Texture Analysis of the Apparent Diffusion Coefficient Focused on Contrast-Enhancing Lesions in Predicting Survival for Bevacizumab-Treated Patients with Recurrent Glioblastoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. MRI Protocol
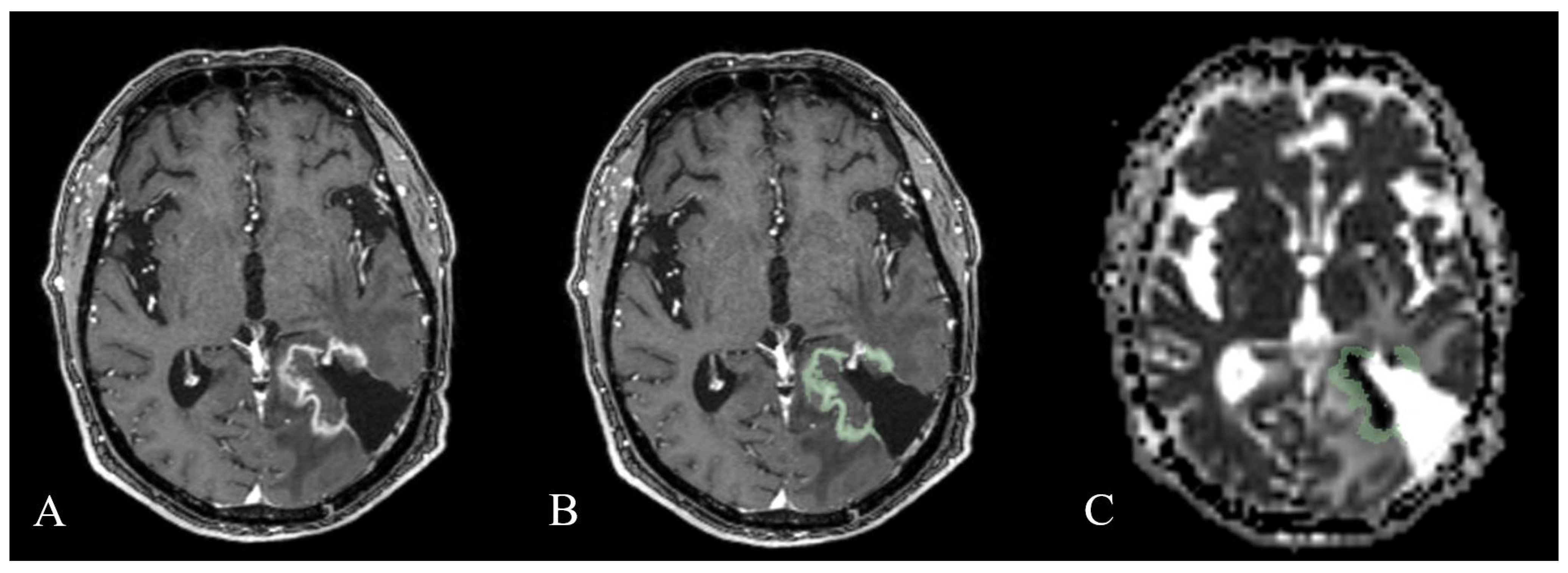
2.3. Image Analysis
2.4. Statistical Analysis
3. Results
3.1. Texture Analysis
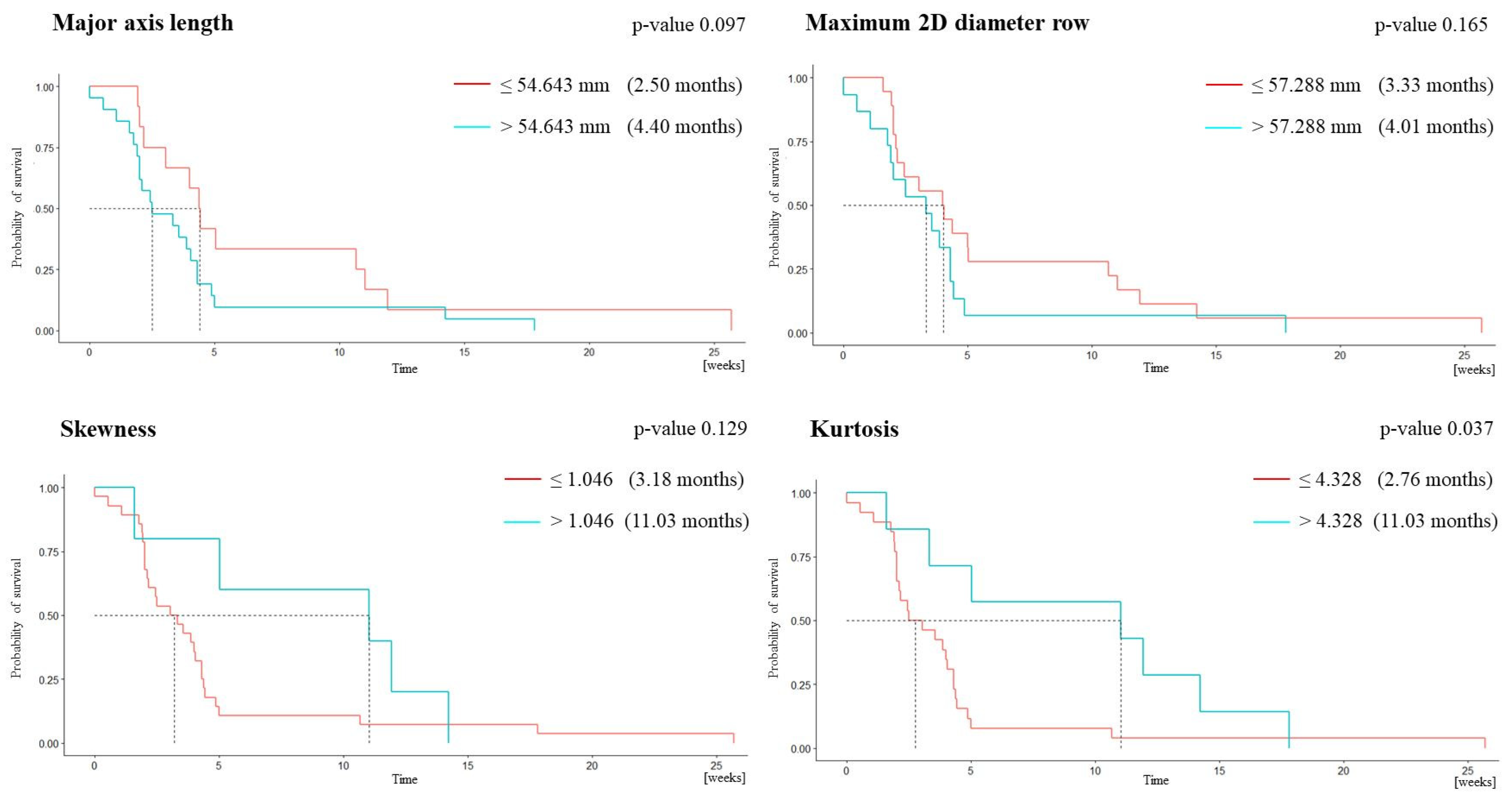
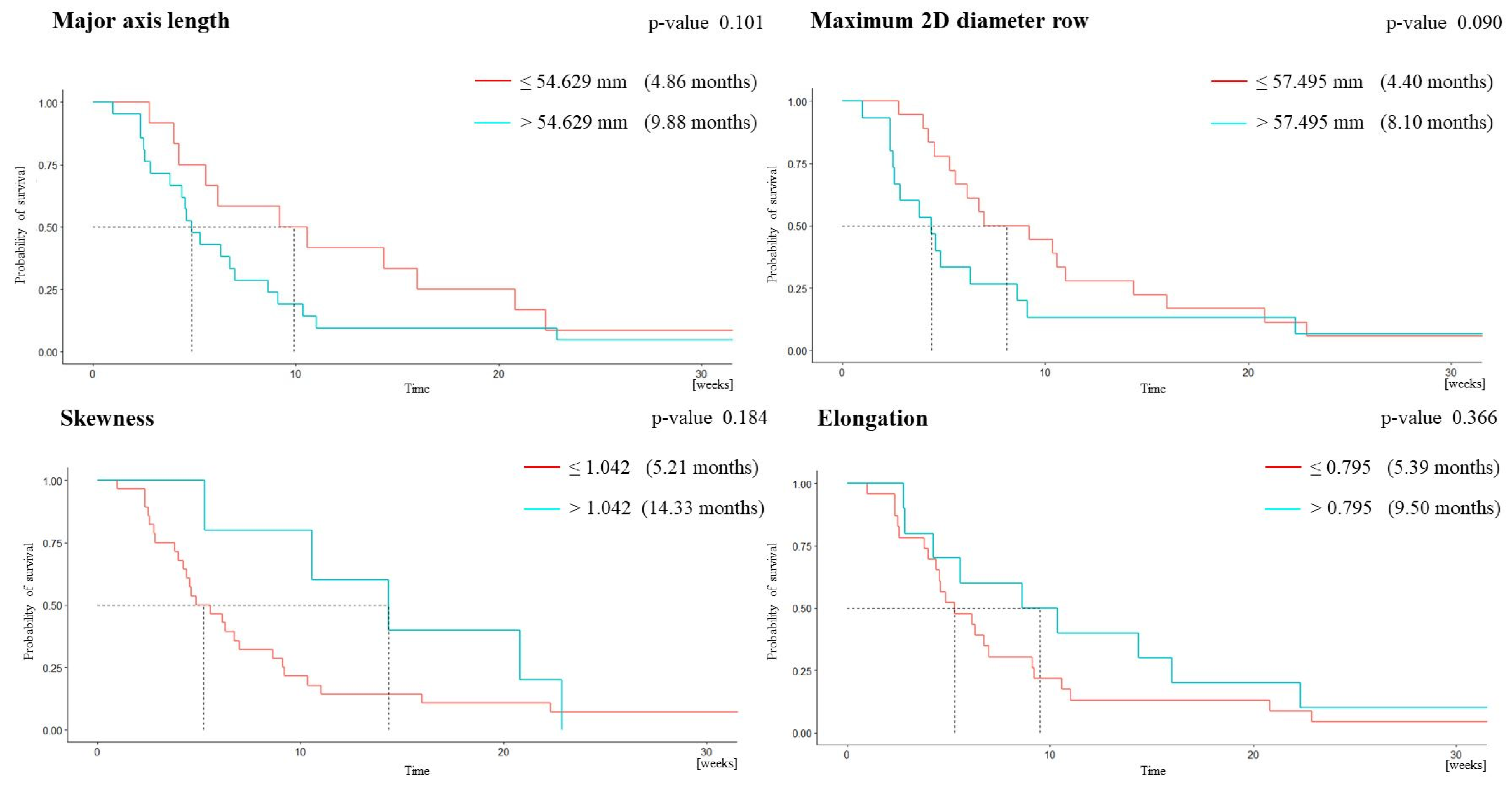
3.2. Survival Analysis
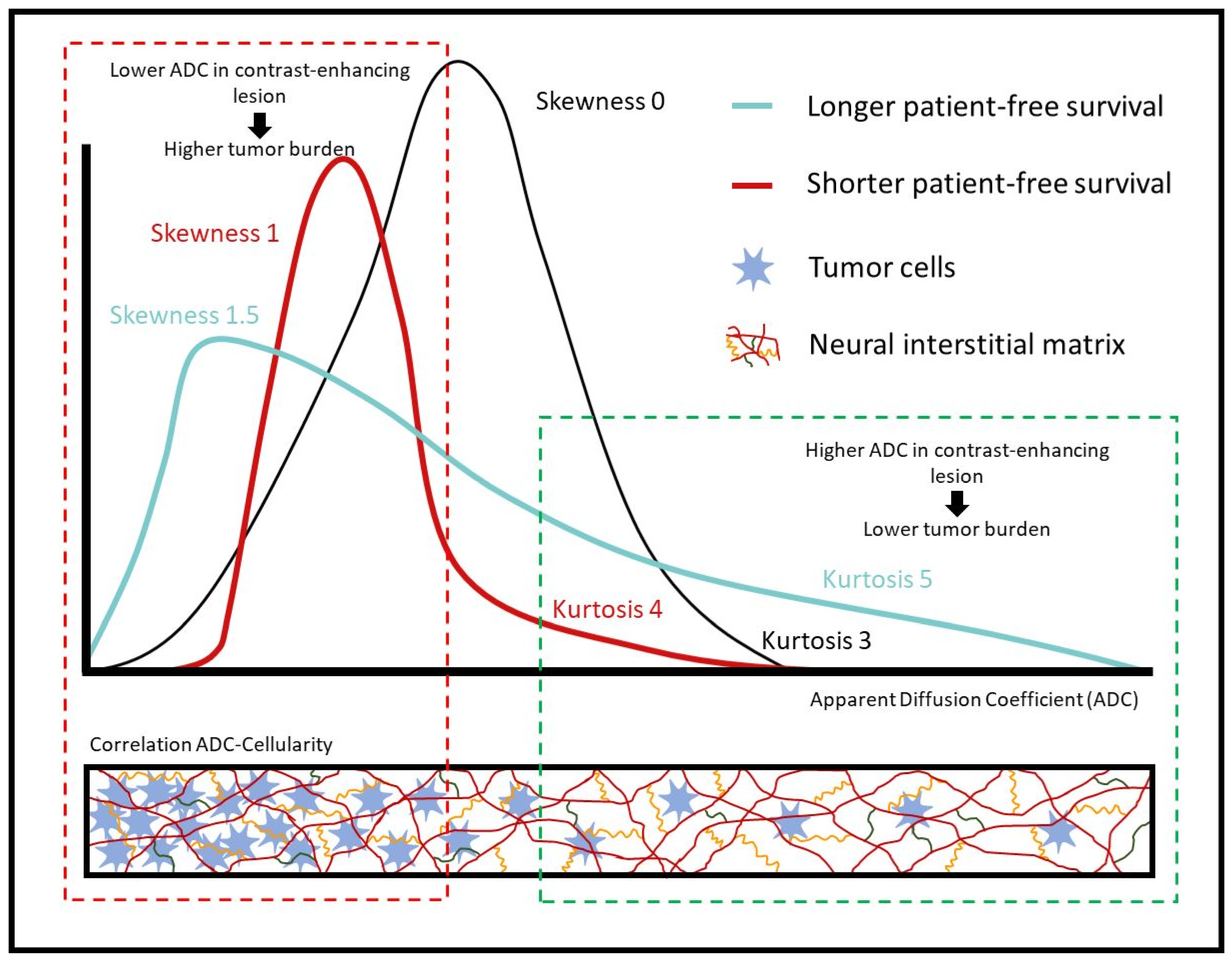
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro-Oncology 2021, 23, iii1–iii105. [Google Scholar] [CrossRef] [PubMed]
- Grochans, S.; Cybulska, A.M.; Simińska, D.; Korbecki, J.; Kojder, K.; Chlubek, D.; Baranowska-Bosiacka, I. Epidemiology of Glioblastoma Multiforme–Literature Review. Cancers 2022, 14, 2412. [Google Scholar] [CrossRef] [PubMed]
- Oronsky, B.; Reid, T.R.; Oronsky, A.; Sandhu, N.; Knox, S.J. A Review of Newly Diagnosed Glioblastoma. Front. Oncol. 2021, 10, 574012. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.-H.; Wang, Z.-F.; Pan, Z.-Y.; Péus, D.; Fernandez, J.D.; Pallud, J.; Li, Z.-Q. A Meta-Analysis of Survival Outcomes Following Reoperation in Recurrent Glioblastoma: Time to Consider the Timing of Reoperation. Front. Neurol. 2019, 10, 286. [Google Scholar] [CrossRef]
- De Lemos, M.L.; Markarian, A.; Chan, E.; Schaff, K.; Walisser, S. Clinical effectiveness of bevacizumab in patients with re-current brain tumours: A population-based evaluation. J. Oncol. Pharm. Pract. 2018, 24, 33–36. [Google Scholar] [CrossRef]
- Prados, M.D.; Byron, S.A.; Tran, N.L.; Phillips, J.J.; Molinaro, A.M.; Ligon, K.L.; Wen, P.Y.; Kuhn, J.G.; Mellinghoff, I.K.; de Groot, J.F.; et al. Toward precision medicine in glioblastoma: The promise and the challenges. Neuro-Oncology 2015, 17, 1051–1063. [Google Scholar] [CrossRef]
- Reardon, D.A.; Ligon, K.L.; Chiocca, E.A.; Wen, P.Y. One size should not fit all: Advancing toward personalized glioblastoma therapy. Discov. Med. 2015, 19, 471–477. [Google Scholar]
- Gerstner, E.R.; Batchelor, T.T. Antiangiogenic Therapy for Glioblastoma. Cancer J. 2012, 18, 45–50. [Google Scholar] [CrossRef]
- Tamura, R.; Tanaka, T.; Akasaki, Y.; Murayama, Y.; Yoshida, K.; Sasaki, H. The role of vascular endothelial growth factor in the hypoxic and immunosuppressive tumor microenvironment: Perspectives for therapeutic implications. Med. Oncol. 2019, 37, 2. [Google Scholar] [CrossRef]
- Friedman, H.S.; Prados, M.D.; Wen, P.Y.; Mikkelsen, T.; Schiff, D.; Abrey, L.E.; Yung, W.A.; Paleologos, N.; Nicholas, M.K.; Jensen, R.; et al. Bevacizumab Alone and in Combination With Irinotecan in Recurrent Glioblastoma. J. Clin. Oncol. 2009, 27, 4733–4740. [Google Scholar] [CrossRef]
- Kreisl, T.N.; Kim, L.; Moore, K.; Duic, P.; Royce, C.; Stroud, I.; Garren, N.; Mackey, M.; Butman, J.; Camphausen, K.; et al. Phase II Trial of Single-Agent Bevacizumab Followed by Bevacizumab Plus Irinotecan at Tumor Progression in Recurrent Glioblastoma. J. Clin. Oncol. 2009, 27, 740–745. [Google Scholar] [CrossRef]
- Nghiemphu, P.L.; Liu, W.; Lee, Y.; Than, T.; Graham, C.; Lai, A.; Green, R.M.; Pope, W.B.; Liau, L.M.; Mischel, P.S.; et al. Bevacizumab and chemotherapy for recurrent glioblastoma: A single-institution experience. Neurology 2009, 72, 1217–1222. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.; Tran, A.; Nghiemphu, P.L.; Pope, W.B.; Solis, O.E.; Selch, M.; Filka, E.; Yong, W.H.; Mischel, P.S.; Liau, L.M.; et al. Phase II study of bevacizumab plus temozolomide during and after radiation therapy for patients with newly diagnosed glioblastoma multi-forme. J. Clin. Oncol. 2011, 29, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Clarke, J.L.; Molinaro, A.M.; Phillips, J.J.; Butowski, N.A.; Chang, S.M.; Perry, A.; Costello, J.F.; DeSilva, A.A.; Rabbitt, J.E.; Prados, M.D. A single-institution phase II trial of radiation, temozolomide, erlotinib, and bevacizumab for initial treatment of glioblastoma. Neuro-Oncology 2014, 16, 984–990. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, M.R.; Dignam, J.J.; Armstrong, T.S.; Wefel, J.S.; Blumenthal, D.T.; Vogelbaum, M.A.; Colman, H.; Chakravarti, A.; Pugh, S.; Won, M.; et al. A Randomized Trial of Bevacizumab for Newly Diagnosed Glioblastoma. N. Engl. J. Med. 2014, 370, 699–708. [Google Scholar] [CrossRef]
- Chinot, O.L.; Wick, W.; Mason, W.; Henriksson, R.; Saran, F.; Nishikawa, R.; Carpentier, A.F.; Hoang-Xuan, K.; Kavan, P.; Cernea, D.; et al. Bevacizumab plus Radiotherapy–Temozolomide for Newly Diagnosed Glioblastoma. N. Engl. J. Med. 2014, 370, 709–722. [Google Scholar] [CrossRef]
- Martinez-Heras, E.; Grussu, F.; Prados, F.; Solana, E.; Llufriu, S. Diffusion-Weighted Imaging: Recent Advances and Applica-tions. Semin. Ultrasound CT MR 2021, 42, 490–506. [Google Scholar] [CrossRef]
- Patterson, D.M.; Padhani, A.R.; Collins, D.J. Technology Insight: Water diffusion MRI—A potential new biomarker of response to cancer therapy. Nat. Clin. Pract. Oncol. 2008, 5, 220–233. [Google Scholar] [CrossRef]
- Hamstra, D.A.; Rehemtulla, A.; Ross, B.D. Diffusion Magnetic Resonance Imaging: A Biomarker for Treatment Response in Oncology. J. Clin. Oncol. 2007, 25, 4104–4109. [Google Scholar] [CrossRef]
- Pope, W.B.; Kim, H.J.; Huo, J.; Alger, J.; Brown, M.S.; Gjertson, D.; Sai, V.; Young, J.R.; Tekchandani, L.; Cloughesy, T.; et al. Recurrent Glioblastoma Multiforme: ADC Histogram Analysis Predicts Response to Bevacizumab Treatment. Radiology 2009, 252, 182–189. [Google Scholar] [CrossRef]
- Jain, R.; Scarpace, L.M.; Ellika, S.; Torcuator, R.; Schultz, L.R.; Hearshen, D.; Mikkelsen, T. Imaging response criteria for recurrent gliomas treated with bevacizumab: Role of diffusion weighted imaging as an imaging biomarker. J. Neuro-Oncology 2009, 96, 423–431. [Google Scholar] [CrossRef]
- Tokgoz, N.; Celik, H.; Gultekin, S.; Tokgoz, N.; Celik, H.; Gultekin, S.; Voyvoda, N.K.; Oner, A.Y.; Tali, E.T. Is there a role for apparent diffusion coefficients in the differential diagnosis of brain tumors? Neuroradiol. J. 2006, 19, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Pope, W.; Lai, A.; Mehta, R.; Kim, H.; Qiao, J.; Young, J.; Xue, X.; Goldin, J.; Brown, M.; Nghiemphu, P.; et al. Apparent Diffusion Coefficient Histogram Analysis Stratifies Progression-Free Survival in Newly Diagnosed Bevacizumab-Treated Glioblastoma. Am. J. Neuroradiol. 2011, 32, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Pope, W.B.; Qiao, X.J.; Kim, H.J.; Lai, A.; Nghiemphu, P.; Xue, X.; Ellingson, B.M.; Schiff, D.; Aregawi, D.; Cha, S.; et al. Apparent diffusion coefficient histogram analysis stratifies progression-free and overall survival in patients with recurrent GBM treated with bevacizumab: A multi-center study. J. Neuro-Oncology 2012, 108, 491–498. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Malkin, M.G.; Rand, S.D.; LaViolette, P.S.; Connelly, J.M.; Mueller, W.M.; Schmainda, K.M. Volumetric analysis of func-tional diffusion maps is a predictive imaging biomarker for cytotoxic and anti-angiogenic treatments in malignant gliomas. J. Neuro-Oncol. 2011, 102, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Ellingson, B.M.; Cloughesy, T.F.; Lai, A.; Mischel, P.S.; Nghiemphu, P.L.; Lalezari, S.; Schmainda, K.M.; Pope, W.B. Graded functional diffusion map-defined characteristics of apparent diffusion coefficients predict overall survival in recurrent glioblastoma treated with bevacizumab. Neuro-Oncology 2011, 13, 1151–1161. [Google Scholar] [CrossRef]
- Paldino, M.J.; Barboriak, D.; Desjardins, A.; Friedman, H.S.; Vredenburgh, J.J. Repeatability of quantitative parameters derived from diffusion tensor imaging in patients with glioblastoma multiforme. J. Magn. Reson. Imaging 2009, 29, 1199–1205. [Google Scholar] [CrossRef]
- Soni, N.; Priya, S.; Bathla, G. Texture Analysis in Cerebral Gliomas: A Review of the Literature. Am. J. Neuroradiol. 2019, 40, 928–934. [Google Scholar] [CrossRef]
- Chaddad, A.; Kucharczyk, M.; Daniel, P.; Sabri, S.; Jean-Claude, B.J.; Niazi, T.; Abdulkarim, B. Radiomics in Glioblastoma: Current Status and Challenges Facing Clinical Implementation. Front. Oncol. 2019, 9, 374. [Google Scholar] [CrossRef]
- Yang, D.; Rao, G.; Martinez, J.; Veeraraghavan, A.; Rao, A. Evaluation of tumor-derived MRI-texture features for discrimination of molecular subtypes and prediction of 12-month survival status in glioblastoma. Med. Phys. 2015, 42, 6725–6735. [Google Scholar] [CrossRef]
- Sasaki, T.; Kinoshita, M.; Fujita, K.; Fukai, J.; Hayashi, N.; Uematsu, Y.; Okita, Y.; Nonaka, M.; Moriuchi, S.; Uda, T.; et al. Radiomics and MGMT promoter methylation for prognostication of newly diagnosed glioblastoma. Sci. Rep. 2019, 9, 14435. [Google Scholar] [CrossRef]
- Boxerman, J.L.; Schmainda, K.M.; Weisskoff, R.M. Relative cerebral blood volume maps corrected for contrast agent extrav-asation significantly correlate with glioma tumor grade, whereas uncorrected maps do not. AJNR Am. J. Neuroradiol. 2006, 27, 859–867. [Google Scholar] [PubMed]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.C.; Pieper, S.; Aerts, H.J.W.L. Computational radiomics system to decode the radiographic phenotype. Can. Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Traverso, A.; van Soest, J.; Dekker, A.; Wee, L. Technical Note: Ontology-guided radiomics analysis workflow (O-RAW). Med. Phys. 2019, 46, 5677–5684. [Google Scholar] [CrossRef]
- Pasqualetti, F.; Giampietro, C.; Montemurro, N.; Giannini, N.; Gadducci, G.; Orlandi, P.; Natali, E.; Chiarugi, P.; Gonnelli, A.; Cantarella, M.; et al. Old and New Systemic Immune-Inflammation Indexes Are Associated with Overall Survival of Glioblastoma Patients Treated with Radio-Chemotherapy. Genes 2022, 13, 1054. [Google Scholar] [CrossRef] [PubMed]
- Pasqualetti, F.; Malfatti, G.; Cantarella, M.; Gonnelli, A.; Montrone, S.; Montemurro, N.; Gadducci, G.; Giannini, N.; Pesaresi, I.; Perrini, P.; et al. Role of magnetic resonance imaging following postoperative radiotherapy in clinical decision-making of patients with high-grade glioma. La Radiol. Med. 2022, 127, 803–808. [Google Scholar] [CrossRef]
- Li, C.; Gan, Y.; Chen, H.; Chen, Y.; Deng, Y.; Zhan, W.; Tan, Q.; Xie, C.; Sharma, H.S.; Zhang, Z. Advanced multimodal imaging in dif-ferentiating glioma recurrence from post-radiotherapy changes. Int. Rev. Neurobiol. 2020, 151, 281–297. [Google Scholar]
- Alexiou, G.A.; Tsiouris, S.; Kyritsis, A.P.; Voulgaris, S.; Argyropoulou, M.I.; Fotopoulos, A.D. Glioma recurrence versus radia-tion necrosis: Accuracy of current imaging modalities. J. Neuro-Oncol. 2009, 95, 1–11. [Google Scholar] [CrossRef]
- Priya, S.; Agarwal, A.; Ward, C.; Locke, T.; Monga, V.; Bathla, G. Survival prediction in glioblastoma on post-contrast magnetic resonance imaging using filtration based first-order texture analysis: Comparison of multiple machine learning models. Neuroradiol. J. 2021, 34, 355–362. [Google Scholar] [CrossRef]
- Choi, Y.; Ahn, K.J.; Nam, Y.; Jang, J.; Shin, N.Y.; Choi, H.S.; Jung, S.L.; Kim, B.S. Analysis of heterogeneity of peritumoral T2 hyperintensity in patients with pre-treatment glioblastoma: Prognostic value of MRI-based radiomics. Eur. J. Radiol. 2019, 120, 108642. [Google Scholar] [CrossRef] [PubMed]
- Ingrisch, M.; Schneider, M.J.; Nörenberg, D.; de Figueiredo, G.N.; Maier-Hein, K.; Suchorska, B.; Schüller, U.; Albert, N.; Brückmann, H.; Reiser, M.; et al. Radiomic Analysis Reveals Prognostic Information in T1-Weighted Baseline Magnetic Resonance Imaging in Patients With Glioblastoma. Investig. Radiol. 2017, 52, 360–366. [Google Scholar] [CrossRef] [PubMed]
- Upadhaya, T.; Morvan, Y.; Stindel, E.; Le Reste, P.J.; Hatt, M. Prognostic value of multimodal MRI tumor features in Glio-blastoma multiforme using textural features analysis. In Proceedings of the 2015 IEEE 12th International Symposium on Bio-medical Imaging (ISBI), Brooklyn, NY, USA, 16–19 April 2015; IEEE: New York, NY, USA, 2015; pp. 50–54. [Google Scholar]
- Grossmann, P.; Narayan, V.; Chang, K.; Rahman, R.; Abrey, L.; Reardon, D.A.; Schwartz, L.H.; Wen, P.Y.; Alexander, B.M.; Huang, R.; et al. Quantitative imaging biomarkers for risk stratification of patients with recurrent glioblastoma treated with bevacizumab. Neuro-Oncology 2017, 19, 1688–1697. [Google Scholar] [CrossRef] [PubMed]
- Vils, A.; Bogowicz, M.; Tanadini-Lang, S.; Vuong, D.; Saltybaeva, N.; Kraft, J.; Wirsching, H.-G.; Gramatzki, D.; Wick, W.; Rushing, E.; et al. Radiomic Analysis to Predict Outcome in Recurrent Glioblastoma Based on Multi-Center MR Imaging From the Prospective DIRECTOR Trial. Front. Oncol. 2021, 11, 636672. [Google Scholar] [CrossRef]
- Huang, S.; Michalek, J.E.; Reardon, D.A.; Wen, P.Y.; Floyd, J.R.; Fox, P.T.; Clarke, G.D.; Jerabek, P.A.; Schmainda, K.M.; Muzi, M. Assessment of tumor hypoxia and perfusion in recurrent glioblastoma following bevacizumab failure using MRI and 18F-FMISO PET. Sci. Rep. 2021, 11, 7632. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Sahebjam, S.; Kim, H.J.; Pope, W.B.; Harris, R.J.; Woodworth, D.C.; Lai, A.; Nghiemphu, P.L.; Mason, W.P.; Cloughesy, T.F. Pretreatment ADC Histogram Analysis Is a Predictive Imaging Biomarker for Bevacizumab Treatment but Not Chemotherapy in Recurrent Glioblastoma. Am. J. Neuroradiol. 2013, 35, 673–679. [Google Scholar] [CrossRef]
- Ellingson, B.M.; Patel, K.; Wang, C.; Raymond, C.; Brenner, A.; de Groot, J.F.; Butowski, N.A.; Zach, L.; Campian, J.L.; Schlossman, J. Val-idation of diffusion MRI as a biomarker for efficacy using randomized phase III trial of bevacizumab with or without VB-111 in recurrent glioblastoma. Neuro-Oncol. Adv. 2021, 3, vdab082. [Google Scholar] [CrossRef]
- Patel, K.S.; Everson, R.G.; Yao, J.; Raymond, C.; Goldman, J.; Schlossman, J.; Tsung, J.; Tan, C.; Pope, W.B.; Ji, M.S.; et al. Diffusion Magnetic Resonance Imaging Phenotypes Predict Overall Survival Benefit From Bevacizumab or Surgery in Recurrent Glioblastoma With Large Tumor Burden. Neurosurgery 2020, 87, 931–938. [Google Scholar] [CrossRef]
- Kurokawa, R.; Baba, A.; Kurokawa, M.; Capizzano, A.; Hassan, O.; Johnson, T.; Ota, Y.; Kim, J.; Hagiwara, A.; Moritani, T. Pretreatment ADC Histogram Analysis as a Prognostic Imaging Biomarker for Patients with Recurrent Glioblastoma Treated with Bevaci-zumab: A Systematic Review and Meta-analysis. AJNR Am. J. Neuroradiol. 2022, 43, 202–206. [Google Scholar] [CrossRef]
- Nougaret, S.; Vargas, H.A.; Lakhman, Y.; Sudre, R.; Do, R.K.G.; Bibeau, F.; Azria, D.; Assenat, E.; Molinari, N.; Pierredon, M.-A.; et al. Intravoxel Incoherent Motion–derived Histogram Metrics for Assessment of Response after Combined Chemotherapy and Radiation Therapy in Rectal Cancer: Initial Experience and Comparison between Single-Section and Volumetric Analyses. Radiology 2016, 280, 446–454. [Google Scholar] [CrossRef]
- Baek, H.J.; Kim, H.S.; Kim, N.; Choi, Y.J.; Kim, Y.J. Percent Change of Perfusion Skewness and Kurtosis: A Potential Imaging Biomarker for Early Treatment Response in Patients with Newly Diagnosed Glioblastomas. Radiology 2012, 264, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Draaisma, K.; Chatzipli, A.; Taphoorn, M.; Kerkhof, M.; Weyerbrock, A.; Sanson, M.; Hoeben, A.; Lukacova, S.; Lombardi, G.; Leenstra, S.; et al. Molecular Evolution of IDH Wild-Type Glioblastomas Treated With Standard of Care Affects Survival and Design of Precision Medicine Trials: A Report From the EORTC 1542 Study. J. Clin. Oncol. 2020, 38, 81–99. [Google Scholar] [CrossRef] [PubMed]
- Kickingereder, P.; Neuberger, U.; Bonekamp, D.; Piechotta, P.L.; Götz, M.; Wick, A.; Sill, M.; Kratz, A.; Shinohara, R.T.; Jones, D.T.W.; et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Neuro-Oncology 2017, 20, 848–857. [Google Scholar] [CrossRef]
- Kickingereder, P.; Götz, M.; Muschelli, J.; Wick, A.; Neuberger, U.; Shinohara, R.T.; Sill, M.; Nowosielski, M.; Schlemmer, H.-P.; Radbruch, A.; et al. Large-scale Radiomic Profiling of Recurrent Glioblastoma Identifies an Imaging Predictor for Stratifying Anti-Angiogenic Treatment Response. Clin. Cancer Res. 2016, 22, 5765–5771. [Google Scholar] [CrossRef] [PubMed]
- Akbari, H.; Rathore, S.; Bakas, S.; Nasrallah, M.P.; Shukla, G.; Mamourian, E.; Rozycki, M.; Bagley, S.J.; Rudie, J.D.; Flanders, A.E. Histopathology-validated machine learning radiographic biomarker for noninvasive discrimination between true progression and pseudo-progression in glioblastoma. Cancer 2020, 126, 2625–2636. [Google Scholar] [CrossRef] [PubMed]
| Age (years) (mean, SD) | 56; 13 |
| Female (n, %) | 13; 39.39% |
| Contrast-enhancing lesion volume (mL) (mean, SD) | 29.36; 22.68 |
| PFS (months) (mean, SD) | 5.14; 5.49 |
| ≤6 months group | 2.89; 1.41 |
| >6 months group | 15.23; 5.77 |
| OS (months) (mean, SD) | 9.23; 8.36 |
| ≤1 year group | 5.49; 2.86 |
| >1 year group | 23.12; 7.30 |
| Feature | Whole Cohort (n = 33) | PFS ≤ 6 Months (n = 27) | PFS > 6 Months (n = 6) | p-Value |
|---|---|---|---|---|
| MAL | 63.11 ± 21.11 | 66.46 ± 21.31 | 48.01 ± 12.54 | 0.027 |
| m2Ddr | 56.51 ± 17.82 | 59.07 ± 18.28 | 45 ± 10.03 | 0.021 |
| Skewness | 0.68 ± 0.46 | 0.60 ± 0.42 | 1.06 ± 0.48 | 0.021 |
| Kurtosis | 3.66 ± 1.89 | 3.36 ± 1.70 | 4.98 ± 2.30 | 0.035 |
| Feature | Whole Cohort (n = 33) | OS ≤ 1 Year (n = 26) | OS > 1 Year (n = 7) | p-Value |
|---|---|---|---|---|
| MAL | 63.11 ± 21.11 | 66.99 ± 21.55 | 48.68 ± 11.58 | 0.021 |
| Elongation | 0.66 ± 0.17 | 0.63 ± 0.18 | 0.77 ± 0.12 | 0.027 |
| m2Ddr | 56.51 ± 17.82 | 59.12 ± 18.64 | 46.83 ± 10.36 | 0.034 |
| Skewness | 0.68 ± 0.46 | 0.60 ± 0.42 | 0.98 ± 0.48 | 0.043 |
| Univariate Analysis | ||||||
|---|---|---|---|---|---|---|
| Feature | AUC | Sensitivity | Specificity | PPV | NPV | Cut-Off Value |
| MAL | 0.790 | 0.667 | 0.704 | 0.333 | 0.905 | 54.643 |
| m2Ddr | 0.747 | 0.833 | 0.519 | 0.278 | 0.933 | 57.288 |
| Skewness | 0.802 | 0.500 | 0.926 | 0.6 | 0.893 | 1.046 |
| Kurtosis | 0.769 | 0.667 | 0.889 | 0.571 | 0.923 | 4.328 |
| Bivariate Analysis | ||||||
| MAL m2Ddr | 0.880 | 0.833 | 0.815 | 0.5 | 0.957 | 96.436 86.200 |
| MAL Skewness | 0.880 | 1.000 | 0.741 | 0.462 | 1.000 | 96.436 0.653 |
| Trivariate Analysis | ||||||
| MAL m2Ddr Skewness | 0.886 | 1.000 | 0.778 | 0.5 | 1.000 | |
| Univariate Analysis | ||||||
|---|---|---|---|---|---|---|
| AUC | Sensibility | Specificity | PPV | NPV | Cut-Off Value | |
| MAL | 0.788 | 0.714 | 0.731 | 0.417 | 0.905 | 54.629 |
| m2Ddr | 0.712 | 0.714 | 0.5 | 0.278 | 0.867 | 57.495 |
| Skewness | 0.742 | 0.429 | 0.923 | 0.6 | 0.857 | 1.042 |
| Elongation | 0.728 | 0.571 | 0.769 | 0.4 | 0.87 | 0.795 |
| Bivariate Analysis | ||||||
| MAL Skewness | 0.880 | 1.000 | 0.741 | 0.462 | 1 | 96.436 0.653 |
| Trivariate Analysis | ||||||
| m2Ddr Elongation Skewness | 0.895 | 0.833 | 0.852 | 0.556 | 0.958 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lopez-Rueda, A.; Puig, J.; Thió-Henestrosa, S.; Moreno-Negrete, J.L.; Zwanzger, C.; Pujol, T.; Aldecoa, I.; Pineda, E.; Valduvieco, I.; González, J.J.; et al. Texture Analysis of the Apparent Diffusion Coefficient Focused on Contrast-Enhancing Lesions in Predicting Survival for Bevacizumab-Treated Patients with Recurrent Glioblastoma. Cancers 2023, 15, 3026. https://doi.org/10.3390/cancers15113026
Lopez-Rueda A, Puig J, Thió-Henestrosa S, Moreno-Negrete JL, Zwanzger C, Pujol T, Aldecoa I, Pineda E, Valduvieco I, González JJ, et al. Texture Analysis of the Apparent Diffusion Coefficient Focused on Contrast-Enhancing Lesions in Predicting Survival for Bevacizumab-Treated Patients with Recurrent Glioblastoma. Cancers. 2023; 15(11):3026. https://doi.org/10.3390/cancers15113026
Chicago/Turabian StyleLopez-Rueda, Antonio, Josep Puig, Santiago Thió-Henestrosa, Javier Luis Moreno-Negrete, Christian Zwanzger, Teresa Pujol, Iban Aldecoa, Estela Pineda, Izaskun Valduvieco, José Juan González, and et al. 2023. "Texture Analysis of the Apparent Diffusion Coefficient Focused on Contrast-Enhancing Lesions in Predicting Survival for Bevacizumab-Treated Patients with Recurrent Glioblastoma" Cancers 15, no. 11: 3026. https://doi.org/10.3390/cancers15113026
APA StyleLopez-Rueda, A., Puig, J., Thió-Henestrosa, S., Moreno-Negrete, J. L., Zwanzger, C., Pujol, T., Aldecoa, I., Pineda, E., Valduvieco, I., González, J. J., & Oleaga, L. (2023). Texture Analysis of the Apparent Diffusion Coefficient Focused on Contrast-Enhancing Lesions in Predicting Survival for Bevacizumab-Treated Patients with Recurrent Glioblastoma. Cancers, 15(11), 3026. https://doi.org/10.3390/cancers15113026







