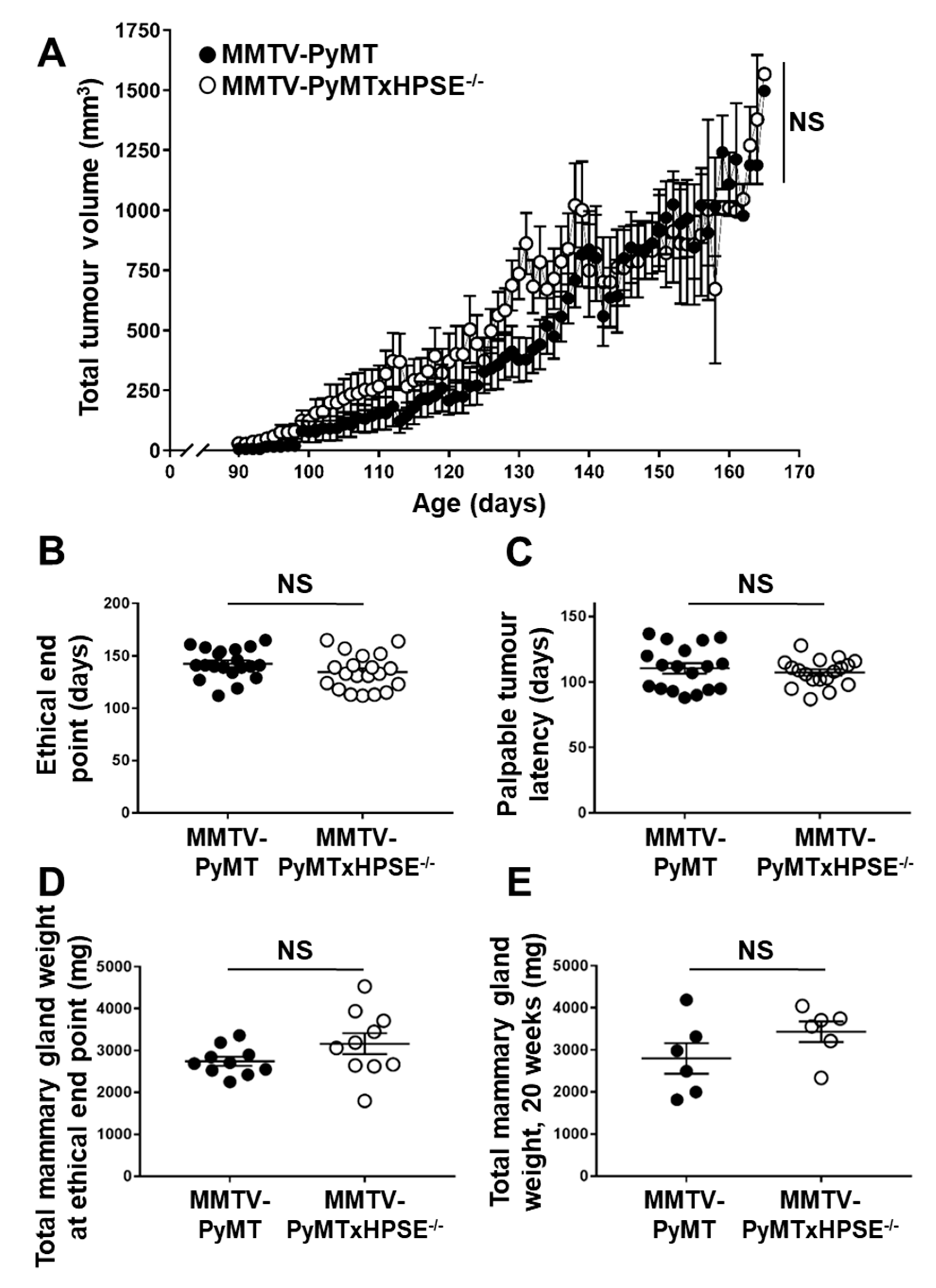
3.2. Evaluation of Spontaneous Mammary Tumour Growth between MMTV-PyMT and MMTV-PyMTxHPSE−/− Mice
3.3. HPSE Expression over Time in MMTV-PyMT Mammary Glands
3.4. The Effect of HPSE on Early- and Late-Stage Tumour Angiogenesis in MMTV-PyMT Mice
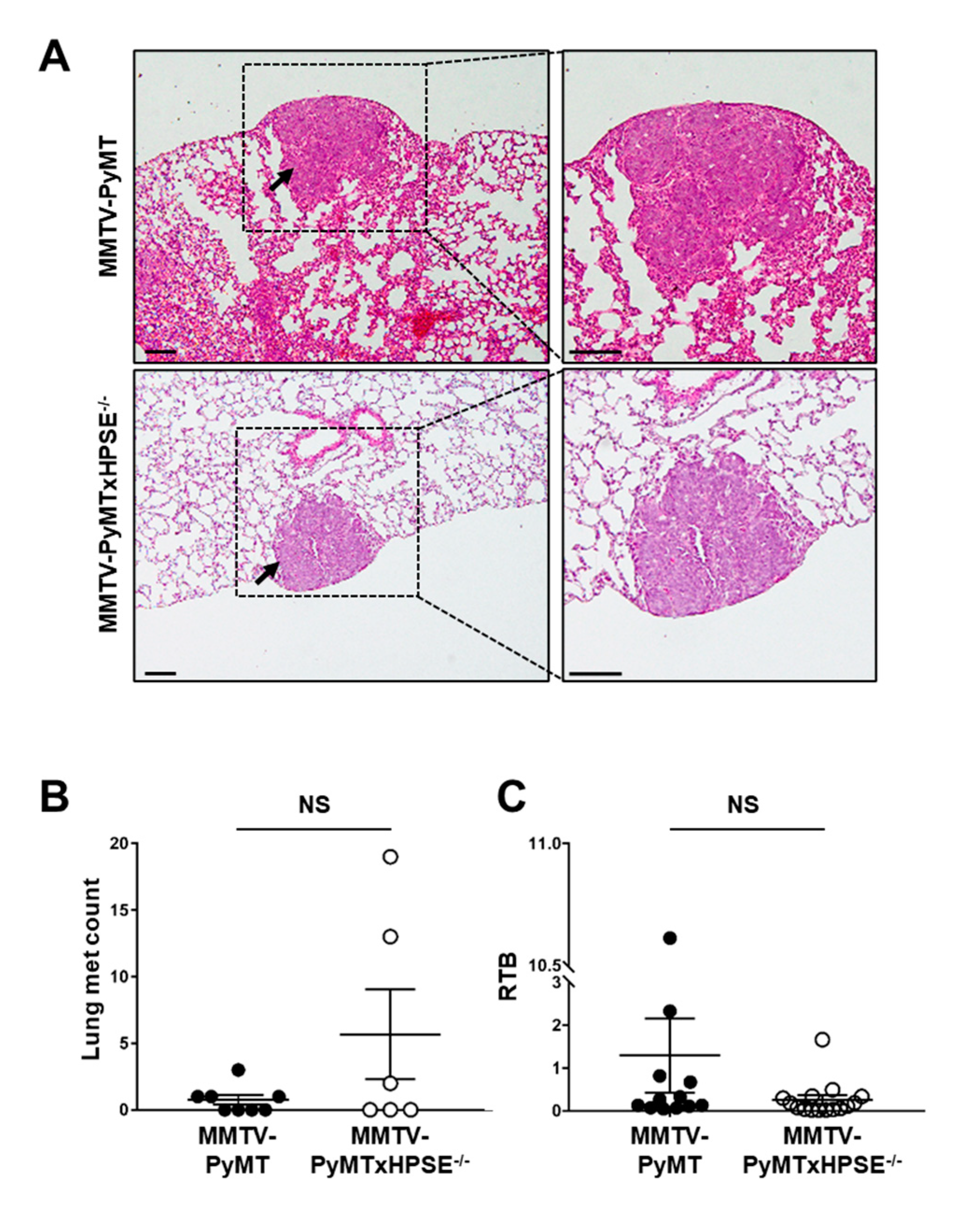
3.5. The Role of Host HPSE in Influencing Lung Metastasis of MMTV-PyMT Mammary Tumours
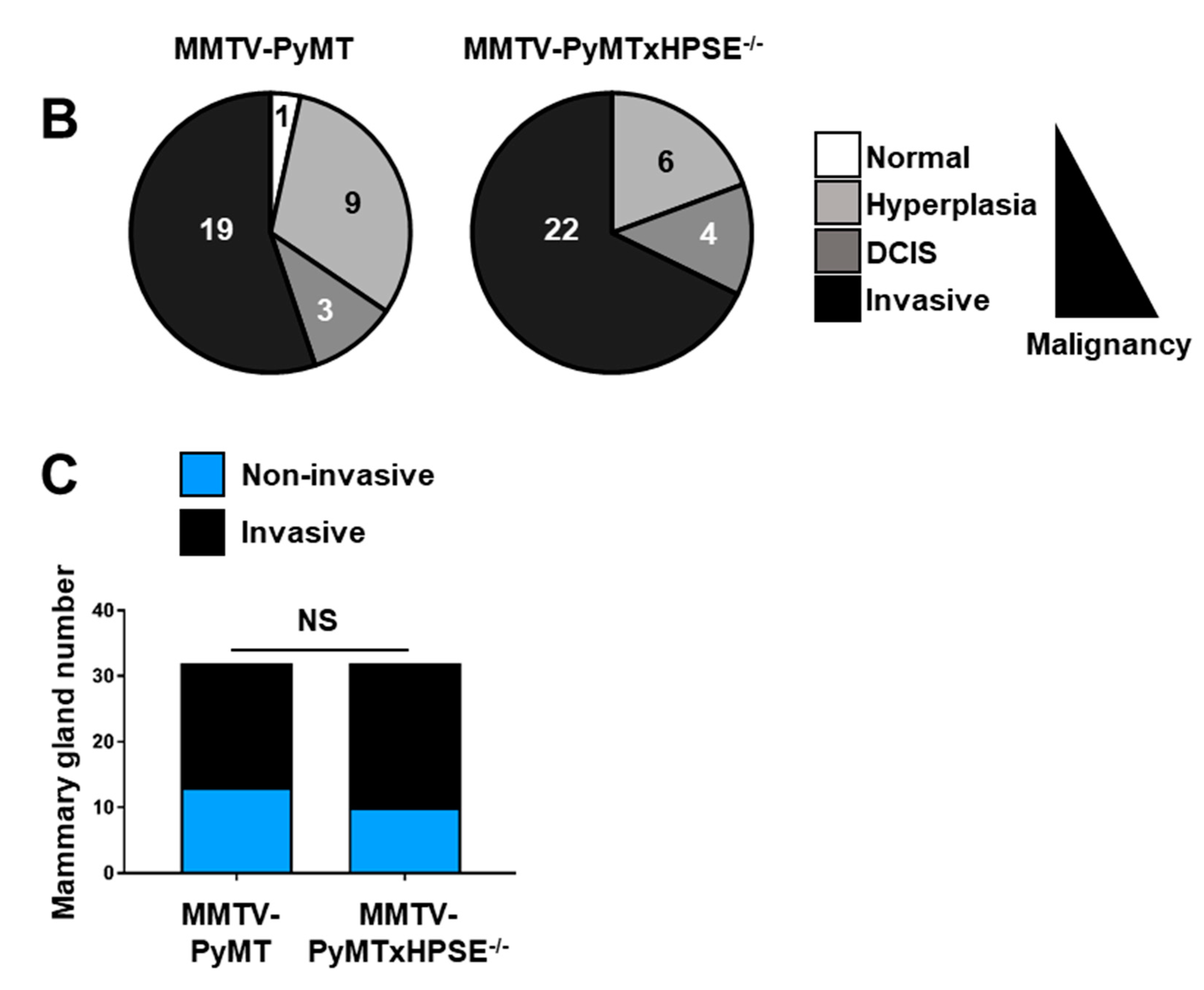
3.6. Evaluation of the Role of HPSE in the Early Stages of Mammary Tumour Development in the MMTV-PyMT Mouse Model
3.7. Investigating the Presence of a Compensatory Mechanism of MMP-2 Expression in MMTV-PyMTxHPSE−/− Mouse Mammary Tumour Lesions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hulett, M.D.; Freeman, C.; Hamdorf, B.J.; Baker, R.T.; Harris, M.J.; Parish, C.R. Cloning of mammalian heparanase, an important enzyme in tumor invasion and metastasis. Nat. Med. 1999, 5, 803–809. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Friedmann, Y.; Elkin, M.; Aingorn, H.; Atzmon, R.; Ishai-Michaeli, R.; Bitan, M.; Pappo, O.; Peretz, T.; Michal, I.; et al. Mammalian heparanase: Gene cloning, expression and function in tumor progression and metastasis. Nat. Med. 1999, 5, 793–802. [Google Scholar] [CrossRef] [PubMed]
- Sarrazin, S.; Lamanna, W.C.; Esko, J.D. Heparan sulfate proteoglycans. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar] [CrossRef] [PubMed]
- De Pasquale, V.; Pavone, L.M. Heparan Sulfate Proteoglycan Signaling in Tumor Microenvironment. Int. J. Mol. Sci. 2020, 21, 6588. [Google Scholar] [CrossRef]
- Knelson, E.H.; Nee, J.C.; Blobe, G.C. Heparan sulfate signaling in cancer. Trends Biochem. Sci. 2014, 39, 277–288. [Google Scholar] [CrossRef]
- Putz, E.M.; Mayfosh, A.J.; Kos, K.; Barkauskas, D.S.; Nakamura, K.; Town, L.; Goodall, K.J.; Yee, D.Y.; Poon, I.K.; Baschuk, N.; et al. NK cell heparanase controls tumor invasion and immune surveillance. J. Clin. Investig. 2017, 127, 2777–2788. [Google Scholar] [CrossRef]
- Poon, I.K.; Goodall, K.J.; Phipps, S.; Chow, J.D.; Pagler, E.B.; Andrews, D.M.; Conlan, C.L.; Ryan, G.F.; White, J.A.; Wong, M.K.; et al. Mice deficient in heparanase exhibit impaired dendritic cell migration and reduced airway inflammation. Eur. J. Immunol. 2014, 44, 1016–1030. [Google Scholar] [CrossRef]
- Wood, R.J.; Hulett, M.D. Cell surface-expressed cation-independent mannose 6-phosphate receptor (CD222) binds enzymatically active heparanase independently of mannose 6-phosphate to promote extracellular matrix degradation. J. Biol. Chem. 2008, 283, 4165–4176. [Google Scholar] [CrossRef]
- Gutter-Kapon, L.; Alishekevitz, D.; Shaked, Y.; Li, J.P.; Aronheim, A.; Ilan, N.; Vlodavsky, I. Heparanase is required for activation and function of macrophages. Proc. Natl. Acad. Sci. USA 2016, 113, E7808–E7817. [Google Scholar] [CrossRef]
- Weissmann, M.; Arvatz, G.; Horowitz, N.; Feld, S.; Naroditsky, I.; Zhang, Y.; Ng, M.; Hammond, E.; Nevo, E.; Vlodavsky, I.; et al. Heparanase-neutralizing antibodies attenuate lymphoma tumor growth and metastasis. Proc. Natl. Acad. Sci. USA 2016, 113, 704–709. [Google Scholar] [CrossRef]
- Vlodavsky, I.; Friedmann, Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J. Clin. Investig. 2001, 108, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Mayfosh, A.; Goodall, K.; Nguyen, T.; Baschuk, N.; Hulett, M. Heparanase is a regulator of natural killer cell activation and cytotoxicity. J. Leukoc. Biol. 2021, 111, 1211–1224. [Google Scholar] [CrossRef] [PubMed]
- Mayfosh, A.J.; Baschuk, N.; Hulett, M.D. Leukocyte Heparanase: A Double-Edged Sword in Tumor Progression. Front. Oncol. 2019, 9, 331. [Google Scholar] [CrossRef] [PubMed]
- Mayfosh, A.J.; Nguyen, T.K.; Hulett, M.D. The Heparanase Regulatory Network in Health and Disease. Int. J. Mol. Sci. 2021, 22, 11096. [Google Scholar] [CrossRef] [PubMed]
- Vlodavsky, I.; Eldor, A.; Haimovitz-Friedman, A.; Matzner, Y.; Ishai-Michaeli, R.; Lider, O.; Naparstek, Y.; Cohen, I.R.; Fuks, Z. Expression of heparanase by platelets and circulating cells of the immune system: Possible involvement in diapedesis and extravasation. Invasion Metastasis 1992, 12, 112–127. [Google Scholar]
- Goshen, R.; Hochberg, A.A.; Korner, G.; Levy, E.; Ishai-Michaeli, R.; Elkin, M.; de Groot, N.; Vlodavsky, I. Purification and characterization of placental heparanase and its expression by cultured cytotrophoblasts. Mol. Hum. Reprod. 1996, 2, 679–684. [Google Scholar] [CrossRef][Green Version]
- Sanderson, R.D.; Elkin, M.; Rapraeger, A.C.; Ilan, N.; Vlodavsky, I. Heparanase regulation of cancer, autophagy and inflammation: New mechanisms and targets for therapy. FEBS J. 2017, 284, 42–55. [Google Scholar] [CrossRef]
- Jayatilleke, K.M.; Hulett, M.D. Heparanase and the hallmarks of cancer. J. Transl. Med. 2020, 18, 453. [Google Scholar] [CrossRef]
- Coombe, D.R.; Gandhi, N.S. Heparanase: A Challenging Cancer Drug Target. Front. Oncol. 2019, 9, 1316. [Google Scholar] [CrossRef]
- Vlodavsky, I.; Ilan, N.; Sanderson, R.D. Forty Years of Basic and Translational Heparanase Research. Adv. Exp. Med. Biol. 2020, 1221, 3–59. [Google Scholar] [CrossRef]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Cohen, I.; Pappo, O.; Elkin, M.; San, T.; Bar-Shavit, R.; Hazan, R.; Peretz, T.; Vlodavsky, I.; Abramovitch, R. Heparanase promotes growth, angiogenesis and survival of primary breast tumors. Int. J. Cancer 2006, 118, 1609–1617. [Google Scholar] [CrossRef] [PubMed]
- Jiao, F.; Bai, S.Y.; Ma, Y.; Yan, Z.H.; Yue, Z.; Yu, Y.; Wang, X.; Wang, J. DNA methylation of heparanase promoter influences its expression and associated with the progression of human breast cancer. PLoS ONE 2014, 9, e92190. [Google Scholar] [CrossRef] [PubMed]
- Elkin, M.; Cohen, I.; Zcharia, E.; Orgel, A.; Guatta-Rangini, Z.; Peretz, T.; Vlodavsky, I.; Kleinman, H.K. Regulation of heparanase gene expression by estrogen in breast cancer. Cancer Res. 2003, 63, 8821–8826. [Google Scholar] [PubMed]
- Wei, R.R.; Sun, D.N.; Yang, H.; Yan, J.; Zhang, X.; Zheng, X.L.; Fu, X.H.; Geng, M.Y.; Huang, X.; Ding, J. CTC clusters induced by heparanase enhance breast cancer metastasis. Acta Pharmacol. Sin. 2018, 39, 1326–1337. [Google Scholar] [CrossRef]
- Boyango, I.; Barash, U.; Naroditsky, I.; Li, J.P.; Hammond, E.; Ilan, N.; Vlodavsky, I. Heparanase cooperates with Ras to drive breast and skin tumorigenesis. Cancer Res. 2014, 74, 4504–4514. [Google Scholar] [CrossRef]
- Boyango, I.; Barash, U.; Fux, L.; Naroditsky, I.; Ilan, N.; Vlodavsky, I. Targeting heparanase to the mammary epithelium enhances mammary gland development and promotes tumor growth and metastasis. Matrix Biol. 2018, 65, 91–103. [Google Scholar] [CrossRef]
- Zhang, L.; Ngo, J.A.; Wetzel, M.D.; Marchetti, D. Heparanase mediates a novel mechanism in lapatinib-resistant brain metastatic breast cancer. Neoplasia 2015, 17, 101–113. [Google Scholar] [CrossRef]
- Teoh, M.L.T.; Fitzgerald, M.P.; Oberley, L.W.; Domann, F.E. Overexpression of Extracellular Superoxide Dismutase Attenuates Heparanase Expression and Inhibits Breast Carcinoma Cell Growth and Invasion. Cancer Res. 2009, 69, 6355–6363. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, X.; Nan, N.; Cao, K.X.; Ma, C.; Yang, G.W.; Yu, M.W.; Yang, L.; Li, J.P.; Wang, X.M.; et al. Elemene inhibits the migration and invasion of 4T1 murine breast cancer cells via heparanase. Mol. Med. Rep. 2017, 16, 794–800. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Chen, Y.; Zhao, H.J.; Xie, C.Y.; Ding, J.; Hou, Y.T. Silencing of heparanase by siRNA inhibits tumor metastasis and angiogenesis of human breast cancer in vitro and in vivo. Cancer Biol. Ther. 2007, 6, 587–595. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hammond, E.; Brandt, R.; Dredge, K. PG545, a heparan sulfate mimetic, reduces heparanase expression in vivo, blocks spontaneous metastases and enhances overall survival in the 4T1 breast carcinoma model. PLoS ONE 2012, 7, e52175. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, G.; Nian, J.; Yu, M.; Chen, S.; Zhang, Y.; Yang, G.; Yang, L.; Cheng, P.; Yan, C.; et al. Elevated heparanase expression is associated with poor prognosis in breast cancer: A study based on systematic review and TCGA data. Oncotarget 2017, 8, 43521–43535. [Google Scholar] [CrossRef] [PubMed]
- Maxhimer, J.B.; Quiros, R.M.; Stewart, R.; Dowlatshahi, K.; Gattuso, P.; Fan, M.; Prinz, R.A.; Xu, X. Heparanase-1 expression is associated with the metastatic potential of breast cancer. Surgery 2002, 132, 326–333. [Google Scholar] [CrossRef]
- Maxhimer, J.B.; Pesce, C.E.; Stewart, R.A.; Gattuso, P.; Prinz, R.A.; Xu, X. Ductal carcinoma in situ of the breast and heparanase-1 expression: A molecular explanation for more aggressive subtypes. J. Am. Coll. Surg. 2005, 200, 328–335. [Google Scholar] [CrossRef]
- Gawthorpe, S.; Brown, J.E.; Arif, M.; Nightingale, P.; Nevill, A.; Carmichael, A.R. Heparanase and COX-2 expression as predictors of lymph node metastasis in large, high-grade breast tumors. Anticancer Res. 2014, 34, 2797–2800. [Google Scholar] [PubMed]
- Zhang, L.; Sullivan, P.S.; Goodman, J.C.; Gunaratne, P.H.; Marchetti, D. MicroRNA-1258 Suppresses Breast Cancer Brain Metastasis by Targeting Heparanase. Cancer Res. 2011, 71, 645–654. [Google Scholar] [CrossRef]
- Cohen, I.; Maly, B.; Simon, I.; Meirovitz, A.; Pikarsky, E.; Zcharia, E.; Peretz, T.; Vlodavsky, I.; Elkin, M. Tamoxifen induces heparanase expression in estrogen receptor-positive breast cancer. Clin. Cancer Res. 2007, 13, 4069–4077. [Google Scholar] [CrossRef]
- Zahavi, T.; Salmon-Divon, M.; Salgado, R.; Elkin, M.; Hermano, E.; Rubinstein, A.M.; Francis, P.A.; Di Leo, A.; Viale, G.; de Azambuja, E.; et al. Heparanase: A potential marker of worse prognosis in estrogen receptor-positive breast cancer. NPJ Breast Cancer 2021, 7, 67. [Google Scholar] [CrossRef]
- Yang, W.J.; Zhang, G.L.; Cao, K.X.; Liu, X.N.; Wang, X.M.; Yu, M.W.; Li, J.P.; Yang, G.W. Heparanase from triple-negative breast cancer and platelets acts as an enhancer of metastasis. Int. J. Oncol. 2020, 57, 890–904. [Google Scholar] [CrossRef]
- Freeman, C.; Parish, C.R. Human platelet heparanase: Purification, characterization and catalytic activity. Biochem. J. 1998, 330 Pt 3, 1341–1350. [Google Scholar] [CrossRef] [PubMed]
- Rautela, J.; Baschuk, N.; Slaney, C.Y.; Jayatilleke, K.M.; Xiao, K.; Bidwell, B.N.; Lucas, E.C.; Hawkins, E.D.; Lock, P.; Wong, C.S.; et al. Loss of Host Type-I IFN Signaling Accelerates Metastasis and Impairs NK-cell Antitumor Function in Multiple Models of Breast Cancer. Cancer Immunol. Res. 2015, 3, 1207–1217. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, K.; Okamoto, H.; Numata, Y.; Takemoto, H. A simple and rapid assay for heparanase activity using homogeneous time-resolved fluorescence. J. Pharm. Biomed. Anal. 2006, 41, 912–917. [Google Scholar] [CrossRef]
- Zhang, Q.; Ming, J.; Li, Y.; Zhang, S.; Li, B.; Qiu, X.; Wang, E. Heparanase expression correlates with angiogenesis and lymphangiogenesis in human lung cancer. Zhongguo Fei Ai Za Zhi 2009, 12, 864–867. [Google Scholar] [CrossRef] [PubMed]
- Ostapoff, K.T.; Awasthi, N.; Cenik, B.K.; Hinz, S.; Dredge, K.; Schwarz, R.E.; Brekken, R.A. PG545, an angiogenesis and heparanase inhibitor, reduces primary tumor growth and metastasis in experimental pancreatic cancer. Mol. Cancer Ther. 2013, 12, 1190–1201. [Google Scholar] [CrossRef]
- Naomoto, Y.; Gunduz, M.; Takaoka, M.; Okawa, T.; Gunduz, E.; Nobuhisa, T.; Kobayashi, M.; Shirakawa, Y.; Yamatsuji, T.; Sonoda, R.; et al. Heparanase promotes angiogenesis through Cox-2 and HIF1alpha. Med. Hypotheses 2007, 68, 162–165. [Google Scholar] [CrossRef]
- Guy, C.T.; Cardiff, R.D.; Muller, W.J. Induction of mammary tumors by expression of polyomavirus middle T oncogene: A transgenic mouse model for metastatic disease. Mol. Cell. Biol. 1992, 12, 954–961. [Google Scholar]
- Duivenvoorden, H.M.; Spurling, A.; O’Toole, S.A.; Parker, B.S. Discriminating the earliest stages of mammary carcinoma using myoepithelial and proliferative markers. PLoS ONE 2018, 13, e0201370. [Google Scholar] [CrossRef]
- Duivenvoorden, H.M.; Rautela, J.; Edgington-Mitchell, L.E.; Spurling, A.; Greening, D.W.; Nowell, C.J.; Molloy, T.J.; Robbins, E.; Brockwell, N.K.; Lee, C.S.; et al. Myoepithelial cell-specific expression of stefin A as a suppressor of early breast cancer invasion. J. Pathol. 2017, 243, 496–509. [Google Scholar] [CrossRef]
- Zcharia, E.; Jia, J.; Zhang, X.; Baraz, L.; Lindahl, U.; Peretz, T.; Vlodavsky, I.; Li, J.P. Newly generated heparanase knock-out mice unravel co-regulation of heparanase and matrix metalloproteinases. PLoS ONE 2009, 4, e5181. [Google Scholar] [CrossRef]
- Steeg, P.S. Targeting metastasis. Nat. Rev. Cancer 2016, 16, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Holen, I.; Speirs, V.; Morrissey, B.; Blyth, K. In vivo models in breast cancer research: Progress, challenges and future directions. Dis. Model. Mech. 2017, 10, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Saxena, M.; Christofori, G. Rebuilding cancer metastasis in the mouse. Mol. Oncol. 2013, 7, 283–296. [Google Scholar] [CrossRef]
- Lin, E.Y.; Jones, J.G.; Li, P.; Zhu, L.; Whitney, K.D.; Muller, W.J.; Pollard, J.W. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003, 163, 2113–2126. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Jia, S.; Zhu, Y.; Utermark, T.; Signoretti, S.; Loda, M.; Schaffhausen, B.; Roberts, T.M. Transgenic Expression of Polyomavirus Middle T Antigen in the Mouse Prostate Gives Rise to Carcinoma. J. Virol. 2011, 85, 5581–5592. [Google Scholar] [CrossRef]
- Kiefer, F.; Anhauser, I.; Soriano, P.; Aguzzi, A.; Courtneidge, S.A.; Wagner, E.F. Endothelial cell transformation by polyomavirus middle T antigen in mice lacking Src-related kinases. Curr. Biol. CB 1994, 4, 100–109. [Google Scholar] [CrossRef]
- Dilworth, S.M. Polyoma virus middle T antigen and its role in identifying cancer-related molecules. Nat. Rev. Cancer 2002, 2, 951. [Google Scholar] [CrossRef]
- Schaffhausen, B.S.; Roberts, T.M. Lessons from polyoma middle T antigen on signaling and transformation: A DNA tumor virus contribution to the war on cancer. Virology 2009, 384, 304–316. [Google Scholar] [CrossRef]
- Zhou, A.Y.; Ichaso, N.; Adamarek, A.; Zila, V.; Forstova, J.; Dibb, N.J.; Dilworth, S.M. Polyomavirus Middle T-Antigen Is a Transmembrane Protein That Binds Signaling Proteins in Discrete Subcellular Membrane Sites. J. Virol. 2011, 85, 3046–3054. [Google Scholar] [CrossRef]
- Raptis, L. Polyomavirus middle tumor antigen increases responsiveness to growth factors. J. Virol. 1991, 65, 2691–2694. [Google Scholar] [CrossRef]
- Courtneidge, S.A.; Smith, A.E. Polyoma virus transforming protein associates with the product of the c-src cellular gene. Nature 1983, 303, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Whitman, M.; Kaplan, D.R.; Schaffhausen, B.; Cantley, L.; Roberts, T.M. Association of phosphatidylinositol kinase activity with polyoma middle-T competent for transformation. Nature 1985, 315, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Ichaso, N.; Dilworth, S.M. Cell transformation by the middle T-antigen of polyoma virus. Oncogene 2001, 20, 7908–7916. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Nogales-Cadenas, R.; Zhang, Q.; Lin, J.R.; Zhang, W.; O’Brien, K.; Montagna, C.; Zhang, Z.D. Transcriptomic dynamics of breast cancer progression in the MMTV-PyMT mouse model. BMC Genom. 2017, 18, 185. [Google Scholar] [CrossRef]
- Nielsen, B.S.; Egeblad, M.; Rank, F.; Askautrud, H.A.; Pennington, C.J.; Pedersen, T.X.; Christensen, I.J.; Edwards, D.R.; Werb, Z.; Lund, L.R. Matrix Metalloproteinase 13 Is Induced in Fibroblasts in Polyomavirus Middle T Antigen-Driven Mammary Carcinoma without Influencing Tumor Progression. PLoS ONE 2008, 3, e2959. [Google Scholar] [CrossRef] [PubMed]
- Whalen, K.A.; Weber, G.F.; Benjamin, T.L.; Schaffhausen, B.S. Polyomavirus Middle T Antigen Induces the Transcription of Osteopontin, a Gene Important for the Migration of Transformed Cells. J. Virol. 2008, 82, 4946–4954. [Google Scholar] [CrossRef][Green Version]
- Jia, R.; Liang, Y.; Chen, R.; Liu, G.; Wang, H.; Tang, M.; Zhou, X.; Wang, H.; Yang, Y.; Wei, H.; et al. Osteopontin facilitates tumor metastasis by regulating epithelial–mesenchymal plasticity. Cell. Death Dis. 2016, 7, e2564. [Google Scholar] [CrossRef]
- Dudley, J.P.; Golovkina, T.V.; Ross, S.R. Lessons Learned from Mouse Mammary Tumor Virus in Animal Models. ILAR J. 2016, 57, 12–23. [Google Scholar] [CrossRef]
- Otten, A.D.; Sanders, M.M.; McKnight, G.S. The MMTV LTR promoter is induced by progesterone and dihydrotestosterone but not by estrogen. Mol. Endocrinol. 1988, 2, 143–147. [Google Scholar] [CrossRef]
- Mink, S.; Ponta, H.; Cato, A.C. The long terminal repeat region of the mouse mammary tumour virus contains multiple regulatory elements. Nucleic Acids Res. 1990, 18, 2017–2024. [Google Scholar] [CrossRef]
- Qin, W.; Golovkina, T.V.; Peng, T.; Nepomnaschy, I.; Buggiano, V.; Piazzon, I.; Ross, S.R. Mammary Gland Expression of Mouse Mammary Tumor Virus Is Regulated by a Novel Element in the Long Terminal Repeat. J. Virol. 1999, 73, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Klarenbeek, S.; van Miltenburg, M.H.; Jonkers, J. Genetically engineered mouse models of PI3K signaling in breast cancer. Mol. Oncol. 2013, 7, 146–164. [Google Scholar] [CrossRef] [PubMed]
- Lin, E.Y.; Nguyen, A.V.; Russell, R.G.; Pollard, J.W. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 2001, 193, 727–740. [Google Scholar] [CrossRef] [PubMed]
- DeNardo, D.G.; Barreto, J.B.; Andreu, P.; Vasquez, L.; Tawfik, D.; Kolhatkar, N.; Coussens, L.M. CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell. 2009, 16, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.I.; Camenisch, T.D.; Stevens, M.V.; Sands, B.J.; McDonald, J.; Schroeder, J.A. CD44 attenuates metastatic invasion during breast cancer progression. Cancer Res. 2005, 65, 6755–6763. [Google Scholar] [CrossRef]
- Schoeffner, D.J.; Matheny, S.L.; Akahane, T.; Factor, V.; Berry, A.; Merlino, G.; Thorgeirsson, U.P. VEGF contributes to mammary tumor growth in transgenic mice through paracrine and autocrine mechanisms. Lab. Investig. 2005, 85, 608–623. [Google Scholar] [CrossRef]
- Muraoka-Cook, R.S.; Kurokawa, H.; Koh, Y.; Forbes, J.T.; Roebuck, L.R.; Barcellos-Hoff, M.H.; Moody, S.E.; Chodosh, L.A.; Arteaga, C.L. Conditional overexpression of active transforming growth factor beta1 in vivo accelerates metastases of transgenic mammary tumors. Cancer Res. 2004, 64, 9002–9011. [Google Scholar] [CrossRef]
- Almholt, K.; Lund, L.R.; Rygaard, J.; Nielsen, B.S.; Dano, K.; Romer, J.; Johnsen, M. Reduced metastasis of transgenic mammary cancer in urokinase-deficient mice. Int. J. Cancer 2005, 113, 525–532. [Google Scholar] [CrossRef]
- Cuevas, B.D.; Winter-Vann, A.M.; Johnson, N.L.; Johnson, G.L. MEKK1 controls matrix degradation and tumor cell dissemination during metastasis of polyoma middle-T driven mammary cancer. Oncogene 2006, 25, 4998–5010. [Google Scholar] [CrossRef]
- Wculek, S.K.; Malanchi, I. Neutrophils support lung colonization of metastasis-initiating breast cancer cells. Nature 2015, 528, 413. [Google Scholar] [CrossRef]
- Bocchinfuso, W.P.; Lindzey, J.K.; Hewitt, S.C.; Clark, J.A.; Myers, P.H.; Cooper, R.; Korach, K.S. Induction of Mammary Gland Development in Estrogen Receptor-α Knockout Mice. Endocrinology 2000, 141, 2982–2994. [Google Scholar] [CrossRef] [PubMed]
- Fata, J.E.; Chaudhary, V.; Khokha, R. Cellular Turnover in the Mammary Gland Is Correlated with Systemic Levels of Progesterone and Not 17β-Estradiol During the Estrous Cycle1. Biol. Reprod. 2001, 65, 680–688. [Google Scholar] [CrossRef] [PubMed]
- Chua, A.C.; Hodson, L.J.; Moldenhauer, L.M.; Robertson, S.A.; Ingman, W.V. Dual roles for macrophages in ovarian cycle-associated development and remodelling of the mammary gland epithelium. Development 2010, 137, 4229–4238. [Google Scholar] [CrossRef] [PubMed]
- Saha, T.; Makar, S.; Swetha, R.; Gutti, G.; Singh, S.K. Estrogen signaling: An emanating therapeutic target for breast cancer treatment. Eur. J. Med. Chem. 2019, 177, 116–143. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.Y.; Yan, J.; Fu, X.; Pan, Q.; Sun, D.; Xu, Y.; Wang, J.; Nie, L.; Tong, L.J.; Shen, A.; et al. Aspirin inhibits cancer metastasis and angiogenesis via targeting heparanase. Clin. Cancer Res. 2017, 23, 6267–6278. [Google Scholar] [CrossRef] [PubMed]
- Deryugina, E.I.; Quigley, J.P. Tumor Angiogenesis: MMP-Mediated Induction of Intravasation- and Metastasis-Sustaining Neovasculature. Matrix Biol. J. Int. Soc. Matrix Biol. 2015, 44–46, 94–112. [Google Scholar] [CrossRef]
- Tam, S.Y.; Wu, V.W.C.; Law, H.K.W. Hypoxia-Induced Epithelial-Mesenchymal Transition in Cancers: HIF-1α and Beyond. Front. Oncol. 2020, 10, 486. [Google Scholar] [CrossRef]
- LaGory, E.L.; Giaccia, A.J. The ever-expanding role of HIF in tumour and stromal biology. Nat. Cell Biol. 2016, 18, 356–365. [Google Scholar] [CrossRef]
- Lifsted, T.; Le Voyer, T.; Williams, M.; Muller, W.; Klein-Szanto, A.; Buetow, K.H.; Hunter, K.W. Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression. Int. J. Cancer 1998, 77, 640–644. [Google Scholar] [CrossRef]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef]
- Iozzo, R.V.; Gubbiotti, M.A. Extracellular matrix: The driving force of mammalian diseases. Matrix Biol. 2018, 71–72, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Bonnans, C.; Chou, J.; Werb, Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell. Biol. 2014, 15, 786–801. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.M.; Bhat, R.; Correia, A.L.; Mott, J.D.; Ilan, N.; Vlodavsky, I.; Pavão, M.S.; Bissell, M. Mammary Branching Morphogenesis Requires Reciprocal Signaling by Heparanase and MMP-14. J. Cell. Biochem. 2015, 116, 1668–1679. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, T.Y.; Zheng, H.; Liu, R.; Wicha, M.S.; Yu, S.M.; Weiss, S.J. Divergent Matrix-Remodeling Strategies Distinguish Developmental from Neoplastic Mammary Epithelial Cell Invasion Programs. Dev. Cell. 2018, 47, 145–160.e146. [Google Scholar] [CrossRef]
- Winer, A.; Adams, S.; Mignatti, P. Matrix Metalloproteinase Inhibitors in Cancer Therapy: Turning Past Failures into Future Successes. Mol. Cancer Ther. 2018, 17, 1147–1155. [Google Scholar] [CrossRef]
- Dove, A. MMP inhibitors: Glimmers of hope amidst clinical failures. Nat. Med. 2002, 8, 95. [Google Scholar] [CrossRef]



| Primer | Sequence (5′ to 3′) | Size (bp) |
|---|---|---|
| MMTV-PyMT (F) | AGG AAC CGG CTT CCA GGT AAG A | |
| MMTV-PyMT (R) | TTG GTG TTC CAA ACC ATT GCA T | 260 |
| HPSE+/+ (F) | GAA GAA CCA TTA TTC ATC TTG CT | |
| HPSE+/+ (R) | CCA AGT GCC AGT CTG CAA GT | 143 |
| HPSE−/− (F) | GGG ATG GAT GCA GGT CTT C | |
| HPSE−/− (R) | CAG ATG GGT GCA GAT TAG ATA T | 300 |
| Fabpi (F) | TGG ACA GGA CTG GAC CTC TGC TTT CCT AGA | |
| Fabpi (R) | TAG AGC TTT CGG ACA TCA CAG GTC ATT CAG | 200 |
| Target | Primary Antibody | Secondary Antibody |
|---|---|---|
| HPSE | Rabbit polyclonal anti-HPSE (10 μg/mL, B85543, Abcam) | Biotinylated goat anti-rabbit IgG (H+L) (6 μg/mL, BA-1000, Vector Laboratories) |
| CD31 | Rabbit polyclonal anti-CD31 (16 μg/mL, AB28364, Abcam) | Biotinylated goat anti-rabbit IgG (H+L) (6 μg/mL, BA-1000, Vector Laboratories) |
| Ki67 | Rabbit polyclonal anti-Ki67 (1 μg/mL, AB15580, Abcam) | Biotinylated goat anti-rabbit IgG (H+L) (6 μg/mL, BA-1000, Vector Laboratories) |
| Smooth muscle myosin heavy chain (SMMHC) | Rabbit monoclonal anti-SMMHC (clone EPR5335, 1.2 μg/mL, AB124679, Abcam) | Biotinylated goat anti-rabbit IgG (H+L) (6 μg/mL, BA-1000, Vector Laboratories) |
| MMP-2 | Rabbit polyclonal anti-MMP-2 (2 μg/mL, AB37150, Abcam) | Biotinylated goat anti-rabbit IgG (H+L) (6 μg/mL, BA-1000, Vector Laboratories) |
| Normal rabbit IgG (isotype control) | IgG from rabbit serum (various working concentrations, 18140, Sigma-Aldrich) | Biotinylated goat anti-rabbit IgG (H+L) (6 μg/mL, BA-1000, Vector Laboratories) |
| Primer | Sequence 5′ to 3′ |
|---|---|
| PyMT cDNA (F) | CCA ACA GAT ACA CCC GCA CAT |
| PyMT cDNA (R) | GGT CTT GGT CGC TTT CTG GAT A |
| Control 18S cDNA (F) | GTA ACC CGT TGA ACC CCA TT |
| Control 18S cDNA (R) | CCA TCC AAT CGG TAG TAG CG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayatilleke, K.M.; Duivenvoorden, H.M.; Ryan, G.F.; Parker, B.S.; Hulett, M.D. Investigating the Role of Heparanase in Breast Cancer Development Utilising the MMTV-PyMT Murine Model of Mammary Carcinoma. Cancers 2023, 15, 3062. https://doi.org/10.3390/cancers15113062
Jayatilleke KM, Duivenvoorden HM, Ryan GF, Parker BS, Hulett MD. Investigating the Role of Heparanase in Breast Cancer Development Utilising the MMTV-PyMT Murine Model of Mammary Carcinoma. Cancers. 2023; 15(11):3062. https://doi.org/10.3390/cancers15113062
Chicago/Turabian StyleJayatilleke, Krishnath M., Hendrika M. Duivenvoorden, Gemma F. Ryan, Belinda S. Parker, and Mark D. Hulett. 2023. "Investigating the Role of Heparanase in Breast Cancer Development Utilising the MMTV-PyMT Murine Model of Mammary Carcinoma" Cancers 15, no. 11: 3062. https://doi.org/10.3390/cancers15113062
APA StyleJayatilleke, K. M., Duivenvoorden, H. M., Ryan, G. F., Parker, B. S., & Hulett, M. D. (2023). Investigating the Role of Heparanase in Breast Cancer Development Utilising the MMTV-PyMT Murine Model of Mammary Carcinoma. Cancers, 15(11), 3062. https://doi.org/10.3390/cancers15113062






