Exploring the Expression of the «Dark Matter» of the Genome in Mesothelioma for Potentially Predictive Biomarkers for Prognosis and Immunotherapy
Abstract
Simple Summary
Abstract
1. Introduction: Why It Is Important to Explore Expressed Genome in Mesothelioma
2. Endogenous Retroviruses Are the TE with Highest Variation in Mesothelioma
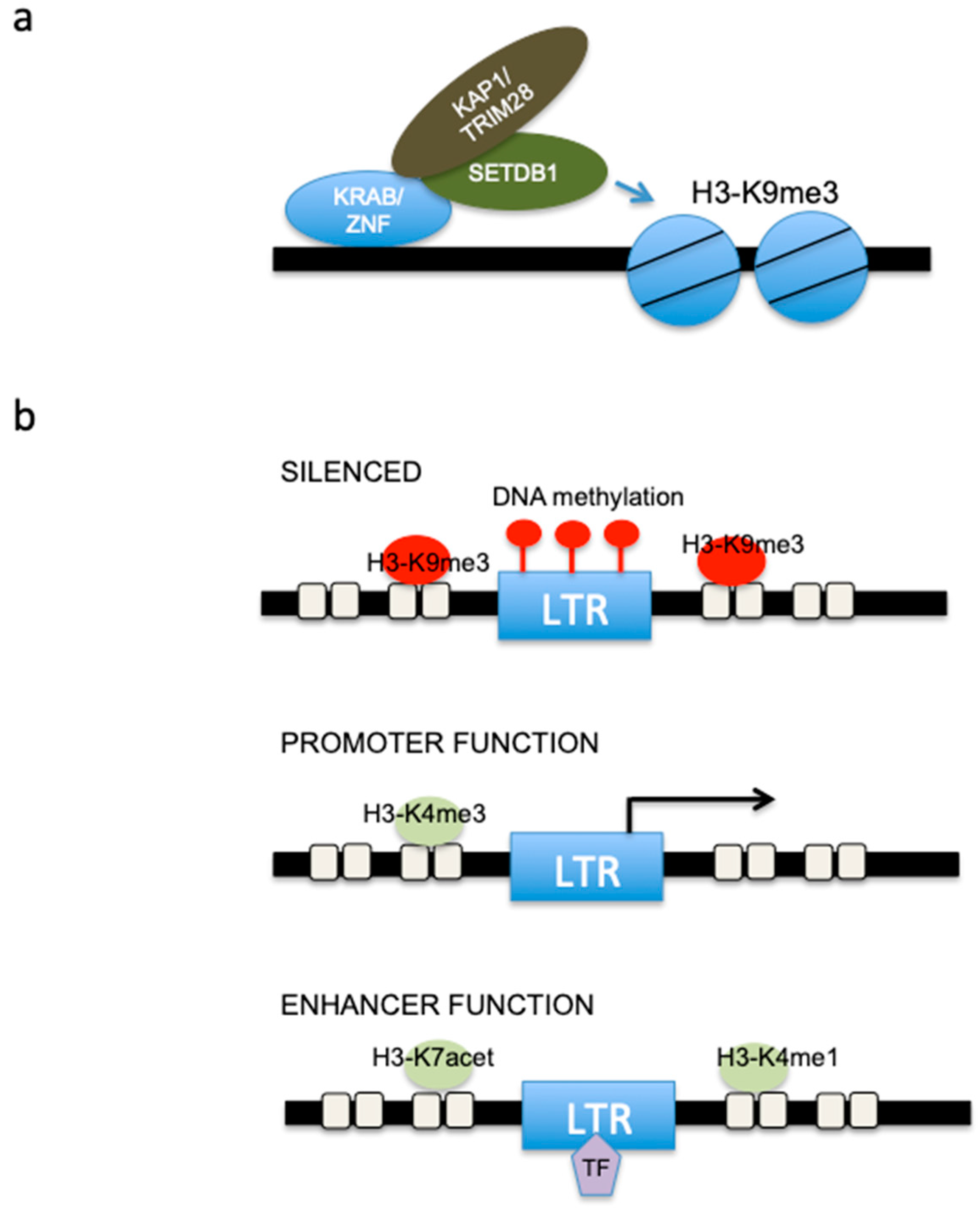
3. ERV and Epigenetic Regulation in Mesothelioma
4. HERV and Type 1 Interferon Activation
5. Conclusions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Felley-Bosco, E.; MacFarlane, M. Asbestos: Modern Insights for Toxicology in the Era of Engineered Nanomaterials. Chem. Res. Toxicol. 2018, 31, 994–1008. [Google Scholar] [CrossRef]
- Carbone, M.; Adusumilli, P.S.; Alexander, H.R., Jr.; Baas, P.; Bardelli, F.; Bononi, A.; Bueno, R.; Felley-Bosco, E.; Galateau-Salle, F.; Jablons, D.; et al. Mesothelioma: Scientific Clues for Prevention, Diagnosis, and Therapy. CA Cancer J. Clin. 2019, 69, 402–429. [Google Scholar] [CrossRef]
- Wagner, J.C. Experimental Production of Mesothelial Tumours of the Pleura by Implantation of Dusts in Laboratory Animals. Nature 1962, 196, 180–181. [Google Scholar] [CrossRef] [PubMed]
- Imielinski, M.; Berger, A.H.; Hammerman, P.S.; Hernandez, B.; Pugh, T.J.; Hodis, E.; Cho, J.; Suh, J.; Capelletti, M.; Sivachenko, A.; et al. Mapping the Hallmarks of Lung Adenocarcinoma with Massively Parallel Sequencing. Cell 2012, 150, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Elsasser, S.J.; Allis, C.D.; Lewis, P.W. Cancer. New Epigenetic Drivers of Cancers. Science 2011, 331, 1145–1146. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Birney, E.; Dunham, I.; Green, E.D.; Gunter, C.; Snyder, M. An Integrated Encyclopedia of DNA Elements in the Human Genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Amaral, P.; Mattick, J. RNA, the Epicenter of Genetic Information; CRC Press: Boca Raton, FL, USA, 2023; ISBN 978-1-00-310924-2. [Google Scholar]
- Smith, J.C.; Sheltzer, J.M. Genome-Wide Identification and Analysis of Prognostic Features in Human Cancers. Cell Rep. 2022, 38, 110569. [Google Scholar] [CrossRef]
- Sun, S.; Frontini, F.; Qi, W.; Hariharan, A.; Ronner, M.; Wipplinger, M.; Blanquart, C.; Rehrauer, H.; Fonteneau, J.F.; Felley-Bosco, E. Endogenous Retrovirus Expression Activates Type-I Interferon Signaling in an Experimental Mouse Model of Mesothelioma Development. Cancer Lett. 2021, 507, 26–38. [Google Scholar] [CrossRef]
- Sun, S. Viral Mimicry Response Is Associated with Clinical Outcome in Pleural Mesothelioma. J. Thorac. Oncol. Clin. Res. Rep. 2022, 3, 100430. [Google Scholar] [CrossRef]
- Mc, C.B. The Origin and Behavior of Mutable Loci in Maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef]
- Britten, R.J.; Davidson, E.H. Gene Regulation for Higher Cells: A Theory. Science 1969, 165, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Lander, E.S.; Linton, L.M.; Birren, B.; Nusbaum, C.; Zody, M.C.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial Sequencing and Analysis of the Human Genome. Nature 2001, 409, 860–921. [Google Scholar] [CrossRef] [PubMed]
- Boeke, J.D.; Garfinkel, D.J.; Styles, C.A.; Fink, G.R. Ty Elements Transpose through an RNA Intermediate. Cell 1985, 40, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Makałowski, W.; Gotea, V.; Pande, A.; Makałowska, I. Transposable Elements: Classification, Identification, and Their Use As a Tool For Comparative Genomics. Methods Mol. Biol. 2019, 1910, 177–207. [Google Scholar] [CrossRef]
- Kramerov, D.A.; Vassetzky, N.S. Origin and Evolution of SINEs in Eukaryotic Genomes. Hered. Edinb. 2011, 107, 487–495. [Google Scholar] [CrossRef]
- Kazazian, H.H., Jr.; Moran, J.V. Mobile DNA in Health and Disease. N. Engl. J. Med. 2017, 377, 361–370. [Google Scholar] [CrossRef]
- Huang, S.; Tao, X.; Yuan, S.; Zhang, Y.; Li, P.; Beilinson, H.A.; Zhang, Y.; Yu, W.; Pontarotti, P.; Escriva, H.; et al. Discovery of an Active RAG Transposon Illuminates the Origins of V(D)J Recombination. Cell 2016, 166, 102–114. [Google Scholar] [CrossRef]
- Kapitonov, V.V.; Jurka, J. RAG1 Core and V(D)J Recombination Signal Sequences Were Derived from Transib Transposons. PLoS Biol. 2005, 3, e181. [Google Scholar] [CrossRef]
- Stocking, C.; Kozak, C.A. Murine Endogenous Retroviruses. Cell Mol. Life Sci. 2008, 65, 3383–3398. [Google Scholar] [CrossRef]
- Hoyt, S.J.; Storer, J.M.; Hartley, G.A.; Grady, P.G.S.; Gershman, A.; de Lima, L.G.; Limouse, C.; Halabian, R.; Wojenski, L.; Rodriguez, M.; et al. From Telomere to Telomere: The Transcriptional and Epigenetic State of Human Repeat Elements. Science 2022, 376, eabk3112. [Google Scholar] [CrossRef]
- Belshaw, R.; Pereira, V.; Katzourakis, A.; Talbot, G.; Paces, J.; Burt, A.; Tristem, M. Long-Term Reinfection of the Human Genome by Endogenous Retroviruses. Proc. Natl. Acad. Sci. USA 2004, 101, 4894–4899. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Perron, H.; Feschotte, C. Variation in Proviral Content among Human Genomes Mediated by LTR Recombination. Mob. DNA 2018, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Vargiu, L.; Rodriguez-Tome, P.; Sperber, G.O.; Cadeddu, M.; Grandi, N.; Blikstad, V.; Tramontano, E.; Blomberg, J. Classification and Characterization of Human Endogenous Retroviruses; Mosaic Forms Are Common. Retrovirology 2016, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Tokuyama, M.; Kong, Y.; Song, E.; Jayewickreme, T.; Kang, I.; Iwasaki, A. ERVmap Analysis Reveals Genome-Wide Transcription of Human Endogenous Retroviruses. Proc. Natl. Acad. Sci. USA 2018, 115, 12565–12572. [Google Scholar] [CrossRef] [PubMed]
- Mager, D.L.; Stoye, J.P. Mammalian Endogenous Retroviruses. Microbiol. Spectr. 2015, 3, Mdna3-0009–2014. [Google Scholar] [CrossRef]
- Gifford, R.; Tristem, M. The Evolution, Distribution and Diversity of Endogenous Retroviruses. Virus Genes 2003, 26, 291–315. [Google Scholar] [CrossRef]
- Hmeljak, J.; Sanchez-Vega, F.; Hoadley, K.A.; Shih, J.; Stewart, C.; Heiman, D.; Tarpey, P.; Danilova, L.; Drill, E.; Gibb, E.A.; et al. Integrative Molecular Characterization of Malignant Pleural Mesothelioma. Cancer Discov. 2018, 8, 1548–1565. [Google Scholar] [CrossRef]
- Bueno, R.; Stawiski, E.W.; Goldstein, L.D.; Durinck, S.; De Rienzo, A.; Modrusan, Z.; Gnad, F.; Nguyen, T.T.; Jaiswal, B.S.; Chirieac, L.R.; et al. Comprehensive Genomic Analysis of Malignant Pleural Mesothelioma Identifies Recurrent Mutations, Gene Fusions and Splicing Alterations. Nat. Genet. 2016, 48, 407–416. [Google Scholar] [CrossRef]
- de Reynies, A.; Jaurand, M.C.; Renier, A.; Couchy, G.; Hysi, I.; Elarouci, N.; Galateau-Salle, F.; Copin, M.C.; Hofman, P.; Cazes, A.; et al. Molecular Classification of Malignant Pleural Mesothelioma: Identification of a Poor Prognosis Subgroup Linked to the Epithelial-to-Mesenchymal Transition. Clin. Cancer Res. 2014, 20, 1323–1334. [Google Scholar] [CrossRef]
- Quetel, L.; Meiller, C.; Assié, J.B.; Blum, Y.; Imbeaud, S.; Montagne, F.; Tranchant, R.; de Wolf, J.; Caruso, S.; Copin, M.C.; et al. Genetic Alterations of Malignant Pleural Mesothelioma: Association with Tumor Heterogeneity and Overall Survival. Mol. Oncol. 2020, 14, 1207–1223. [Google Scholar] [CrossRef]
- Colunga, T.; Hayworth, M.; Kress, S.; Reynolds, D.M.; Chen, L.; Nazor, K.L.; Baur, J.; Singh, A.M.; Loring, J.F.; Metzger, M.; et al. Human Pluripotent Stem Cell-Derived Multipotent Vascular Progenitors of the Mesothelium Lineage Have Utility in Tissue Engineering and Repair. Cell Rep. 2019, 26, 2566–2579.e10. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhou, Z.; Fei, L.; Sun, H.; Wang, R.; Chen, Y.; Chen, H.; Wang, J.; Tang, H.; Ge, W.; et al. Construction of a Human Cell Landscape at Single-Cell Level. Nature 2020, 581, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Solovyov, A.; Vabret, N.; Arora, K.S.; Snyder, A.; Funt, S.A.; Bajorin, D.F.; Rosenberg, J.E.; Bhardwaj, N.; Ting, D.T.; Greenbaum, B.D. Global Cancer Transcriptome Quantifies Repeat Element Polarization between Immunotherapy Responsive and T Cell Suppressive Classes. Cell Rep. 2018, 23, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Grosso, S.; Marini, A.; Gyuraszova, K.; Voorde, J.V.; Sfakianos, A.; Garland, G.D.; Tenor, A.R.; Mordue, R.; Chernova, T.; Morone, N.; et al. The Pathogenesis of Mesothelioma Is Driven by a Dysregulated Translatome. Nat. Commun. 2021, 12, 4920. [Google Scholar] [CrossRef]
- Kasperek, A.; Béguin, A.; Bawa, O.; De Azevedo, K.; Job, B.; Massard, C.; Scoazec, J.; Heidmann, T.; Heidmann, O. Therapeutic Potential of the Human Endogenous Retroviral Envelope Protein HEMO: A Pan-cancer Analysis. Mol. Oncol. 2022, 16, 1451–1473. [Google Scholar] [CrossRef]
- Goke, J.; Lu, X.; Chan, Y.S.; Ng, H.H.; Ly, L.H.; Sachs, F.; Szczerbinska, I. Dynamic Transcription of Distinct Classes of Endogenous Retroviral Elements Marks Specific Populations of Early Human Embryonic Cells. Cell Stem. Cell 2015, 16, 135–141. [Google Scholar] [CrossRef]
- Geis, F.K.; Goff, S.P. Silencing and Transcriptional Regulation of Endogenous Retroviruses: An Overview. Viruses 2020, 12, 884. [Google Scholar] [CrossRef]
- Xue, B.; Sechi, L.A.; Kelvin, D.J. Human Endogenous Retrovirus K (HML-2) in Health and Disease. Front. Microbiol. 2020, 11, 1690. [Google Scholar] [CrossRef]
- Garcia-Montojo, M.; Doucet-O’Hare, T.; Henderson, L.; Nath, A. Human Endogenous Retrovirus-K (HML-2): A Comprehensive Review. Crit. Rev. Microbiol. 2018, 44, 715–738. [Google Scholar] [CrossRef]
- Hughes, J.F.; Coffin, J.M. Human Endogenous Retrovirus K Solo-LTR Formation and Insertional Polymorphisms: Implications for Human and Viral Evolution. Proc. Natl. Acad. Sci. USA 2004, 101, 1668–1672. [Google Scholar] [CrossRef]
- Schmitt, K.; Reichrath, J.; Roesch, A.; Meese, E.; Mayer, J. Transcriptional Profiling of Human Endogenous Retrovirus Group HERV-K(HML-2) Loci in Melanoma. Genome Biol. Evol. 2013, 5, 307–328. [Google Scholar] [CrossRef] [PubMed]
- Rowe, H.M.; Trono, D. Dynamic Control of Endogenous Retroviruses during Development. Virology 2011, 411, 273–287. [Google Scholar] [CrossRef]
- Friedli, M.; Trono, D. The Developmental Control of Transposable Elements and the Evolution of Higher Species. Annu. Rev. Cell Dev. Biol. 2015, 31, 429–451. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.C.; Lorincz, M.C. Silencing of Endogenous Retroviruses: When and Why Do Histone Marks Predominate? Trends Biochem. Sci. 2012, 37, 127–133. [Google Scholar] [CrossRef]
- Pisano, M.P.; Grandi, N.; Tramontano, E. High-Throughput Sequencing Is a Crucial Tool to Investigate the Contribution of Human Endogenous Retroviruses (HERVs) to Human Biology and Development. Viruses 2020, 12, 633. [Google Scholar] [CrossRef]
- Hurst, T.P.; Magiorkinis, G. Epigenetic Control of Human Endogenous Retrovirus Expression: Focus on Regulation of Long-Terminal Repeats (LTRs). Viruses 2017, 9, 130. [Google Scholar] [CrossRef] [PubMed]
- Howard, G.; Eiges, R.; Gaudet, F.; Jaenisch, R.; Eden, A. Activation and Transposition of Endogenous Retroviral Elements in Hypomethylation Induced Tumors in Mice. Oncogene 2008, 27, 404–408. [Google Scholar] [CrossRef]
- Jaenisch, R.; Schnieke, A.; Harbers, K. Treatment of Mice with 5-Azacytidine Efficiently Activates Silent Retroviral Genomes in Different Tissues. Proc. Natl. Acad. Sci. USA 1985, 82, 1451–1455. [Google Scholar] [CrossRef]
- Bourc’his, D.; Bestor, T.H. Meiotic Catastrophe and Retrotransposon Reactivation in Male Germ Cells Lacking Dnmt3L. Nature 2004, 431, 96–99. [Google Scholar] [CrossRef]
- Seifarth, W.; Frank, O.; Zeilfelder, U.; Spiess, B.; Greenwood, A.D.; Hehlmann, R.; Leib-Mösch, C. Comprehensive Analysis of Human Endogenous Retrovirus Transcriptional Activity in Human Tissues with a Retrovirus-Specific Microarray. J. Virol. 2005, 79, 341–352. [Google Scholar] [CrossRef]
- de Cubas, A.A.; Dunker, W.; Zaninovich, A.; Hongo, R.A.; Bhatia, A.; Panda, A.; Beckermann, K.E.; Bhanot, G.; Ganesan, S.; Karijolich, J.; et al. DNA Hypomethylation Promotes Transposable Element Expression and Activation of Immune Signaling in Renal Cell Cancer. JCI Insight 2020, 5, 137569. [Google Scholar] [CrossRef] [PubMed]
- Lavie, L.; Kitova, M.; Maldener, E.; Meese, E.; Mayer, J. CpG Methylation Directly Regulates Transcriptional Activity of the Human Endogenous Retrovirus Family HERV-K(HML-2). J. Virol. 2005, 79, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Stengel, S.; Fiebig, U.; Kurth, R.; Denner, J. Regulation of Human Endogenous Retrovirus-K Expression in Melanomas by CpG Methylation. Genes. Chromosomes Cancer 2010, 49, 401–411. [Google Scholar] [CrossRef]
- Xie, Q.; Chen, S.; Tian, R.; Huang, X.; Deng, R.; Xue, B.; Qin, Y.; Xu, Y.; Wang, J.; Guo, M.; et al. Long Noncoding RNA ITPRIP-1 Positively Regulates the Innate Immune Response through Promotion of Oligomerization and Activation of MDA5. J. Virol. 2018, 92, e00507-18. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Rose, C.M.; Cass, A.A.; Williams, A.G.; Darwish, M.; Lianoglou, S.; Haverty, P.M.; Tong, A.J.; Blanchette, C.; Albert, M.L.; et al. Transposable Element Expression in Tumors Is Associated with Immune Infiltration and Increased Antigenicity. Nat. Commun. 2019, 10, 5228. [Google Scholar] [CrossRef]
- Ren, Y.R.; Patel, K.; Paun, B.C.; Kern, S.E. Structural Analysis of the Cancer-Specific Promoter in Mesothelin and in Other Genes Overexpressed in Cancers. J. Biol. Chem. 2011, 286, 11960–11969. [Google Scholar] [CrossRef]
- Nelson, H.H.; Almquist, L.M.; LaRocca, J.L.; Plaza, S.L.; Lambert-Messerlian, G.M.; Sugarbaker, D.J.; Bueno, R.; Godleski, J.J.; Marsit, C.J.; Christensen, B.C.; et al. The Relationship between Tumor MSLN Methylation and Serum Mesothelin (SMRP) in Mesothelioma. Epigenetics 2011, 6, 1029–1034. [Google Scholar] [CrossRef]
- Robinson, C.; van Bruggen, I.; Segal, A.; Dunham, M.; Sherwood, A.; Koentgen, F.; Robinson, B.W.; Lake, R.A. A Novel SV40 TAg Transgenic Model of Asbestos-Induced Mesothelioma: Malignant Transformation Is Dose Dependent. Cancer Res. 2006, 66, 10786–10794. [Google Scholar] [CrossRef]
- Zhang, S.M.; Cai, W.L.; Liu, X.; Thakral, D.; Luo, J.; Chan, L.H.; McGeary, M.K.; Song, E.; Blenman, K.R.M.; Micevic, G.; et al. KDM5B Promotes Immune Evasion by Recruiting SETDB1 to Silence Retroelements. Nature 2021, 598, 682–687. [Google Scholar] [CrossRef]
- Karimi, M.M.; Goyal, P.; Maksakova, I.A.; Bilenky, M.; Leung, D.; Tang, J.X.; Shinkai, Y.; Mager, D.L.; Jones, S.; Hirst, M.; et al. DNA Methylation and SETDB1/H3K9me3 Regulate Predominantly Distinct Sets of Genes, Retroelements, and Chimeric Transcripts in MESCs. Cell Stem Cell 2011, 8, 676–687. [Google Scholar] [CrossRef]
- Fukuda, K.; Shinkai, Y. SETDB1-Mediated Silencing of Retroelements. Viruses 2020, 12, 596. [Google Scholar] [CrossRef]
- Pontis, J.; Planet, E.; Offner, S.; Turelli, P.; Duc, J.; Coudray, A.; Theunissen, T.W.; Jaenisch, R.; Trono, D. Hominoid-Specific Transposable Elements and KZFPs Facilitate Human Embryonic Genome Activation and Control Transcription in Naive Human ESCs. Cell Stem Cell 2019, 24, 724–735.e5. [Google Scholar] [CrossRef]
- Lorenzini, E.; Torricelli, F.; Zamponi, R.; Donati, B.; Manicardi, V.; Sauta, E.; Faria do Valle, I.; Reggiani, F.; Gugnoni, M.; Manzotti, G.; et al. KAP1 Is a New Non-Genetic Vulnerability of Malignant Pleural Mesothelioma (MPM). NAR Cancer 2022, 4, zcac024. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Strogantsev, R.; Takahashi, N.; Kazachenka, A.; Lorincz, M.C.; Hemberger, M.; Ferguson-Smith, A.C. ZFP57 Regulation of Transposable Elements and Gene Expression within and beyond Imprinted Domains. Epigenet. Chromatin 2019, 12, 49. [Google Scholar] [CrossRef] [PubMed]
- Turelli, P.; Castro-Diaz, N.; Marzetta, F.; Kapopoulou, A.; Raclot, C.; Duc, J.; Tieng, V.; Quenneville, S.; Trono, D. Interplay of TRIM28 and DNA Methylation in Controlling Human Endogenous Retroelements. Genome Res. 2014, 24, 1260–1270. [Google Scholar] [CrossRef] [PubMed]
- Frietze, S.; O’Geen, H.; Blahnik, K.R.; Jin, V.X.; Farnham, P.J. ZNF274 Recruits the Histone Methyltransferase SETDB1 to the 3’ Ends of ZNF Genes. PLoS ONE 2010, 5, e15082. [Google Scholar] [CrossRef]
- Carter, T.A.; Singh, M.; Dumbovic, G.; Chobirko, J.D.; Rinn, J.L.; Feschotte, C. Mosaic Cis-Regulatory Evolution Drives Transcriptional Partitioning of HERVH Endogenous Retrovirus in the Human Embryo. eLife 2022, 11, 76257. [Google Scholar] [CrossRef]
- Cuellar, T.L.; Herzner, A.M.; Zhang, X.; Goyal, Y.; Watanabe, C.; Friedman, B.A.; Janakiraman, V.; Durinck, S.; Stinson, J.; Arnott, D.; et al. Silencing of Retrotransposons by SETDB1 Inhibits the Interferon Response in Acute Myeloid Leukemia. J. Cell Biol. 2017, 216, 3535–3549. [Google Scholar] [CrossRef]
- Pagano, M.; Ceresoli, L.G.; Zucali, P.A.; Pasello, G.; Garassino, M.; Grosso, F.; Tiseo, M.; Soto Parra, H.; Zanelli, F.; Cappuzzo, F.; et al. Mutational Profile of Malignant Pleural Mesothelioma (MPM) in the Phase II RAMES Study. Cancers 2020, 12, 2948. [Google Scholar] [CrossRef]
- Kang, H.C.; Kim, H.K.; Lee, S.; Mendez, P.; Kim, J.W.; Woodard, G.; Yoon, J.H.; Jen, K.Y.; Fang, L.T.; Jones, K.; et al. Whole Exome and Targeted Deep Sequencing Identify Genome-Wide Allelic Loss and Frequent SETDB1 Mutations in Malignant Pleural Mesotheliomas. Oncotarget 2016, 7, 8321–8331. [Google Scholar] [CrossRef]
- Rajagopalan, D.; Tirado-Magallanes, R.; Bhatia, S.S.; Teo, W.S.; Sian, S.; Hora, S.; Lee, K.K.; Zhang, Y.; Jadhav, S.P.; Wu, Y.; et al. TIP60 Represses Activation of Endogenous Retroviral Elements. Nucleic Acids Res. 2018, 46, 9456–9470. [Google Scholar] [CrossRef] [PubMed]
- Huda, A.; Bowen, N.J.; Conley, A.B.; Jordan, I.K. Epigenetic Regulation of Transposable Element Derived Human Gene Promoters. Gene 2011, 475, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Hong, C.; Zhang, B.; Lowdon, R.F.; Xing, X.; Li, D.; Zhou, X.; Lee, H.J.; Maire, C.L.; Ligon, K.L.; et al. DNA Hypomethylation within Specific Transposable Element Families Associates with Tissue-Specific Enhancer Landscape. Nat. Genet. 2013, 45, 836–841. [Google Scholar] [CrossRef] [PubMed]
- Rowe, H.M.; Kapopoulou, A.; Corsinotti, A.; Fasching, L.; Macfarlan, T.S.; Tarabay, Y.; Viville, S.; Jakobsson, J.; Pfaff, S.L.; Trono, D. TRIM28 Repression of Retrotransposon-Based Enhancers Is Necessary to Preserve Transcriptional Dynamics in Embryonic Stem Cells. Genome Res. 2013, 23, 452–461. [Google Scholar] [CrossRef]
- Grandi, N.; Pisano, M.P.; Tramontano, E. The Emerging Field of Human Endogenous Retroviruses: Understanding Their Physiological Role and Contribution to Diseases. Future Virol. 2019, 14, 441–444. [Google Scholar] [CrossRef]
- Blond, J.L.; Lavillette, D.; Cheynet, V.; Bouton, O.; Oriol, G.; Chapel-Fernandes, S.; Mandrand, B.; Mallet, F.; Cosset, F.L. An Envelope Glycoprotein of the Human Endogenous Retrovirus HERV-W Is Expressed in the Human Placenta and Fuses Cells Expressing the Type D Mammalian Retrovirus Receptor. J. Virol. 2000, 74, 3321–3329. [Google Scholar] [CrossRef]
- Mi, S.; Lee, X.; Li, X.; Veldman, G.M.; Finnerty, H.; Racie, L.; LaVallie, E.; Tang, X.Y.; Edouard, P.; Howes, S.; et al. Syncytin Is a Captive Retroviral Envelope Protein Involved in Human Placental Morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef]
- Lavialle, C.; Cornelis, G.; Dupressoir, A.; Esnault, C.; Heidmann, O.; Vernochet, C.; Heidmann, T. Paleovirology of “Syncytins”, Retroviral Env Genes Exapted for a Role in Placentation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20120507. [Google Scholar] [CrossRef]
- Dupressoir, A.; Lavialle, C.; Heidmann, T. From Ancestral Infectious Retroviruses to Bona Fide Cellular Genes: Role of the Captured Syncytins in Placentation. Placenta 2012, 33, 663–671. [Google Scholar] [CrossRef]
- Griffiths, D.J. Endogenous Retroviruses in the Human Genome Sequence. Genome Biol. 2001, 2, REVIEWS1017. [Google Scholar] [CrossRef]
- Grow, E.J.; Flynn, R.A.; Chavez, S.L.; Bayless, N.L.; Wossidlo, M.; Wesche, D.J.; Martin, L.; Ware, C.B.; Blish, C.A.; Chang, H.Y.; et al. Intrinsic Retroviral Reactivation in Human Preimplantation Embryos and Pluripotent Cells. Nature 2015, 522, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Broad Institute TCGA Genome Data Analysis Center (2014): Correlation between Aggregated Molecular Cancer Subtypes and Selected Clinical Features. Broad Institute of MIT and Harvard. Available online: http://gdac.broadinstitute.org/runs/analyses__2014_10_17/reports/cancer/MESO-TP/Correlate_Clinical_vs_Molecular_Subtypes/nozzle.html (accessed on 12 May 2015).
- Hiltbrunner, S.; Fleischmann, Z.; Sokol, E.S.; Zoche, M.; Felley-Bosco, E.; Curioni Fontecedro, A. Genomic Landscape of Pleural and Peritoneal Mesothelioma Tumors. Br. J. Cancer 2022, 127, 1997–2005. [Google Scholar] [CrossRef] [PubMed]
- Mangiante, L.; Alcala, N.; Sexton-Oates, A.; Di Genova, A.; Gonzalez-Perez, A.; Khandekar, A.; Bergstrom, E.N.; Kim, J.; Liu, X.; Blazquez-Encinas, R.; et al. Multiomic Analysis of Malignant Pleural Mesothelioma Identifies Molecular Axes and Specialized Tumor Profiles Driving Intertumor Heterogeneity. Nat. Genet. 2023, 55, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Delaunay, T.; Achard, C.; Boisgerault, N.; Grard, M.; Petithomme, T.; Chatelain, C.; Dutoit, S.; Blanquart, C.; Royer, P.J.; Minvielle, S.; et al. Frequent Homozygous Deletions of Type I Interferon Genes in Pleural Mesothelioma Confer Sensitivity to Oncolytic Measles Virus. J. Thorac. Oncol. 2020, 15, 827–842. [Google Scholar] [CrossRef]
- Roulois, D.; Loo Yau, H.; Singhania, R.; Wang, Y.; Danesh, A.; Shen, S.Y.; Han, H.; Liang, G.; Jones, P.A.; Pugh, T.J.; et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell 2015, 162, 961–973. [Google Scholar] [CrossRef] [PubMed]
- Chiappinelli, K.B.; Strissel, P.L.; Desrichard, A.; Li, H.; Henke, C.; Akman, B.; Hein, A.; Rote, N.S.; Cope, L.M.; Snyder, A.; et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via DsRNA Including Endogenous Retroviruses. Cell 2015, 162, 974–986. [Google Scholar] [CrossRef]
- Felley-Bosco, E. Non-Coding RNA Regulatory Networks in Mesothelioma: A Narrative Review of Their Implication in Innate Immune Signaling Pathway. Precis Cancer Med. 2021, 4, pcm-21-4. [Google Scholar] [CrossRef]
- Chen, R.; Ishak, C.A.; De Carvalho, D.D. Endogenous Retroelements and the Viral Mimicry Response in Cancer Therapy and Cellular Homeostasis. Cancer Discov. 2021, 11, 2707–2725. [Google Scholar] [CrossRef]
- Peters, S.; Scherpereel, A.; Cornelissen, R.; Oulkhouir, Y.; Greillier, L.; Kaplan, M.A.; Talbot, T.; Monnet, I.; Hiret, S.; Baas, P.; et al. First-Line Nivolumab plus Ipilimumab versus Chemotherapy in Patients with Unresectable Malignant Pleural Mesothelioma: 3-Year Outcomes from CheckMate 743. Ann. Oncol. 2022, 33, 488–499. [Google Scholar] [CrossRef]
- Mayer, J.; Blomberg, J.; Seal, R.L. A Revised Nomenclature for Transcribed Human Endogenous Retroviral Loci. Mob. DNA 2011, 2, 7. [Google Scholar] [CrossRef]
- Panda, A.; de Cubas, A.A.; Stein, M.; Riedlinger, G.; Kra, J.; Mayer, T.; Smith, C.C.; Vincent, B.G.; Serody, J.S.; Beckermann, K.E.; et al. Endogenous Retrovirus Expression Is Associated with Response to Immune Checkpoint Blockade in Clear Cell Renal Cell Carcinoma. JCI Insight 2018, 3, 121522. [Google Scholar] [CrossRef] [PubMed]
- Au, L.; Hatipoglu, E.; Robert de Massy, M.; Litchfield, K.; Beattie, G.; Rowan, A.; Schnidrig, D.; Thompson, R.; Byrne, F.; Horswell, S.; et al. Determinants of Anti-PD-1 Response and Resistance in Clear Cell Renal Cell Carcinoma. Cancer Cell 2021, 39, 1497–1518.e11. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.C.; Beckermann, K.E.; Bortone, D.S.; De Cubas, A.A.; Bixby, L.M.; Lee, S.J.; Panda, A.; Ganesan, S.; Bhanot, G.; Wallen, E.M.; et al. Endogenous Retroviral Signatures Predict Immunotherapy Response in Clear Cell Renal Cell Carcinoma. J. Clin. Investig. 2018, 128, 4804–4820. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.A.; Hou, Y.; Bakouny, Z.; Ficial, M.; Sant’ Angelo, M.; Forman, J.; Ross-Macdonald, P.; Berger, A.C.; Jegede, O.A.; Elagina, L.; et al. Interplay of Somatic Alterations and Immune Infiltration Modulates Response to PD-1 Blockade in Advanced Clear Cell Renal Cell Carcinoma. Nat. Med. 2020, 26, 909–918. [Google Scholar] [CrossRef]
- Wolf, M.M.; Rathmell, W.K.; De Cubas, A.A. Immunogenicity in Renal Cell Carcinoma: Shifting Focus to Alternative Sources of Tumour-Specific Antigens. Nat. Rev. Nephrol. 2023; Online ahead of print. [Google Scholar] [CrossRef]
- Badal, B.; Solovyov, A.; Di Cecilia, S.; Chan, J.M.; Chang, L.W.; Iqbal, R.; Aydin, I.T.; Rajan, G.S.; Chen, C.; Abbate, F.; et al. Transcriptional Dissection of Melanoma Identifies a High-Risk Subtype Underlying TP53 Family Genes and Epigenome Deregulation. JCI Insight 2017, 2, 92102. [Google Scholar] [CrossRef]
- Lecuelle, J.; Favier, L.; Fraisse, C.; Lagrange, A.; Kaderbhai, C.; Boidot, R.; Chevrier, S.; Joubert, P.; Routy, B.; Truntzer, C.; et al. MER4 Endogenous Retrovirus Correlated with Better Efficacy of Anti-PD1/PD-L1 Therapy in Non-Small Cell Lung Cancer. J. Immunother. Cancer 2022, 10, e004241. [Google Scholar] [CrossRef]
- Ng, K.W.; Boumelha, J.; Enfield, K.S.S.; Almagro, J.; Cha, H.; Pich, O.; Karasaki, T.; Moore, D.A.; Salgado, R.; Sivakumar, M.; et al. Antibodies against Endogenous Retroviruses Promote Lung Cancer Immunotherapy. Nature 2023, 616, 563–573. [Google Scholar] [CrossRef]
- Sterman, D.H.; Recio, A.; Haas, A.R.; Vachani, A.; Katz, S.I.; Gillespie, C.T.; Cheng, G.; Sun, J.; Moon, E.; Pereira, L.; et al. A Phase I Trial of Repeated Intrapleural Adenoviral-Mediated Interferon-Beta Gene Transfer for Mesothelioma and Metastatic Pleural Effusions. Mol. Ther. 2010, 18, 852–860. [Google Scholar] [CrossRef]
- Vanbervliet-Defrance, B.; Delaunay, T.; Daunizeau, T.; Kepenekian, V.; Glehen, O.; Weber, K.; Estornes, Y.; Ziverec, A.; Djemal, L.; Delphin, M.; et al. Cisplatin Unleashes Toll-like Receptor 3-Mediated Apoptosis through the Downregulation of c-FLIP in Malignant Mesothelioma. Cancer Lett. 2020, 472, 29–39. [Google Scholar] [CrossRef]
- Achard, C.; Boisgerault, N.; Delaunay, T.; Roulois, D.; Nedellec, S.; Royer, P.J.; Pain, M.; Combredet, C.; Mesel-Lemoine, M.; Cellerin, L.; et al. Sensitivity of Human Pleural Mesothelioma to Oncolytic Measles Virus Depends on Defects of the Type I Interferon Response. Oncotarget 2015, 6, 44892–44904. [Google Scholar] [CrossRef]
- Baas, P.; Scherpereel, A.; Nowak, A.K.; Fujimoto, N.; Peters, S.; Tsao, A.S.; Mansfield, A.S.; Popat, S.; Jahan, T.; Antonia, S.; et al. First-Line Nivolumab plus Ipilimumab in Unresectable Malignant Pleural Mesothelioma (CheckMate 743): A Multicentre, Randomised, Open-Label, Phase 3 Trial. Lancet 2021, 397, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.M.; Jang, H.J.; Liang, Y.; Maeng, J.H.; Tzeng, S.-C.; Wu, A.; Basri, N.L.; Qu, X.; Fan, C.; Li, A.; et al. Pan-Cancer Analysis Identifies Tumor-Specific Antigens Derived from Transposable Elements. Nat. Genet. 2023, 55, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Dumoulin, D.W.; Cornelissen, R.; Bezemer, K.; Baart, S.J.; Aerts, J.G.J.V. Long-Term Follow-Up of Mesothelioma Patients Treated with Dendritic Cell Therapy in Three Phase I/II Trials. Vaccines 2021, 9, 525. [Google Scholar] [CrossRef] [PubMed]
- Jansz, N.; Faulkner, G.J. Endogenous Retroviruses in the Origins and Treatment of Cancer. Genome Biol. 2021, 22, 147. [Google Scholar] [CrossRef]
- Warming, S.; Liu, P.; Suzuki, T.; Akagi, K.; Lindtner, S.; Pavlakis, G.N.; Jenkins, N.A.; Copeland, N.G. Evi3, a Common Retroviral Integration Site in Murine B-Cell Lymphoma, Encodes an EBFAZ-Related Krüppel-like Zinc Finger Protein. Blood 2003, 101, 1934–1940. [Google Scholar] [CrossRef] [PubMed]
- Tomlins, S.A.; Laxman, B.; Dhanasekaran, S.M.; Helgeson, B.E.; Cao, X.; Morris, D.S.; Menon, A.; Jing, X.; Cao, Q.; Han, B.; et al. Distinct Classes of Chromosomal Rearrangements Create Oncogenic ETS Gene Fusions in Prostate Cancer. Nature 2007, 448, 595–599. [Google Scholar] [CrossRef] [PubMed]
- Shahid, S.; Slotkin, R.K. The Current Revolution in Transposable Element Biology Enabled by Long Reads. Curr. Opin. Plant Biol. 2020, 54, 49–56. [Google Scholar] [CrossRef]
- Perrino, M.; De Vincenzo, F.; Cordua, N.; Borea, F.; Aliprandi, M.; Santoro, A.; Zucali, P.A. Immunotherapy with Immune Checkpoint Inhibitors and Predictive Biomarkers in Malignant Mesothelioma: Work Still in Progress. Front. Immunol. 2023, 14, 1121557. [Google Scholar] [CrossRef]

| Class | Related Exogenous Group | Group |
|---|---|---|
| I | Gamma | HERV1, HERV-H *, HERV-W *, HERV-E *, ERVMER34-1 |
| II | Beta | HERV-K * |
| III | Spuma | HERV-L *, MaLR |
| ERV | Cancer | Biomarker Type | Reference |
|---|---|---|---|
| ERVmap_1248 | Mesothelioma | Prognostic | [10] |
| ERV3-2 | Clear cell renal cell carcinoma | Immunotherapy response | [93,94] |
| ERV_4700 | Clear cell renal cell carcinoma | Immunotherapy response | [95] |
| ERV_2282 | Clear cell renal cell carcinoma | Prognostic | [96] |
| ERVmap_2637 | Melanoma | Immunotherapy response | [60] |
| MER4 | Non-small cell lung cancer | Prognostic and immunotherapy response | [99] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Felley-Bosco, E. Exploring the Expression of the «Dark Matter» of the Genome in Mesothelioma for Potentially Predictive Biomarkers for Prognosis and Immunotherapy. Cancers 2023, 15, 2969. https://doi.org/10.3390/cancers15112969
Felley-Bosco E. Exploring the Expression of the «Dark Matter» of the Genome in Mesothelioma for Potentially Predictive Biomarkers for Prognosis and Immunotherapy. Cancers. 2023; 15(11):2969. https://doi.org/10.3390/cancers15112969
Chicago/Turabian StyleFelley-Bosco, Emanuela. 2023. "Exploring the Expression of the «Dark Matter» of the Genome in Mesothelioma for Potentially Predictive Biomarkers for Prognosis and Immunotherapy" Cancers 15, no. 11: 2969. https://doi.org/10.3390/cancers15112969
APA StyleFelley-Bosco, E. (2023). Exploring the Expression of the «Dark Matter» of the Genome in Mesothelioma for Potentially Predictive Biomarkers for Prognosis and Immunotherapy. Cancers, 15(11), 2969. https://doi.org/10.3390/cancers15112969





