Effects of Co-Culture EBV-miR-BART1-3p on Proliferation and Invasion of Gastric Cancer Cells Based on Exosomes
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. EBER In Situ Hybridization
2.3. Preparation of Exosome-Free Fetal Bovine Serum
2.4. Isolation of Exosomes
2.5. Exosome Morphology Observed by Transmission Electron Microscopy
2.6. Exosome Size and Concentration by Nanoparticle Tracking Analysis (NTA)
2.7. Extraction of Cell Protein
2.8. Extraction of Exosomal Protein
2.9. Determination of Cellular and Exosomal Protein Concentrations
2.10. Expression Analysis of Exosomal Protein
2.11. Extraction of Ribonucleic Acid
2.12. Library Construction and Small RNA Sequencing
2.13. Expression Profile Analysis of EBV-miRNA
2.14. Construction of SNU-719-Inhibitor Cell Line with Low Expression of EBV-miR-BART1-3p
2.15. Verification of the Expression Level of miR-BART1-3p in Cell Lines and the Expression of Target Genes in AGS Cells after Co-Culture of Exosomes for 24 h
2.16. CCK8 Cell Proliferation Experiment
2.17. Cell Scratch Experiment
2.18. Transwell Migration Assay
2.19. Transwell Invasion
2.20. Absorption of Exosomes by Living Cells Observed by Laser Confocal Microscopy
2.21. Expression of miR-BART1-3p after Co-Culture with GC Cells in Exosomes with High Expression or Silence of miR-BART1-3p and Its Effect on Proliferation and Invasion
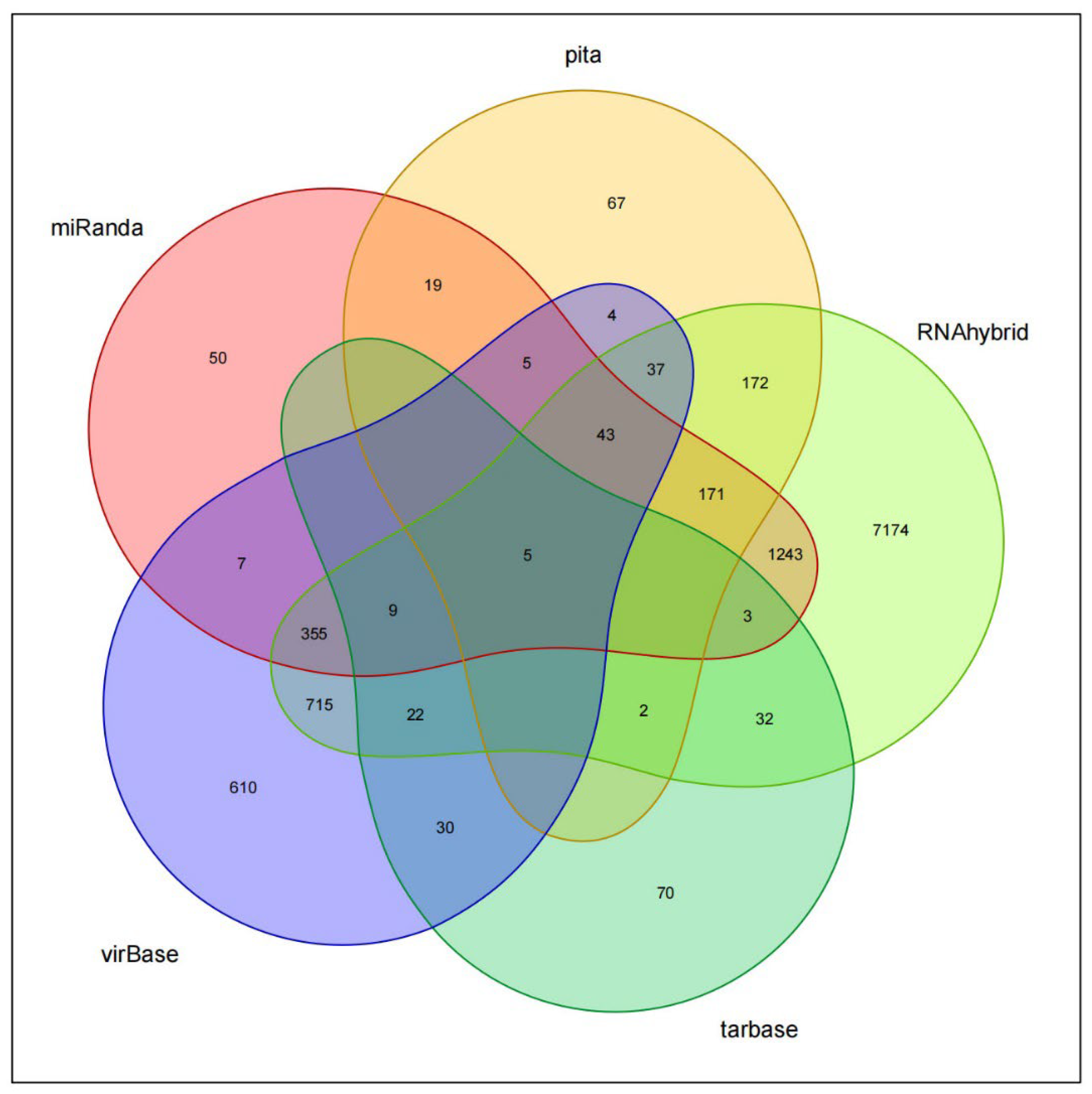
2.22. Target Gene Analysis of miR-BART1-3p
2.23. Statistical Analysis
3. Results
3.1. Characterization of miRNA in Exosomes of EBV-Positive GC Cells
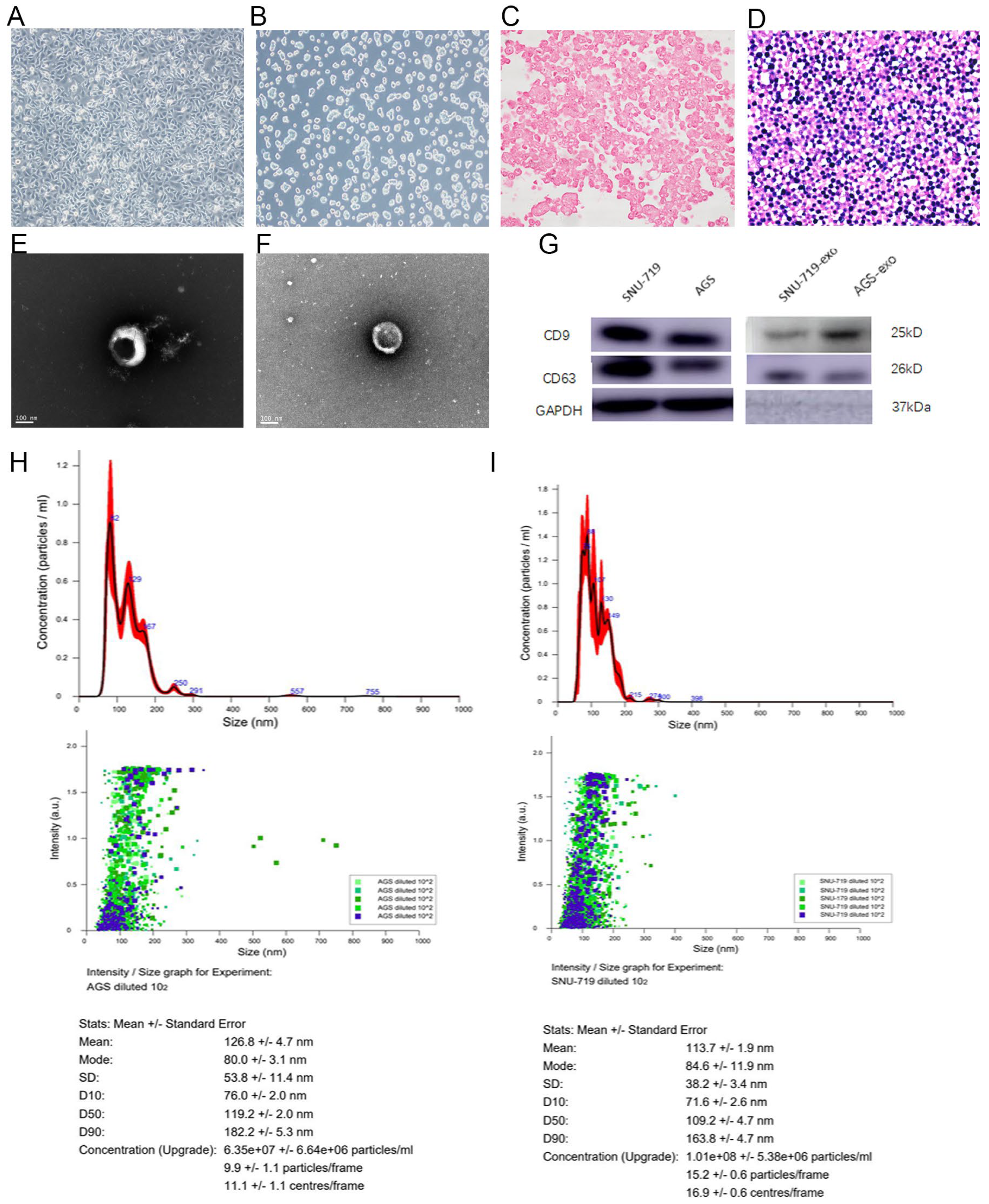
3.1.1. Morphology of AGS and SNU-719 GC Cells
3.1.2. Structure of Exosomes Secreted by SNU-719 and AGS Cells
3.1.3. Size of Exosomes Secreted by SNU-719 and AGS Cells
3.1.4. Expression Analysis of Exosomal Proteins Secreted by SNU-719 and AGS Cells
3.1.5. Distribution of Small RNAs in Cells and Exosomes
Annotation of ncRNAs in SNU-719 and AGS Cells and Exosomes
Small RNA Sequence Length Distribution in SNU-719 and AGS Cells and Exosomes
sRNA Analysis of Genomic Repeat Regions in SNU-719 and AGS Cells and Exosomes
Analysis of Functional Elements in SNU-719 and AGS Cells and Exosomes
Common and Unique Sequence Analysis of SNU-719 and AGS Cells and Exosomes
Correlation Analysis between SNU-719 and AGS Cells and Exosomes
Differential Expression of miRNA in SNU-719 and AGS Cells and Exosomes
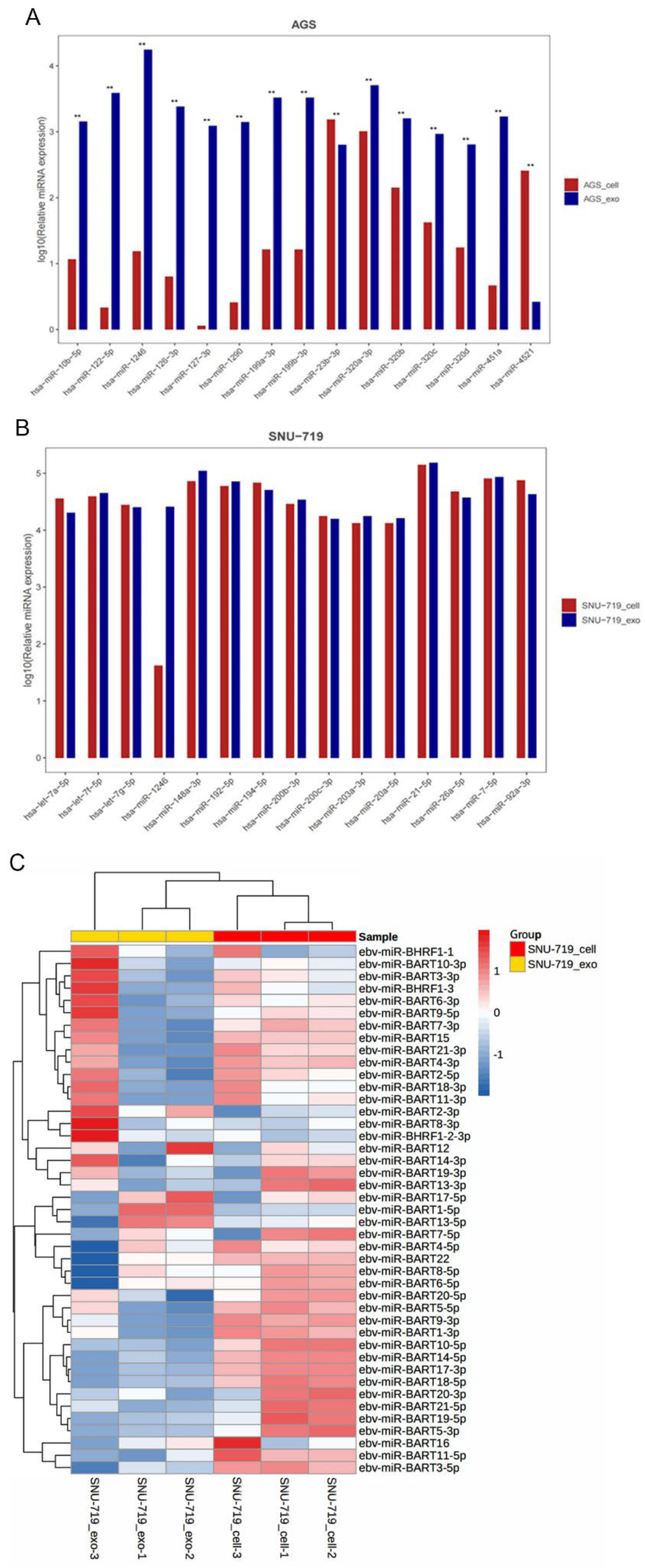
Expression Analysis of the Top 15 miRNAs in Cells and Exosomes
Differential Expression Analysis of EBV-Related miRNAs in SNU-719 Cells and Their Exosomes
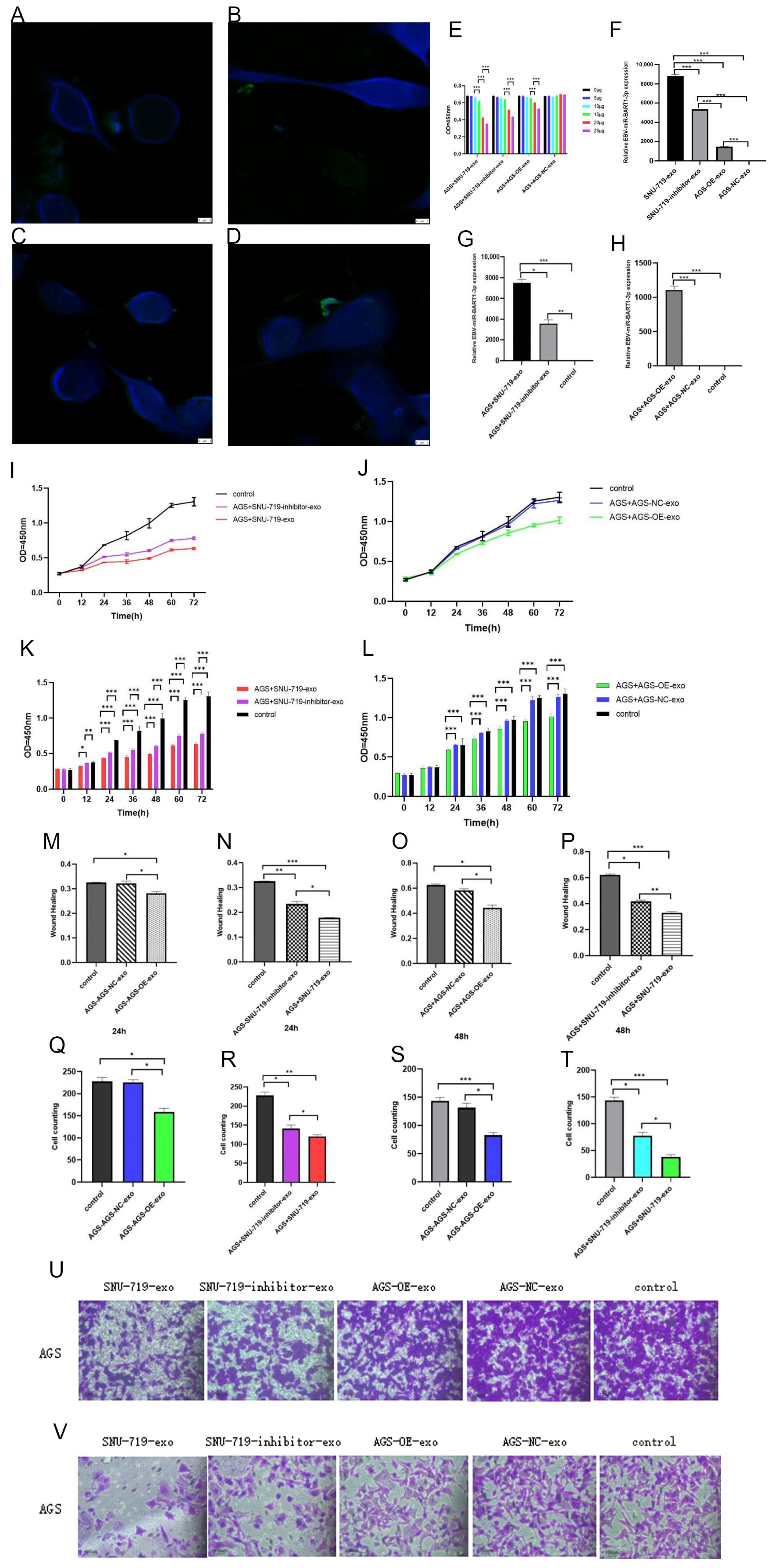
3.2. Effects of High and Low Expression of EBV-miR-BART1-3p on Proliferation and Invasion Ability of GC Cells
3.3. Effects of Co-Culture with Exosomes with High and Low Expression of EBV-miR-BART1-3p on Proliferation and Invasion of GC Cells
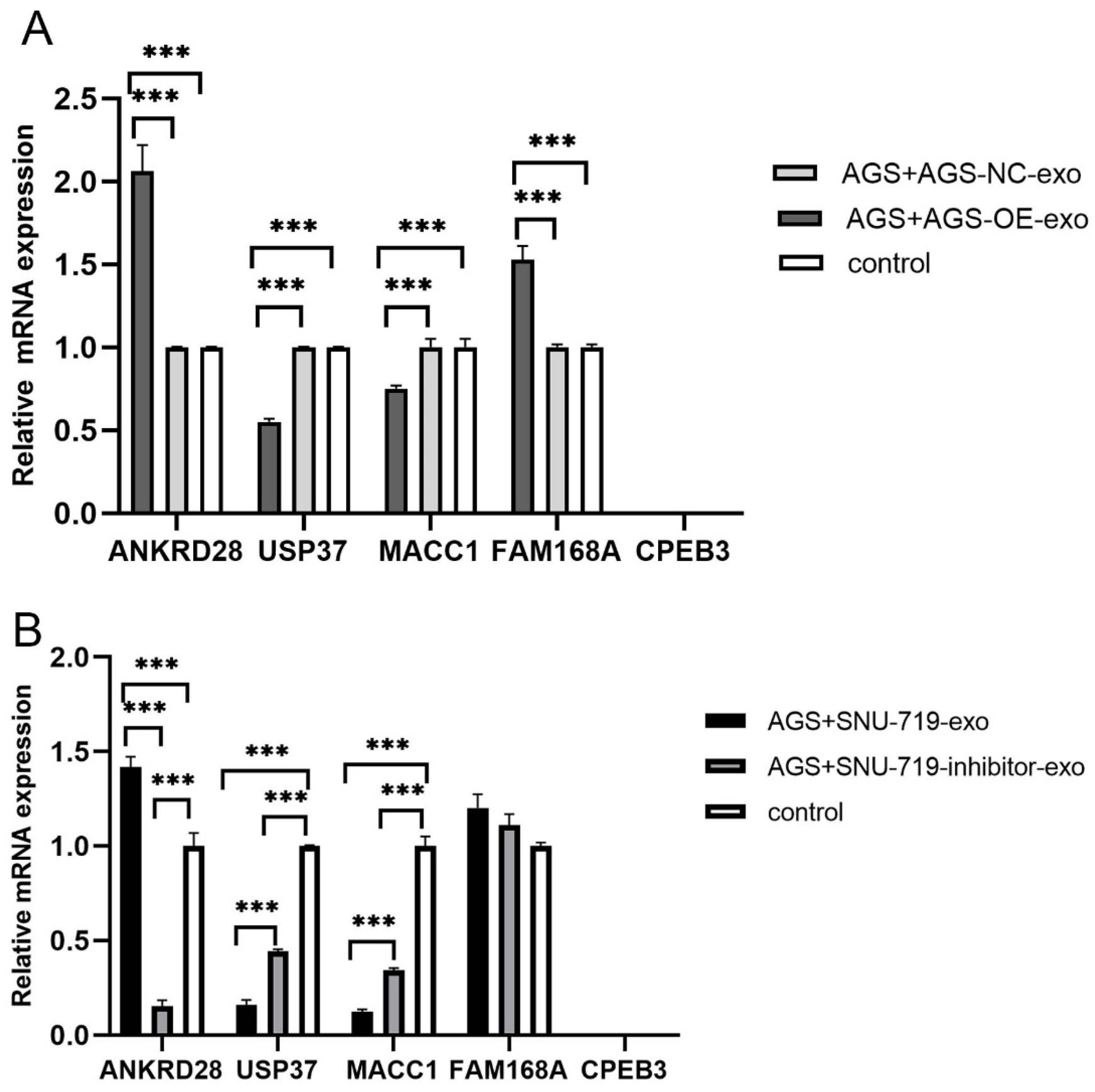
3.4. Changes of Target Genes in Gastric Cancer Cells after Co-Culture with High and Low Expression of EBV-miR-BART1-3p in Exosomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Xia, C.; Dong, X.; Li, H.; Cao, M.; Sun, D.; He, S.; Yang, F.; Yan, X.; Zhang, S.; Li, N.; et al. Cancer statistics in China and United States, 2022: Profiles, trends, and determinants. Chin. Med. J. 2022, 135, 584–590. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Song, H.J.; Srivastava, A.; Lee, J.; Kim, Y.S.; Kim, K.M.; Ki Kang, W.; Kim, M.; Kim, S.; Park, C.K.; Kim, S. Host Inflammatory Response Predicts Survival of Patients with Epstein-Barr Virus–Associated Gastric Carcinoma. Gastroenterology 2010, 139, 84–92.e2. [Google Scholar] [CrossRef] [PubMed]
- Kanda, T. EBV-Encoded Latent Genes. Adv. Exp. Med. Biol. 2018, 1045, 377–394. [Google Scholar] [CrossRef]
- Jadli, A.S.; Ballasy, N.; Edalat, P.; Patel, V.B. Inside (sight) of tiny communicator: Exosome biogenesis, secretion, and uptake. Mol. Cell. Biochem. 2020, 467, 77–94. [Google Scholar] [CrossRef]
- Ailawadi, S.; Wang, X.; Gu, H.; Fan, G.C. Pathologic function and therapeutic potential of exosomes in cardiovascular disease. Biochim. Biophys. Acta 2015, 1852, 1–11. [Google Scholar] [CrossRef]
- Skalsky, R.L.; Cullen, B.R. EBV Noncoding RNAs. Curr. Top. Microbiol. Immunol. 2015, 391, 181–217. [Google Scholar] [CrossRef]
- Aga, M.; Bentz, G.L.; Raffa, S.; Torrisi, M.R.; Kondo, S.; Wakisaka, N.; Yoshizaki, T.; Pagano, J.S.; Shackelford, J. Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene 2014, 33, 4613–4622. [Google Scholar] [CrossRef]
- Zomer, A.; Maynard, C.; Verweij, F.J.; Kamermans, A.; Schäfer, R.; Beerling, E.; Schiffelers, R.M.; de Wit, E.; Berenguer, J.; Ellenbroek, S.I.J.; et al. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell 2015, 161, 1046–1057. [Google Scholar] [CrossRef]
- Syn, N.; Wang, L.; Sethi, G.; Thiery, J.P.; Goh, B.C. Exosome-Mediated Metastasis: From Epithelial–Mesenchymal Transition to Escape from Immunosurveillance. Trends Pharm. Sci. 2016, 37, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Soldevilla, B.; Rodríguez, M.; San Millán, C.; García, V.; Fernández-Periañez, R.; Gil-Calderón, B.; Martín, P.; García-Grande, A.; Silva, J.; Bonilla, F.; et al. Tumor-derived exosomes are enriched in DNp73, which promotes oncogenic potential in acceptor cells and correlates with patient survival. Hum. Mol. Genet. 2014, 23, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Chahar, H.S.; Bao, X.; Casola, A. Exosomes and Their Role in the Life Cycle and Pathogenesis of RNA Viruses. Viruses 2015, 7, 3204–3225. [Google Scholar] [CrossRef] [PubMed]
- Kurywchak, P.; Tavormina, J.; Kalluri, R. The emerging roles of exosomes in the modulation of immune responses in cancer. Genome Med. 2018, 10, 23. [Google Scholar] [CrossRef]
- Shinozaki-Ushiku, A.; Kunita, A.; Isogai, M.; Hibiya, T.; Ushiku, T.; Takada, K.; Fukayama, M. Profiling of Virus-Encoded MicroRNAs in Epstein-Barr Virus-Associated Gastric Carcinoma and Their Roles in Gastric Carcinogenesis. J. Virol. 2015, 89, 5581–5591. [Google Scholar] [CrossRef]
- Karimzadeh, M.; Tabibzadeh, A.; Moghoofei, M.; Abbasi, S.; Sadri Nahand, J.; Sadeghii, F.; Karbalaie Niya, M.H.; Keyvani, H.; Bokharaei-Salim, F.; Esghaei, M.; et al. As Evidence-Based Tumorigenic Role of Epstein-Barr Virus miR-BART1-3p in Neurological Tumors. Asian Pac. J. Cancer Prev. 2021, 22, 257–266. [Google Scholar] [CrossRef]
- Park, M.C.; Kim, H.; Choi, H.; Chang, M.S.; Lee, S.K. Epstein-Barr Virus miR-BART1-3p Regulates the miR-17-92 Cluster by Targeting E2F3. Int. J. Mol. Sci. 2021, 22, 10936. [Google Scholar] [CrossRef]
- Min, K.; Kim, J.Y.; Lee, S.K. Epstein-Barr virus miR-BART1-3p suppresses apoptosis and promotes migration of gastric carcinoma cells by targeting DAB2. Int. J. Biol. Sci. 2020, 16, 694–707. [Google Scholar] [CrossRef]
- Iizasa, H.; Nanbo, A.; Nishikawa, J.; Jinushi, M.; Yoshiyama, H. Epstein-Barr Virus (EBV)-associated Gastric Carcinoma. Viruses 2012, 4, 3420–3439. [Google Scholar] [CrossRef]
- Meckes, D.G. Exosomal Communication Goes Viral. J. Virol. 2015, 89, 5200–5203. [Google Scholar] [CrossRef]
- Iwakiri, D. Multifunctional non-coding Epstein-Barr virus encoded RNAs (EBERs) contribute to viral pathogenesis. Virus Res. 2016, 212, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Barth, S.; Meister, G.; Grässer, F.A. EBV-encoded miRNAs. Biochim. Biophys. Acta 2011, 1809, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Feederle, R.; Haar, J.; Bernhardt, K.; Linnstaedt, S.D.; Bannert, H.; Lips, H.; Cullen, B.R.; Delecluse, H.J. The members of an Epstein-Barr virus microRNA cluster cooperate to transform B lymphocytes. J. Virol. 2011, 85, 9801–9810. [Google Scholar] [CrossRef]
- Marquitz, A.R.; Mathur, A.; Shair, K.H.; Raab-Traub, N. Infection of Epstein-Barr virus in a gastric carcinoma cell line induces anchorage independence and global changes in gene expression. Proc. Natl. Acad. Sci. USA 2012, 109, 9593–9598. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Pfuhl, T.; Motsch, N.; Barth, S.; Nicholls, J.; Grässer, F.; Meister, G. Identification of novel Epstein-Barr virus microRNA genes from nasopharyngeal carcinomas. J. Virol. 2009, 83, 3333–3341. [Google Scholar] [CrossRef]
- Hoshina, S.; Sekizuka, T.; Kataoka, M.; Hasegawa, H.; Hamada, H.; Kuroda, M.; Katano, H. Profile of Exosomal and Intracellular microRNA in Gamma-Herpesvirus-Infected Lymphoma Cell Lines. PLoS ONE 2016, 11, e0162574. [Google Scholar] [CrossRef] [PubMed]
- Wortzel, I.; Dror, S.; Kenific, C.M.; Lyden, D. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell. 2019, 49, 347–360. [Google Scholar] [CrossRef]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell. Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Gurung, S.; Perocheau, D.; Touramanidou, L.; Baruteau, J. The exosome journey: From biogenesis to uptake and intracellular signalling. Cell. Commun. Signal. 2021, 19, 47. [Google Scholar] [CrossRef]
- Deng, G.; Qu, J.; Zhang, Y.; Che, X.; Cheng, Y.; Fan, Y.; Zhang, S.; Na, D.; Liu, Y.; Qu, X. Gastric cancer-derived exosomes promote peritoneal metastasis by destroying the mesothelial barrier. FEBS Lett. 2017, 591, 2167–2179. [Google Scholar] [CrossRef]
- Guduric-Fuchs, J.; O’Connor, A.; Camp, B.; O’Neill, C.L.; Medina, R.J.; Simpson, D.A. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genom. 2012, 13, 357. [Google Scholar] [CrossRef]
- Purushothaman, A. Exosomes from Cell Culture-Conditioned Medium: Isolation by Ultracentrifugation and Characterization. Methods Mol. Biol. 2019, 1952, 233–244. [Google Scholar] [CrossRef]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Wu, X.; Showiheen, S.A.A.; Sun, A.R.; Crawford, R.; Xiao, Y.; Mao, X.; Prasadam, I. Exosomes Extraction and Identification. Methods Mol. Biol. 2019, 2054, 81–91. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Treece, A.L.; Duncan, D.L.; Tang, W.; Elmore, S.; Morgan, D.R.; Dominguez, R.L.; Speck, O.; Meyers, M.O.; Gulley, M.L. Gastric adenocarcinoma microRNA profiles in fixed tissue and in plasma reveal cancer-associated and Epstein-Barr virus-related expression patterns. Lab. Investig. 2016, 96, 661–671. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, X.; Qin, Z.; Wei, L.; Lu, Y.; Peng, Q.; Gao, Y.; Zhang, X.; Zhang, X.; Li, Z.; et al. Epstein-Barr virus miR-BART3-3p promotes tumorigenesis by regulating senescence pathway in gastric cancer. J. Biol. Chem. 2019, 294, 4854–4866. [Google Scholar] [CrossRef]
- Torres Crigna, A.; Fricke, F.; Nitschke, K.; Worst, T.; Erb, U.; Karremann, M.; Buschmann, D.; Elvers-Hornung, S.; Tucher, C.; Schiller, M.; et al. Inter-Laboratory Comparison of Extracellular Vesicle Isolation Based on Ultracentrifugation. Transfus. Med. Hemother 2021, 48, 48–59. [Google Scholar] [CrossRef]
- Wang, D.; Wang, X.; Si, M.; Yang, J.; Sun, S.; Wu, H.; Cui, S.; Qu, X.; Yu, X. Exosome-encapsulated miRNAs contribute to CXCL12/CXCR4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett. 2020, 474, 36–52. [Google Scholar] [CrossRef]
- Kim, H.; Choi, H.; Lee, S.K. Epstein-Barr Virus MicroRNA miR-BART20-5p suppresses lytic induction by inhibiting BAD-mediated caspase-3-dependent apoptosis. J. Virol. 2015, 90, 1359–1368. [Google Scholar] [CrossRef]
- Jung, Y.J.; Choi, H.; Kim, H.; Lee, S.K. MicroRNA miR-BART20-5p stabilizes Epstein-Barr virus latency by directly targeting BZLF1 and BRLF1. J. Virol. 2014, 88, 9027–9037. [Google Scholar] [CrossRef]
- Yoon, J.H.; Ham, I.H.; Kim, O.; Ashktorab, H.; Smoot, D.T.; Nam, S.W.; Lee, J.Y.; Hur, H.; Park, W.S. Gastrokine 1 protein is a potential theragnostic target for gastric cancer. Gastric Cancer 2018, 21, 956–967. [Google Scholar] [CrossRef]
- Yoon, J.H.; Park, Y.G.; Nam, S.W.; Park, W.S. The diagnostic value of serum gastrokine 1 (GKN1) protein in gastric cancer. Cancer Med. 2019, 8, 5507–5514. [Google Scholar] [CrossRef]
- Yoon, J.H.; Ashktorab, H.; Smoot, D.T.; Nam, S.W.; Hur, H.; Park, W.S. Uptake and tumor-suppressive pathways of exosome-associated GKN1 protein in gastric epithelial cells. Gastric Cancer 2020, 23, 848–862. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, M.; Hu, S.; Zhang, T.; Li, J.; Gao, F.; Zhang, Z.; Zheng, K.; Li, G.; Ren, C.; Chen, X.; et al. Effects of Co-Culture EBV-miR-BART1-3p on Proliferation and Invasion of Gastric Cancer Cells Based on Exosomes. Cancers 2023, 15, 2841. https://doi.org/10.3390/cancers15102841
Lin M, Hu S, Zhang T, Li J, Gao F, Zhang Z, Zheng K, Li G, Ren C, Chen X, et al. Effects of Co-Culture EBV-miR-BART1-3p on Proliferation and Invasion of Gastric Cancer Cells Based on Exosomes. Cancers. 2023; 15(10):2841. https://doi.org/10.3390/cancers15102841
Chicago/Turabian StyleLin, Mengyao, Shun Hu, Tianyi Zhang, Jiezhen Li, Feng Gao, Zhenzhen Zhang, Ke Zheng, Guoping Li, Caihong Ren, Xiangna Chen, and et al. 2023. "Effects of Co-Culture EBV-miR-BART1-3p on Proliferation and Invasion of Gastric Cancer Cells Based on Exosomes" Cancers 15, no. 10: 2841. https://doi.org/10.3390/cancers15102841
APA StyleLin, M., Hu, S., Zhang, T., Li, J., Gao, F., Zhang, Z., Zheng, K., Li, G., Ren, C., Chen, X., Guo, F., & Zhang, S. (2023). Effects of Co-Culture EBV-miR-BART1-3p on Proliferation and Invasion of Gastric Cancer Cells Based on Exosomes. Cancers, 15(10), 2841. https://doi.org/10.3390/cancers15102841






