A Reconstructed Human Melanoma-in-Skin Model to Study Immune Modulatory and Angiogenic Mechanisms Facilitating Initial Melanoma Growth and Invasion
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Blood and Tissue Collection
2.2. Cell Isolation and Culture
2.2.1. Primary Skin Cells
2.2.2. Melanoma Cell Lines
2.2.3. Endothelial Cells
2.2.4. Monocytes
2.3. Construction of Reconstructed Human Skin with or without Melanoma Cells
2.4. (Immuno)histochemistry
2.5. Measurement of RhS and Mel-RhS Contraction
2.6. Measurement of Cytokine Secretion in Culture Supernatant
2.7. Sprouting Assay
2.8. Monocyte Exposure to RhS- and Mel-RhS-Derived Culture Supernatants
2.9. Flow Cytometry
2.10. Statistical Analysis
3. Results
3.1. Melanoma Cell Lines Recapitulate Different Stages of the Disease in a 3D Human Melanoma-in-Skin Model
3.2. Cytokine and Chemokine Release Profiles Differ between Mel-RhS Models
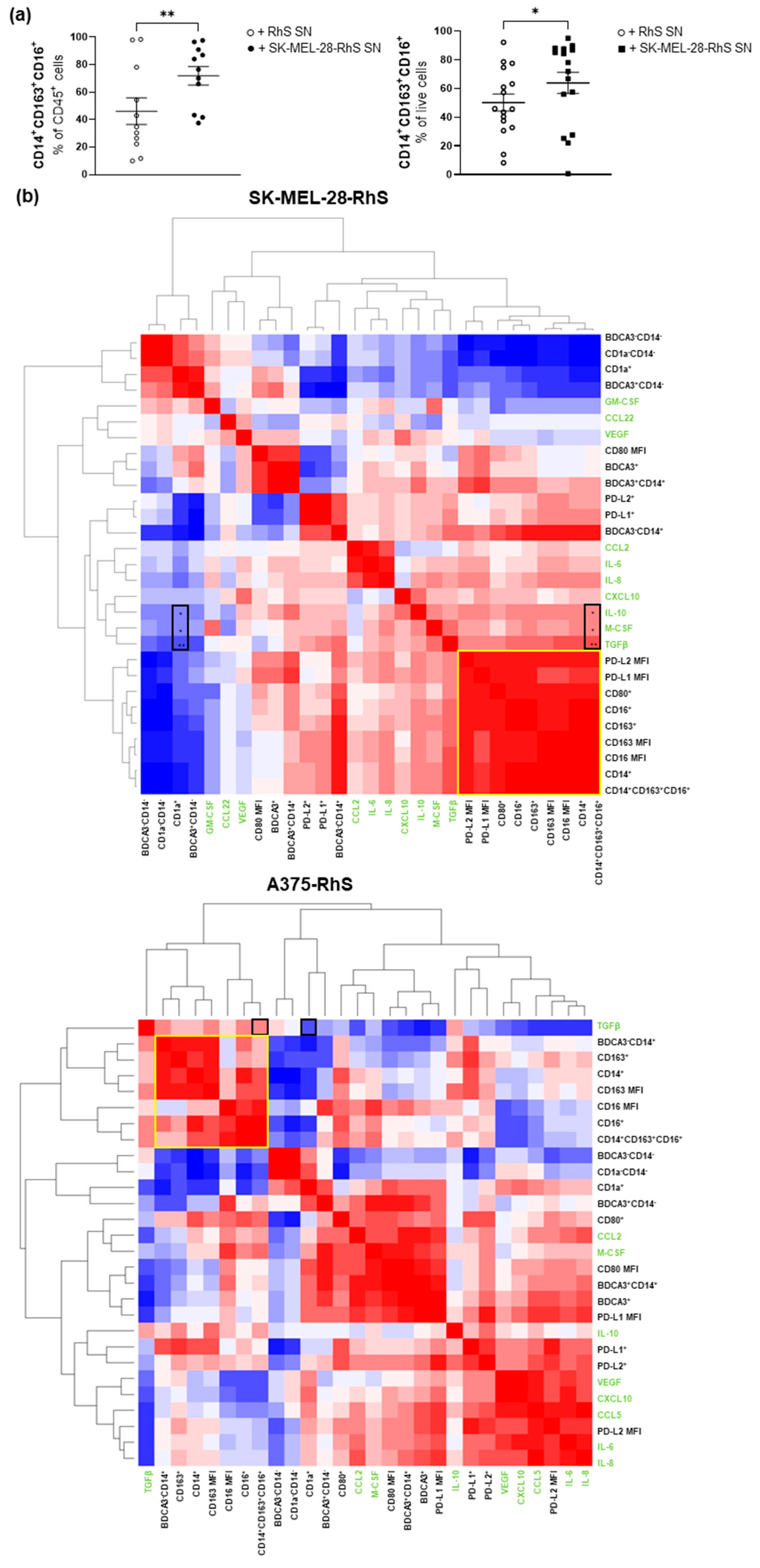
3.3. SK-MEL-28-RhS and A375-RhS Suppress Monocyte-to-Dendritic Cell Differentiation through the Release of Soluble Factors: Relative Contributions of IL-10, M-CSF, and TGFβ
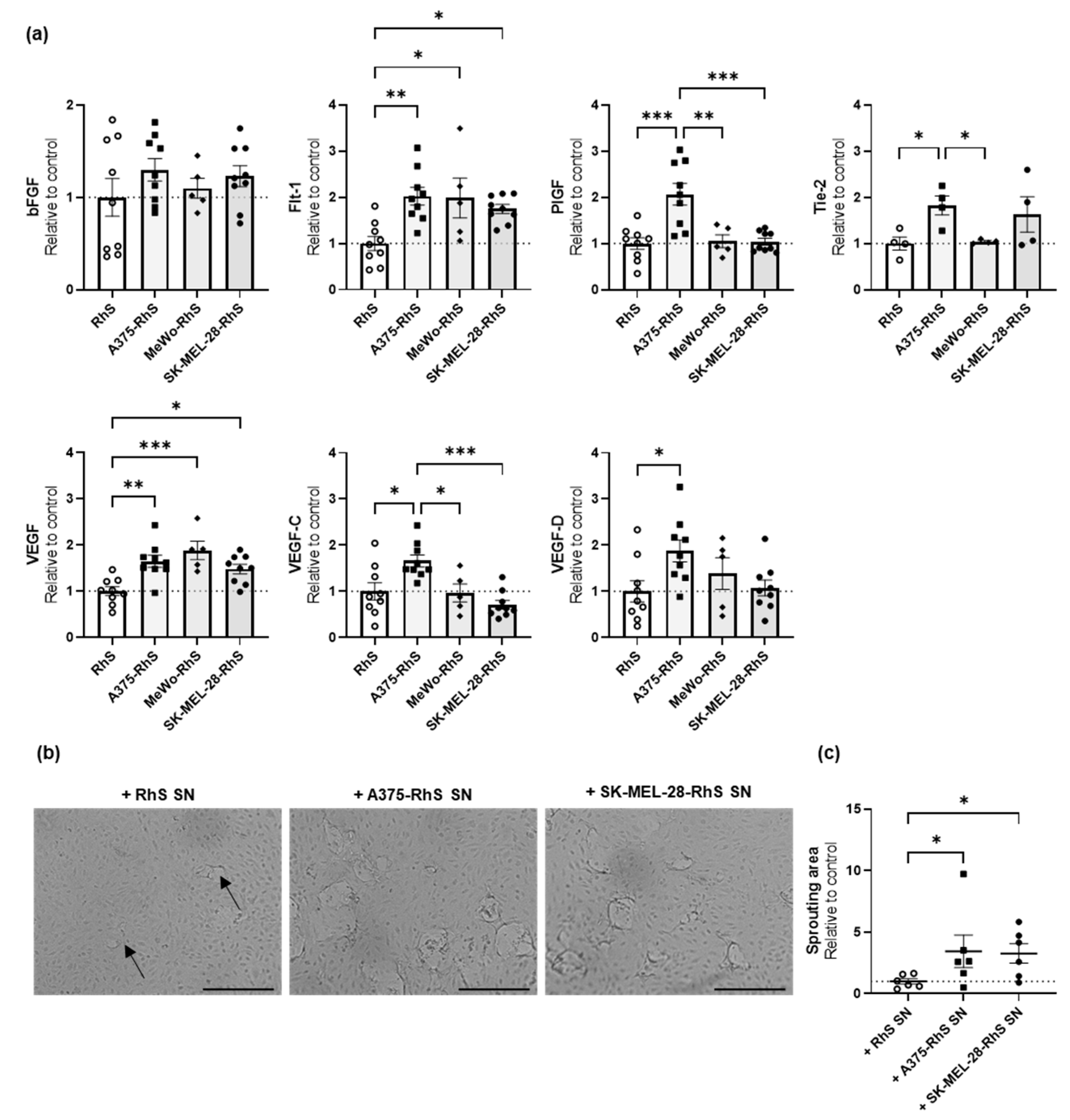
3.4. A375-RhS and SK-MEL-28-RhS Induce Angiogenesis In Vitro
3.5. A375-RhS Represents a More Advanced Stage of Melanoma Progression and Induces Fibroblast Activation in a TGFβ-Dependent Fashion
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tsao, H.; Chin, L.; Garraway, L.A.; Fisher, D.E. Melanoma: From mutations to medicine. Genes Dev. 2012, 26, 1131–1155. [Google Scholar] [CrossRef] [PubMed]
- Eddy, K.; Shah, R.; Chen, S. Decoding Melanoma Development and Progression: Identification of Therapeutic Vulnerabilities. Front. Oncol. 2020, 10, 626129. [Google Scholar] [CrossRef] [PubMed]
- Davis, L.E.; Shalin, S.C.; Tackett, A.J. Current state of melanoma diagnosis and treatment. Cancer Biol. 2019, 20, 1366–1379. [Google Scholar] [CrossRef] [PubMed]
- Braeuer, R.R.; Zigler, M.; Villares, G.J.; Dobroff, A.S.; Bar-Eli, M. Transcriptional control of melanoma metastasis: The importance of the tumor microenvironment. Semin. Cancer Biol. 2011, 21, 83–88. [Google Scholar] [CrossRef]
- Berraondo, P.; Sanmamed, M.F.; Ochoa, M.C.; Etxeberria, I.; Aznar, M.A.; Perez-Gracia, J.L.; Rodriguez-Ruiz, M.E.; Ponz-Sarvise, M.; Castanon, E.; Melero, I. Cytokines in clinical cancer immunotherapy. Br. J. Cancer 2019, 120, 6–15. [Google Scholar] [CrossRef] [PubMed]
- Lindenberg, J.J.; van de Ven, R.; Lougheed, S.M.; Zomer, A.; Santegoets, S.J.A.M.; Griffioen, A.W.; Hooijberg, E.; van den Eertwegh, A.J.M.; Thijssen, V.L.; Scheper, R.J.; et al. Functional characterization of a STAT3-dependent dendritic cell-derived CD14+ cell population arising upon IL-10-driven maturation. Oncoimmunology 2013, 2, e23837. [Google Scholar] [CrossRef]
- Tcyganov, E.; Mastio, J.; Chen, E.; Gabrilovich, D.I. Plasticity of myeloid-derived suppressor cells in cancer. Curr. Opin. Immunol. 2018, 51, 76–82. [Google Scholar] [CrossRef]
- Monteran, L.; Erez, N. The Dark Side of Fibroblasts: Cancer-Associated Fibroblasts as Mediators of Immunosuppression in the Tumor Microenvironment. Front. Immunol. 2019, 10, 1835. [Google Scholar] [CrossRef]
- Zhou, L.; Yang, K.; Andl, T.; Wickett, R.R.; Zhang, Y. Perspective of Targeting Cancer-Associated Fibroblasts in Melanoma. J. Cancer 2015, 6, 717–726. [Google Scholar] [CrossRef]
- Bielenberg, D.R.; Zetter, B.R. The Contribution of Angiogenesis to the Process of Metastasis. Cancer J. 2015, 21, 267–273. [Google Scholar] [CrossRef]
- Schaaf, M.B.; Garg, A.D.; Agostinis, P. Defining the role of the tumor vasculature in antitumor immunity and immunotherapy. Cell Death Dis. 2018, 9, 115. [Google Scholar] [CrossRef]
- Cho, W.C.; Jour, G.; Aung, P.P. Role of angiogenesis in melanoma progression: Update on key angiogenic mechanisms and other associated components. Semin. Cancer Biol. 2019, 59, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Streit, M.; Detmar, M. Angiogenesis, lymphangiogenesis, and melanoma metastasis. Oncogene 2003, 22, 3172–3179. [Google Scholar] [CrossRef] [PubMed]
- Di Blasio, S.; van Wigcheren, G.F.; Becker, A.; van Duffelen, A.; Gorris, M.; Verrijp, K.; Stefanini, I.; Bakker, G.J.; Bloemendal, M.; Halilovic, A.; et al. The tumour microenvironment shapes dendritic cell plasticity in a human organotypic melanoma culture. Nat. Commun. 2020, 11, 2749. [Google Scholar] [CrossRef] [PubMed]
- Michielon, E.; Lopez Gonzalez, M.; Burm, J.L.A.; Waaijman, T.; Jordanova, E.S.; de Gruijl, T.D.; Gibbs, S. Micro-environmental cross-talk in an organotypic human melanoma-in-skin model directs M2-like monocyte differentiation via IL-10. Cancer Immunol. Immunother. 2020, 69, 2319–2331. [Google Scholar] [CrossRef] [PubMed]
- Haridas, P.; McGovern, J.A.; McElwain, S.D.L.; Simpson, M.J. Quantitative comparison of the spreading and invasion of radial growth phase and metastatic melanoma cells in a three-dimensional human skin equivalent model. PeerJ 2017, 5, e3754. [Google Scholar] [CrossRef]
- Bourland, J.; Fradette, J.; Auger, F.A. Tissue-engineered 3D melanoma model with blood and lymphatic capillaries for drug development. Sci. Rep. 2018, 8, 13191. [Google Scholar] [CrossRef]
- Vorsmann, H.; Groeber, F.; Walles, H.; Busch, S.; Beissert, S.; Walczak, H.; Kulms, D. Development of a human three-dimensional organotypic skin-melanoma spheroid model for in vitro drug testing. Cell Death Dis. 2013, 4, e719. [Google Scholar] [CrossRef]
- Hill, D.S.; Robinson, N.D.; Caley, M.P.; Chen, M.; O’Toole, E.A.; Armstrong, J.L.; Przyborski, S.; Lovat, P.E. A Novel Fully Humanized 3D Skin Equivalent to Model Early Melanoma Invasion. Mol. Cancer 2015, 14, 2665–2673. [Google Scholar] [CrossRef]
- Marconi, A.; Quadri, M.; Farnetani, F.; Ciardo, S.; Palazzo, E.; Lotti, R.; Cesinaro, A.M.; Fabbiani, L.; Vaschieri, C.; Puviani, M.; et al. In Vivo Melanoma Cell Morphology Reflects Molecular Signature and Tumor Aggressiveness. J. Investig. Dermatol. 2022, 142, 2205–2216.e6. [Google Scholar] [CrossRef]
- Commandeur, S.; Sparks, S.J.; Chan, H.L.; Gao, L.; Out, J.J.; Gruis, N.A.; van Doorn, R.; El Ghalbzouri, A. In-vitro melanoma models: Invasive growth is determined by dermal matrix and basement membrane. Melanoma Res. 2014, 24, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Ecker, B.L.; Douglass, S.M.; Kugel, C.H., 3rd; Webster, M.R.; Almeida, F.V.; Somasundaram, R.; Hayden, J.; Ban, E.; Ahmadzadeh, H.; et al. Remodeling of the Collagen Matrix in Aging Skin Promotes Melanoma Metastasis and Affects Immune Cell Motility. Cancer Discov. 2019, 9, 64–81. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Fukunaga-Kalabis, M.; Herlyn, M. The three-dimensional human skin reconstruct model: A tool to study normal skin and melanoma progression. J. Vis. Exp. 2011, 54, e2937. [Google Scholar] [CrossRef]
- Muller, I.; Kulms, D. A 3D Organotypic Melanoma Spheroid Skin Model. J. Vis. Exp. 2018, 54, e57500. [Google Scholar] [CrossRef] [PubMed]
- Waaijman, T.; Breetveld, M.; Ulrich, M.; Middelkoop, E.; Scheper, R.J.; Gibbs, S. Use of a Collagen–Elastin Matrix as Transport Carrier System to Transfer Proliferating Epidermal Cells to Human Dermis In Vitro. Cell Transplant. 2010, 19, 1339–1348. [Google Scholar] [CrossRef] [PubMed]
- Kroeze, K.L.; Jurgens, W.J.; Doulabi, B.Z.; van Milligen, F.J.; Scheper, R.J.; Gibbs, S. Chemokine-mediated migration of skin-derived stem cells: Predominant role for CCL5/RANTES. J. Investig. Dermatol. 2009, 129, 1569–1581. [Google Scholar] [CrossRef]
- Tsao, H.; Goel, V.; Wu, H.; Yang, G.; Haluska, F.G. Genetic interaction between NRAS and BRAF mutations and PTEN/MMAC1 inactivation in melanoma. J. Investig. Dermatol. 2004, 122, 337–341. [Google Scholar] [CrossRef]
- Sapkota, B.; Hill, C.E.; Pollack, B.P. Vemurafenib enhances MHC induction in BRAF(V600E) homozygous melanoma cells. Oncoimmunology 2013, 2, e22890. [Google Scholar] [CrossRef]
- Nissan, M.H.; Pratilas, C.A.; Jones, A.M.; Ramirez, R.; Won, H.; Liu, C.; Tiwari, S.; Kong, L.; Hanrahan, A.J.; Yao, Z.; et al. Loss of NF1 in cutaneous melanoma is associated with RAS activation and MEK dependence. Cancer Res. 2014, 74, 2340–2350. [Google Scholar] [CrossRef]
- Davies, M.A.; Stemke-Hale, K.; Lin, E.; Tellez, C.; Deng, W.; Gopal, Y.N.; Woodman, S.E.; Calderone, T.C.; Ju, Z.; Lazar, A.J.; et al. Integrated Molecular and Clinical Analysis of AKT Activation in Metastatic Melanoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 7538–7546. [Google Scholar] [CrossRef]
- Ji, Z.; Njauw, C.N.; Taylor, M.; Neel, V.; Flaherty, K.T.; Tsao, H. p53 rescue through HDM2 antagonism suppresses melanoma growth and potentiates MEK inhibition. J. Investig. Dermatol. 2012, 132, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Monsuur, H.N.; Weijers, E.M.; Niessen, F.B.; Gefen, A.; Koolwijk, P.; Gibbs, S.; van den Broek, L.J. Extensive Characterization and Comparison of Endothelial Cells Derived from Dermis and Adipose Tissue: Potential Use in Tissue Engineering. PLoS ONE 2016, 11, e0167056. [Google Scholar] [CrossRef]
- Koning, J.J.; Rodrigues Neves, C.T.; Schimek, K.; Thon, M.; Spiekstra, S.W.; Waaijman, T.; de Gruijl, T.D.; Gibbs, S. A Multi-Organ-on-Chip Approach to Investigate How Oral Exposure to Metals Can Cause Systemic Toxicity Leading to Langerhans Cell Activation in Skin. Front. Toxicol. 2021, 3, 824825. [Google Scholar] [CrossRef] [PubMed]
- van de Ven, R.; Lindenberg, J.J.; Oosterhoff, D.; de Gruijl, T.D. Dendritic Cell Plasticity in Tumor-Conditioned Skin: CD14(+) Cells at the Cross-Roads of Immune Activation and Suppression. Front. Immunol. 2013, 4, 403. [Google Scholar] [CrossRef]
- Quaresmini, D.; Guida, M. Neoangiogenesis in Melanoma: An Issue in Biology and Systemic Treatment. Front. Immunol. 2020, 11, 584903. [Google Scholar] [CrossRef]
- Erdogan, B.; Webb, D.J. Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochem. Soc. Trans. 2017, 45, 229–236. [Google Scholar] [CrossRef]
- Sahai, E.; Astsaturov, I.; Cukierman, E.; DeNardo, D.G.; Egeblad, M.; Evans, R.M.; Fearon, D.; Greten, F.R.; Hingorani, S.R.; Hunter, T.; et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Limandjaja, G.C.; van den Broek, L.J.; Breetveld, M.; Waaijman, T.; Monstrey, S.; de Boer, E.M.; Scheper, R.J.; Niessen, F.B.; Gibbs, S. Characterization of In Vitro Reconstructed Human Normotrophic, Hypertrophic, and Keloid Scar Models. Tissue Eng. Part C Methods 2018, 24, 242–253. [Google Scholar] [CrossRef]
- Limandjaja, G.C.; van den Broek, L.J.; Waaijman, T.; Breetveld, M.; Monstrey, S.; Scheper, R.J.; Niessen, F.B.; Gibbs, S. Reconstructed human keloid models show heterogeneity within keloid scars. Arch. Dermatol. Res. 2018, 310, 815–826. [Google Scholar] [CrossRef]
- Scharenberg, M.A.; Pippenger, B.E.; Sack, R.; Zingg, D.; Ferralli, J.; Schenk, S.; Martin, I.; Chiquet-Ehrismann, R. TGF-beta-induced differentiation into myofibroblasts involves specific regulation of two MKL1 isoforms. J. Cell Sci. 2014, 127, 1079–1091. [Google Scholar] [CrossRef]
- Singhal, M.; Augustin, H.G. Beyond Angiogenesis: Exploiting Angiocrine Factors to Restrict Tumor Progression and Metastasis. Cancer Res. 2020, 80, 659–662. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.E.; Harney, A.S.; Pollard, J.W. The Multifaceted Role of Perivascular Macrophages in Tumors. Cancer Cell 2016, 30, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wang, H.; Wang, X.; Jiang, G.; Liu, H.; Zhang, G.; Wang, H.; Fang, R.; Bu, X.; Cai, S.; et al. TGF-β induces M2-like macrophage polarization via SNAIL-mediated suppression of a pro-inflammatory phenotype. Oncotarget 2016, 7, 52294–52306. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958. [Google Scholar] [CrossRef] [PubMed]
- Linde, N.; Gutschalk, C.M.; Hoffmann, C.; Yilmaz, D.; Mueller, M.M. Integrating macrophages into organotypic co-cultures: A 3D in vitro model to study tumor-associated macrophages. PLoS ONE 2012, 7, e40058. [Google Scholar] [CrossRef]
- Lindenberg, J.J.; Oosterhoff, D.; Sombroek, C.C.; Lougheed, S.M.; Hooijberg, E.; Stam, A.G.M.; Santegoets, S.J.A.M.; Tijssen, H.J.; Buter, J.; Pinedo, H.M.; et al. IL-10 conditioning of human skin affects the distribution of migratory dendritic cell subsets and functional T cell differentiation. PLoS ONE 2013, 8, e70237. [Google Scholar] [CrossRef]
- Serini, G.; Gabbiani, G. Mechanisms of Myofibroblast Activity and Phenotypic Modulation. Exp. Cell Res. 1999, 250, 273–283. [Google Scholar] [CrossRef]
- Avagliano, A.; Granato, G.; Ruocco, M.R.; Romano, V.; Belviso, I.; Carfora, A.; Montagnani, S.; Arcucci, A. Metabolic Reprogramming of Cancer Associated Fibroblasts: The Slavery of Stromal Fibroblasts. Biomed. Res. Int. 2018, 2018, 6075403. [Google Scholar] [CrossRef]
- Boyano, M.D.; Garcia-Vázquez, M.D.; López-Michelena, T.; Gardeazabal, J.; Bilbao, J.; Cañavate, M.L.; De Galdeano, A.G.; Izu, R.; Díaz-Ramón, L.; Raton, J.A.; et al. Soluble interleukin-2 receptor, intercellular adhesion molecule-1 and interleukin-10 serum levels in patients with melanoma. Br. J. Cancer 2000, 83, 847. [Google Scholar] [CrossRef]
- Dummer, W.; Bastian, B.C.; Ernst, N.; Schänzle, C.; Schwaaf, A.; Bröcker, E.-B. Interleukin-10 production in malignant melanoma: Preferential detection of IL-10-secreting tumor cells in metastatic lesions. Int. J. Cancer 1996, 66, 607–610. [Google Scholar] [CrossRef]
- Dummer, W.; Becker, J.C.; Schwaaf, A.; Leverkus, M.; Moll, T.; Bröcker, E.B. Elevated serum levels of interleukin-10 in patients with metastatic malignant melanoma. Melanoma Res. 1995, 5, 67–68. [Google Scholar] [CrossRef] [PubMed]
- Neubert, N.J.; Schmittnaegel, M.; Bordry, N.; Nassiri, S.; Wald, N.; Martignier, C.; Tille, L.; Homicsko, K.; Damsky, W.; Maby-El Hajjami, H.; et al. T cell-induced CSF1 promotes melanoma resistance to PD1 blockade. Sci. Transl. Med. 2018, 10, eaan3311. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer 2016, 16, 582–598. [Google Scholar] [CrossRef]
- Tobin, R.P.; Jordan, K.R.; Kapoor, P.; Spongberg, E.; Davis, D.; Vorwald, V.M.; Couts, K.L.; Gao, D.; Smith, D.E.; Borgers, J.S.W.; et al. IL-6 and IL-8 Are Linked with Myeloid-Derived Suppressor Cell Accumulation and Correlate with Poor Clinical Outcomes in Melanoma Patients. Front. Oncol. 2019, 9, 1223. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Bournazou, E.; Sansone, P.; Berishaj, M.; Gao, S.P.; Daly, L.; Wels, J.; Theilen, T.; Granitto, S.; Zhang, X.; et al. The IL-6/JAK/Stat3 feed-forward loop drives tumorigenesis and metastasis. Neoplasia 2013, 15, 848–862. [Google Scholar] [CrossRef] [PubMed]
- Sanmamed, M.F.; Perez-Gracia, J.L.; Schalper, K.A.; Fusco, J.P.; Gonzalez, A.; Rodriguez-Ruiz, M.E.; Onate, C.; Perez, G.; Alfaro, C.; Martin-Algarra, S.; et al. Changes in serum interleukin-8 (IL-8) levels reflect and predict response to anti-PD-1 treatment in melanoma and non-small-cell lung cancer patients. Ann. Oncol. 2017, 28, 1988–1995. [Google Scholar] [CrossRef]
- Young, H.L.; Rowling, E.J.; Bugatti, M.; Giurisato, E.; Luheshi, N.; Arozarena, I.; Acosta, J.C.; Kamarashev, J.; Frederick, D.T.; Cooper, Z.A.; et al. An adaptive signaling network in melanoma inflammatory niches confers tolerance to MAPK signaling inhibition. J. Exp. Med. 2017, 214, 1691–1710. [Google Scholar] [CrossRef] [PubMed]
- Sos, M.L.; Levin, R.S.; Gordan, J.D.; Oses-Prieto, J.A.; Webber, J.T.; Salt, M.; Hann, B.; Burlingame, A.L.; McCormick, F.; Bandyopadhyay, S.; et al. Oncogene mimicry as a mechanism of primary resistance to BRAF inhibitors. Cell Rep. 2014, 8, 1037–1048. [Google Scholar] [CrossRef]
- Tsukamoto, H.; Fujieda, K.; Miyashita, A.; Fukushima, S.; Ikeda, T.; Kubo, Y.; Senju, S.; Ihn, H.; Nishimura, Y.; Oshiumi, H. Combined Blockade of IL6 and PD-1/PD-L1 Signaling Abrogates Mutual Regulation of Their Immunosuppressive Effects in the Tumor Microenvironment. Cancer Res. 2018, 78, 5011–5022. [Google Scholar] [CrossRef]
- Alfaro, C.; Teijeira, A.; Onate, C.; Perez, G.; Sanmamed, M.F.; Andueza, M.P.; Alignani, D.; Labiano, S.; Azpilikueta, A.; Rodriguez-Paulete, A.; et al. Tumor-Produced Interleukin-8 Attracts Human Myeloid-Derived Suppressor Cells and Elicits Extrusion of Neutrophil Extracellular Traps (NETs). Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 3924–3936. [Google Scholar] [CrossRef]





| Cell Line | BRAF Status | PTEN Status | NRAS Status | Origin | Supplier |
|---|---|---|---|---|---|
| A375 | c.1799T>A | WT [27] | WT [27] | skin | ATCC |
| COLO829 | c.1799T>A | c.493_634del142 | WT | skin | ATCC |
| G361 | c.1799T>A | skin | ATCC | ||
| MeWo | WT [28] | WT [29] | WT [30] | lymph node | ATCC |
| RPMI-7951 | c.1799T>A | c.1_79del79 | WT [30] | lymph node | ATCC |
| SK-MEL-28 | c.1799T>A [28,31] | A499G [27] | WT [27,30] | lymph node | CLS Cell Lines Service GmbH |
| Mel-RhS | Melanoma Stage | Cytokines | Immune Modulation | Angiogenic Factors | Sprouting |
|---|---|---|---|---|---|
| RPMI-7951-RhS | None | None | N.D. | None | N.D. |
| COLO829-RhS | Very early stage | None | N.D. | None | N.D. |
| G361-RhS | Very early stage | None | N.D. | None | N.D. |
| MeWo-RhS | RGP | None | N.D. | ↑ Flt-1, VEGF | No |
| SK-MEL-28-RhS | Early invasive stage | ↑ CCL5, CXCL10, GM-CSF, IL-10, TGFβ | Via IL-10, M-CSF, TGFβ | ↑ Flt-1, VEGF | Yes |
| A375-RhS | Late invasive stage | ↑ CCL2, GM-CSF, IL-6, IL-8, IL-10, M-CSF, TGFβ | Needs to be further investigated | ↑ Flt-1, PlGF, Tie-2, VEGF, VEGF-C, VEGF-D | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Michielon, E.; López González, M.; Stolk, D.A.; Stolwijk, J.G.C.; Roffel, S.; Waaijman, T.; Lougheed, S.M.; de Gruijl, T.D.; Gibbs, S. A Reconstructed Human Melanoma-in-Skin Model to Study Immune Modulatory and Angiogenic Mechanisms Facilitating Initial Melanoma Growth and Invasion. Cancers 2023, 15, 2849. https://doi.org/10.3390/cancers15102849
Michielon E, López González M, Stolk DA, Stolwijk JGC, Roffel S, Waaijman T, Lougheed SM, de Gruijl TD, Gibbs S. A Reconstructed Human Melanoma-in-Skin Model to Study Immune Modulatory and Angiogenic Mechanisms Facilitating Initial Melanoma Growth and Invasion. Cancers. 2023; 15(10):2849. https://doi.org/10.3390/cancers15102849
Chicago/Turabian StyleMichielon, Elisabetta, Marta López González, Dorian A. Stolk, Joeke G. C. Stolwijk, Sanne Roffel, Taco Waaijman, Sinéad M. Lougheed, Tanja D. de Gruijl, and Susan Gibbs. 2023. "A Reconstructed Human Melanoma-in-Skin Model to Study Immune Modulatory and Angiogenic Mechanisms Facilitating Initial Melanoma Growth and Invasion" Cancers 15, no. 10: 2849. https://doi.org/10.3390/cancers15102849
APA StyleMichielon, E., López González, M., Stolk, D. A., Stolwijk, J. G. C., Roffel, S., Waaijman, T., Lougheed, S. M., de Gruijl, T. D., & Gibbs, S. (2023). A Reconstructed Human Melanoma-in-Skin Model to Study Immune Modulatory and Angiogenic Mechanisms Facilitating Initial Melanoma Growth and Invasion. Cancers, 15(10), 2849. https://doi.org/10.3390/cancers15102849







