Biomarker Reproducibility Challenge: A Review of Non-Nucleotide Biomarker Discovery Protocols from Body Fluids in Breast Cancer Diagnosis
Abstract
Simple Summary
Abstract
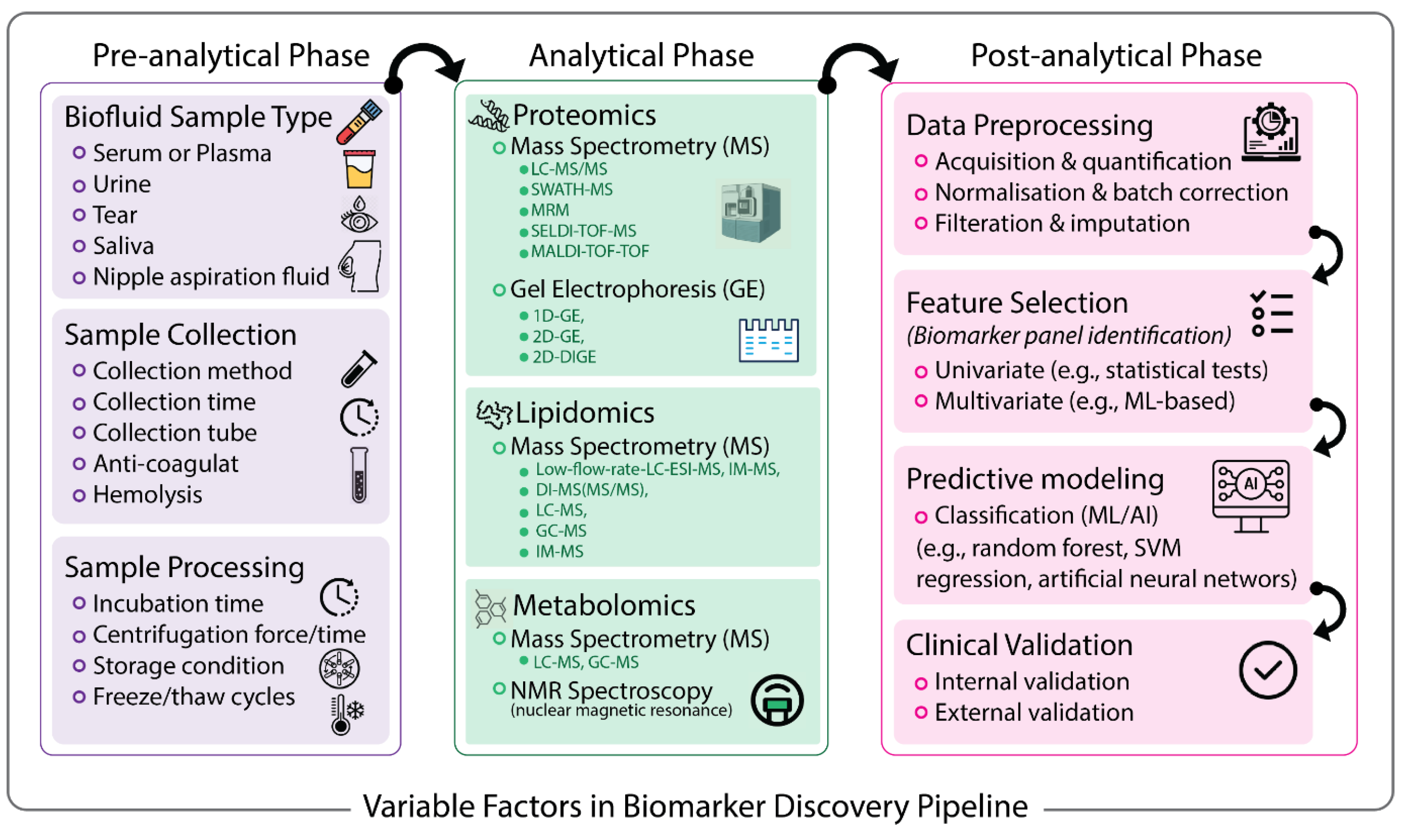
1. Background
2. Pre-Analytical Variables
2.1. Biofluids Are Excellent Sources of Biomarkers
2.1.1. Serum and Plasma
2.1.2. Urine
2.1.3. Tears
2.1.4. Nipple Aspiration Fluid
2.1.5. Saliva
2.1.6. Extracellular Vesicles
2.2. Sample Collection and Processing Variables Impact the Discovery of Accurate Biomarkers
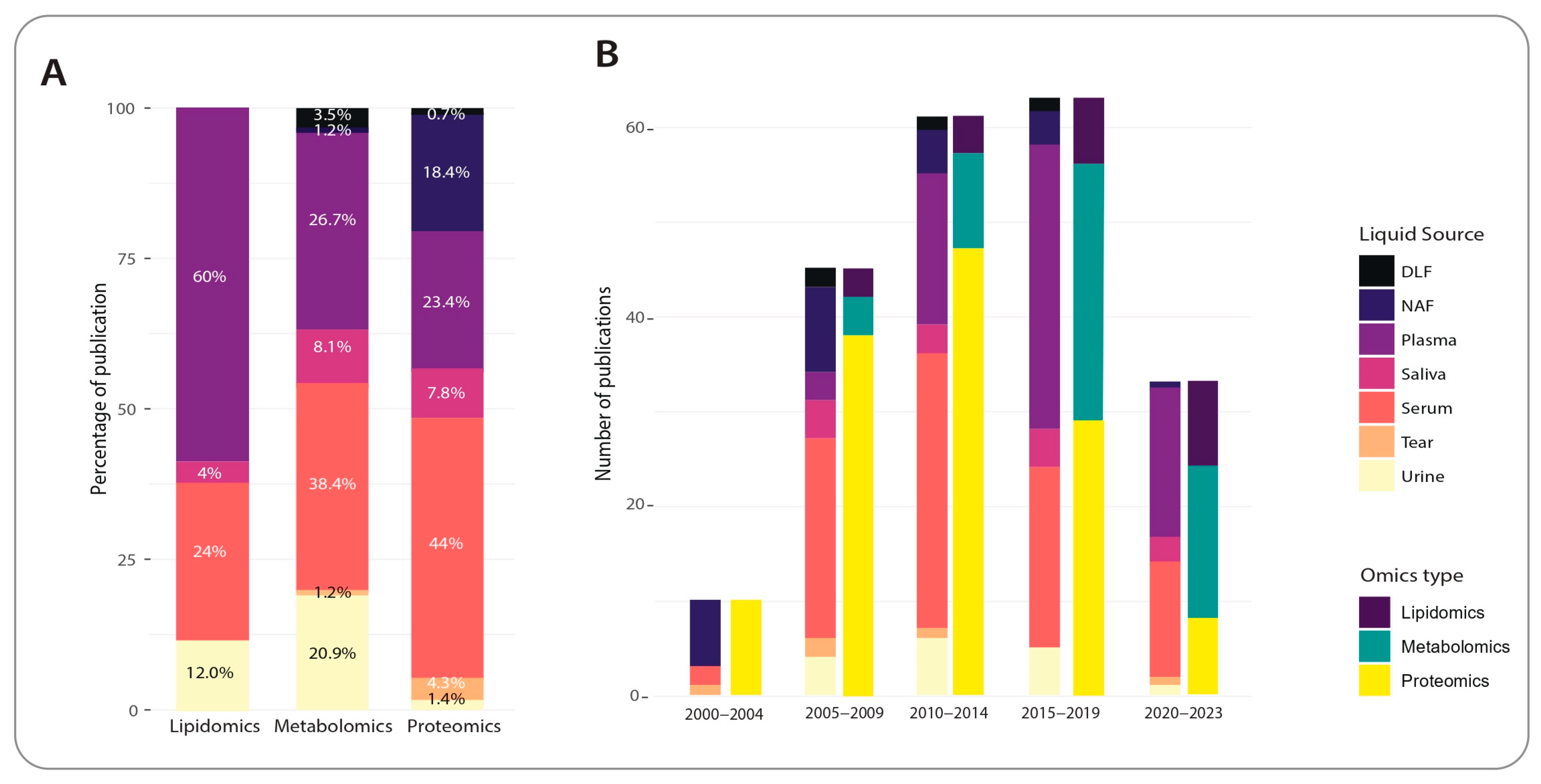
2.3. Trends in Non-Invasive, Non-Nucleotide Biomarker Discovery for Breast Cancer
3. Analytical Techniques for Biomarker Discovery
3.1. Proteomic Approaches
3.2. Metabolomic Approaches
3.3. Lipidomic Approaches
4. Post-Analytical Steps and Variations
4.1. Data Pre-Processing
4.2. Biomarker Signature Panel Identification (Feature Selection)
4.3. Biomarker Predictive Modelling (Classification)
4.4. Clinical Validation
5. Conclusions and Future Perspective
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perry, S.; Kowalski, T.L.; Chang, C.H. Quality of Life Assessment in Women with Breast Cancer: Benefits, Acceptability and Utilization. Health Qual. Life Outcomes 2007, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R. Global, Regional, National Burden of Breast Cancer in 185 Countries: Evidence from GLOBOCAN 2018. Breast Cancer Res. Treat. 2021, 187, 557–567. [Google Scholar] [CrossRef]
- Autier, P.; Boniol, M. Mammography Screening: A Major Issue in Medicine. Eur. J. Cancer 2018, 90, 34–62. [Google Scholar] [CrossRef]
- Løberg, M.; Lousdal, M.L.; Bretthauer, M.; Kalager, M. Benefits and Harms of Mammography Screening. Breast Cancer Res. 2015, 17, 63. [Google Scholar] [CrossRef] [PubMed]
- Bartkowiak, K.; Heidrich, I.; Kwiatkowski, M.; Banys-Paluchowski, M.; Andreas, A.; Wurlitzer, M.; Geffken, M.; Voß, H.; Zeller, T.; Blankenberg, S.; et al. Circulating Cellular Communication Network Factor 1 Protein as a Sensitive Liquid Biopsy Marker for Early Detection of Breast Cancer. Clin. Chem. 2022, 68, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Orlandi, R.; De Bortoli, M.; Ciniselli, C.M.; Vaghi, E.; Caccia, D.; Garrisi, V.; Pizzamiglio, S.; Veneroni, S.; Bonini, C.; Agresti, R.; et al. Hepcidin and Ferritin Blood Level as Noninvasive Tools for Predicting Breast Cancer. Ann. Oncol. 2014, 25, 352–357. [Google Scholar] [CrossRef]
- Jasbi, P.; Wang, D.; Cheng, S.L.; Fei, Q.; Cui, J.Y.; Liu, L.; Wei, Y.; Raftery, D.; Gu, H. Breast Cancer Detection Using Targeted Plasma Metabolomics. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1105, 26–37. [Google Scholar] [CrossRef]
- Beretov, J.; Wasinger, V.C.; Millar, E.K.A.; Schwartz, P.; Graham, P.H.; Li, Y. Proteomic Analysis of Urine to Identify Breast Cancer Biomarker Candidates Using a Label-Free LC-MS/MS Approach. PLoS ONE 2015, 10, e0141876. [Google Scholar] [CrossRef]
- Gajbhiye, A.; Dabhi, R.; Taunk, K.; Vannuruswamy, G.; RoyChoudhury, S.; Adhav, R.; Seal, S.; Mane, A.; Bayatigeri, S.; Santra, M.K.; et al. Urinary Proteome Alterations in HER2 Enriched Breast Cancer Revealed by Multipronged Quantitative Proteomics. Proteomics 2016, 16, 2403–2418. [Google Scholar] [CrossRef]
- Soydinc, H.O.; Duranyildiz, D.; Guney, N.; Derin, D.; Yasasever, V. Utility of Serum and Urine Upar Levels for Diagnosis of Breast Cancer. Asian Pac. J. Cancer Prev. 2012, 13, 2887–2889. [Google Scholar] [CrossRef] [PubMed]
- Takayama, T.; Tsutsui, H.; Shimizu, I.; Toyama, T.; Yoshimoto, N.; Endo, Y.; Inoue, K.; Todoroki, K.; Min, J.Z.; Mizuno, H.; et al. Diagnostic Approach to Breast Cancer Patients Based on Target Metabolomics in Saliva by Liquid Chromatography with Tandem Mass Spectrometry. Clin. Chim. Acta 2016, 452, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Castagnola, M.; Picciotti, P.M.; Messana, I.; Fanali, C.; Fiorita, A.; Cabras, T.; Calò, L.; Pisano, E.; Passali, G.C.; Iavarone, F.; et al. Potential Applications of Human Saliva as Diagnostic Fluid. Acta Otorhinolaryngol. Ital. 2011, 31, 347–357. [Google Scholar] [PubMed]
- Li, J.; Zhao, J.; Yu, X.; Lange, J.; Kuerer, H.; Krishnamurthy, S.; Schilling, E.; Khan, S.A.; Sukumar, S.; Chan, D.W. Identification of Biomarkers for Breast Cancer in Nipple Aspiration and Ductal Lavage Fluid. Clin. Cancer Res. 2005, 11, 8312–8320. [Google Scholar] [CrossRef] [PubMed]
- Noble, J.L.; Dua, R.S.; Coulton, G.R.; Isacke, C.M.; Gui, G.P.H. A Comparative Proteinomic Analysis of Nipple Aspiration Fluid from Healthy Women and Women with Breast Cancer. Eur. J. Cancer 2007, 43, 2315–2320. [Google Scholar] [CrossRef] [PubMed]
- Pawlik, T.M.; Fritsche, H.; Coombes, K.R.; Xiao, L.; Krishnamurthy, S.; Hunt, K.K.; Pusztai, L.; Chen, J.N.; Clarke, C.H.; Arun, B.; et al. Significant Differences in Nipple Aspirate Fluid Protein Expression between Healthy Women and Those with Breast Cancer Demonstrated by Time-of-Flight Mass Spectrometry. Breast Cancer Res. Treat. 2005, 89, 149–157. [Google Scholar] [CrossRef]
- Lebrecht, A.; Boehm, D.; Schmidt, M.; Koelbl, H.; Schwirz, R.L.; Grus, F.H. Diagnosis of Breast Cancer by Tear Proteomic Pattern. Cancer Genom. Proteom. 2009, 6, 177–182. [Google Scholar]
- Böhm, D.; Keller, K.; Pieter, J.; Boehm, N.; Wolters, D.; Siggelkow, W.; Lebrecht, A.; Schmidt, M.; Kölbl, H.; Pfeiffer, N.; et al. Comparison of Tear Protein Levels in Breast Cancer Patients and Healthy Controls Using a de Novo Proteomic Approach. Oncol. Rep. 2012, 28, 429–438. [Google Scholar] [CrossRef]
- Tiwari, E.; Pallipady, A.; Mishra, S. Pre-analytical, Analytical and Postanalytical Errors in Chemical Laboratory. Int. J. Sci. Res. 2013, 4, 2279–2281. [Google Scholar]
- Klont, F.; Horvatovich, P.; Govorukhina, N.; Bischoff, R. Pre- and Post-Analytical Factors in Biomarker Discovery. In Methods in Molecular Biology; Humana Press: New York, NY, USA, 2019; Volume 1959, pp. 1–22. [Google Scholar]
- Zhuang, W.; Camacho, L.; Silva, C.S.; Hong, H. Reproducibility Challenges for Biomarker Detection with Uncertain but Informative Experimental Data. Biomark Med. 2020, 14, 1255–1263. [Google Scholar] [CrossRef]
- Li, J.; Guan, X.; Fan, Z.; Ching, L.M.; Li, Y.; Wang, X.; Cao, W.M.; Liu, D.X. Non-Invasive Biomarkers for Early Detection of Breast Cancer. Cancers 2020, 12, 2767. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Lai, W.; Fan, D.; Fang, Q. Protein Biomarkers in Breast Cancer-Derived Extracellular Vesicles for Use in Liquid Biopsies. Am. J. Physiol. Cell Physiol. 2021, 321, C779–C797. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Chu, P.Y. Current and Developing Liquid Biopsy Techniques for Breast Cancer. Cancers 2022, 14, 2052. [Google Scholar] [CrossRef]
- Seale, K.N.; Tkaczuk, K.H.R. Circulating Biomarkers in Breast Cancer. Clin. Breast Cancer 2022, 22, e319–e331. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zhang, J.; Guo, F.; Zhao, W.; Zhan, Y.; Liu, C.; Fan, Y.; Wang, J. Identification of Apolipoprotein C-I Peptides as a Potential Biomarker and Its Biological Roles in Breast Cancer. Med. Sci. Monit. 2016, 22, 1152–1160. [Google Scholar] [CrossRef]
- Böhm, D.; Keller, K.; Wehrwein, N.; Lebrecht, A.; Schmidt, M.; Kölbl, H.; Grus, F.H. Serum Proteome Profiling of Primary Breast Cancer Indicates a Specific Biomarker Profile. Oncol. Rep. 2011, 26, 1051–1056. [Google Scholar] [CrossRef]
- Scumaci, D.; Tammè, L.; Fiumara, C.V.; Pappaianni, G.; Concolino, A.; Leone, E.; Faniello, M.C.; Quaresima, B.; Ricevuto, E.; Costanzo, F.S.; et al. Plasma Proteomic Profiling in Hereditary Breast Cancer Reveals a BRCA1-Specific Signature: Diagnostic and Functional Implications. PLoS ONE 2015, 10, e0129762. [Google Scholar] [CrossRef]
- Chen, I.H.; Xue, L.; Hsu, C.C.; Paez, J.S.P.; Panb, L.; Andaluz, H.; Wendt, M.K.; Iliuk, A.B.; Zhu, J.K.; Tao, W.A. Phosphoproteins in Extracellular Vesicles as Candidate Markers for Breast Cancer. Proc. Natl. Acad. Sci. USA 2017, 114, 3175–3180. [Google Scholar] [CrossRef]
- Corrêa, S.; Panis, C.; Binato, R.; Herrera, A.C.; Pizzatti, L.; Abdelhay, E. Identifying Potential Markers in Breast Cancer Subtypes Using Plasma Label-Free Proteomics. J. Proteom. 2017, 151, 33–42. [Google Scholar] [CrossRef]
- George, A.L.; Shaheed, S.U.; Sutton, C.W. High-throughput Proteomic Profiling of Nipple Aspirate Fluid from Breast Cancer Patients Compared with Non-cancer Controls: A Step Closer to Clinical Feasibility. J. Clin. Med. 2021, 10, 2243. [Google Scholar] [CrossRef]
- Park, J.; Shin, Y.; Kim, T.H.; Kim, D.H.; Lee, A. Plasma Metabolites as Possible Biomarkers for Diagnosis of Breast Cancer. PLoS ONE 2019, 14, e0225129. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Jasbi, P.; Shi, X.; Turner, C.; Hrovat, J.; Liu, L.; Rabena, Y.; Porter, P.; Gu, H. Early Breast Cancer Detection Using Untargeted and Targeted Metabolomics. J. Proteome Res. 2021, 20, 3124–3133. [Google Scholar] [CrossRef] [PubMed]
- Eniu, D.T.; Romanciuc, F.; Moraru, C.; Goidescu, I.; Eniu, D.; Staicu, A.; Rachieriu, C.; Buiga, R.; Socaciu, C. The Decrease of Some Serum Free Amino Acids Can Predict Breast Cancer Diagnosis and Progression. Scand. J. Clin. Lab. Investig. 2019, 79, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Cala, M.; Aldana, J.; Sánchez, J.; Guio, J.; Meesters, R.J.W. Urinary Metabolite and Lipid Alterations in Colombian Hispanic Women with Breast Cancer: A Pilot Study. J. Pharm. Biomed. Anal. 2018, 152, 234–241. [Google Scholar] [CrossRef]
- Murata, T.; Yanagisawa, T.; Kurihara, T.; Kaneko, M.; Ota, S.; Enomoto, A.; Tomita, M.; Sugimoto, M.; Sunamura, M.; Hayashida, T.; et al. Salivary Metabolomics with Alternative Decision Tree-Based Machine Learning Methods for Breast Cancer Discrimination. Breast Cancer Res. Treat. 2019, 177, 591–601. [Google Scholar] [CrossRef]
- Zhong, L.; Cheng, F.; Lu, X.; Duan, Y.; Wang, X. Untargeted Saliva Metabonomics Study of Breast Cancer Based on Ultra Performance Liquid Chromatography Coupled to Mass Spectrometry with HILIC and RPLC Separations. Talanta 2016, 158, 351–360. [Google Scholar] [CrossRef]
- Jiang, N.; Zhang, G.; Pan, L.; Yan, C.; Zhang, L.; Weng, Y.; Wang, W.; Chen, X.; Yang, G. Potential Plasma Lipid Biomarkers in Early-Stage Breast Cancer. Biotechnol. Lett. 2017, 39, 1657–1666. [Google Scholar] [CrossRef]
- Chen, X.; Chen, H.; Dai, M.; Ai, J.; Li, Y.; Mahon, B.; Dai, S.; Deng, Y. Plasma Lipidomics Profiling Identified Lipid Biomarkers in Distinguishing Early-Stage Breast Cancer from Benign Lesions. Oncotarget 2016, 7, 36622–36631. [Google Scholar] [CrossRef]
- Gumà, J.; Adriá-Cebrián, J.; Ruiz-Aguado, B.; Albacar, C.; Girona, J.; Rodríguez-Calvo, R.; Martínez-Micaelo, N.; Lam, E.W.F.; Masana, L.; Guaita-Esteruelas, S. Altered Serum Metabolic Profile Assessed by Advanced 1h-Nmr in Breast Cancer Patients. Cancers 2021, 13, 4281. [Google Scholar] [CrossRef]
- Bel’Skaya, L.V.; Sarf, E.A.; Kosenok, V.K. Analysis of Saliva Lipids in Breast and Prostate Cancer by IR Spectroscopy. Diagnostics 2021, 11, 1325. [Google Scholar] [CrossRef]
- Tuli, L.; Ressom, H.W. LC-MS Based Detection of Differential Protein Expression. J. Proteom. Bioinform. 2009, 2, 416–438. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.E. Sample Preparation and Fractionation for Proteome Analysis and Cancer Biomarker Discovery by Mass Spectrometry. J. Sep. Sci. 2009, 32, 771–798. [Google Scholar] [CrossRef] [PubMed]
- Luque-Garcia, J.L.; Neubert, T.A. Sample Preparation for Serum/Plasma Profiling and Biomarker Identification by Mass Spectrometry. J. Chromatogr. A 2007, 1153, 259–276. [Google Scholar] [CrossRef]
- Urabe, F.; Kosaka, N.; Ito, K.; Kimura, T.; Egawa, S.; Ochiya, T. Extracellular Vesicles as Biomarkers and Therapeutic Targets for Cancer. Am. J. Physiol. Cell Physiol. 2020, 318, C29–C39. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, Y.; Guo, Z.; Shao, C.; Sun, H.; Zhang, Y.; Sun, Y.; Liu, Y.; Song, Y.; Zhang, L.; et al. A Comparative Proteomics Analysis of Five Body Fluids: Plasma, Urine, Cerebrospinal Fluid, Amniotic Fluid, and Saliva. Proteom. Clin. Appl. 2018, 12, e1800008. [Google Scholar] [CrossRef] [PubMed]
- Athanasatou, A.; Kandyliari, A.; Malisova, O.; Kapsokefalou, M. Fluctuation of Water Intake and of Hydration Indices during the Day in a Sample of Healthy Greek Adults. Nutrients 2019, 11, 793. [Google Scholar] [CrossRef] [PubMed]
- Katsani, K.R.; Sakellari, D. Saliva Proteomics Updates in Biomedicine. J. Biol. Res. 2019, 26, 17. [Google Scholar] [CrossRef]
- Günther, U.L. Metabolomics Biomarkers for Breast Cancer. Pathobiology 2015, 82, 153–165. [Google Scholar] [CrossRef]
- Kaczor-Urbanowicz, K.E.; Martin Carreras-Presas, C.; Aro, K.; Tu, M.; Garcia-Godoy, F.; Wong, D.T.W. Saliva Diagnostics—Current Views and Directions. Exp. Biol. Med. 2017, 242, 459–472. [Google Scholar] [CrossRef]
- Nanjappa, V.; Thomas, J.K.; Marimuthu, A.; Muthusamy, B.; Radhakrishnan, A.; Sharma, R.; Ahmad Khan, A.; Balakrishnan, L.; Sahasrabuddhe, N.A.; Kumar, S.; et al. Plasma Proteome Database as a Resource for Proteomics Research: 2014 Update. Nucleic Acids Res. 2013, 42, D959–D965. [Google Scholar] [CrossRef]
- Quehenberger, O.; Armando, A.M.; Brown, A.H.; Milne, S.B.; Myers, D.S.; Merrill, A.H.; Bandyopadhyay, S.; Jones, K.N.; Kelly, S.; Shaner, R.L.; et al. Lipidomics Reveals a Remarkable Diversity of Lipids in Human Plasma1. J. Lipid Res. 2010, 51, 3299–3305. [Google Scholar] [CrossRef] [PubMed]
- Lawton, K.A.; Berger, A.; Mitchell, M.; Milgram, K.E.; Evans, A.M.; Guo, L.; Hanson, R.W.; Kalhan, S.C.; Ryals, J.A.; Milburn, M.V. Analysis of the Adult Human Plasma Metabolome. Pharmacogenomics 2008, 9, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Aa, J.; Wang, G.; Yan, B.; Zhang, Y.; Wang, X.; Zhao, C.; Cao, B.; Shi, J.; Li, M.; et al. Differences in Metabolite Profile between Blood Plasma and Serum. Anal. Biochem. 2010, 406, 105–112. [Google Scholar] [CrossRef]
- Boyanton, B.L.; Blick, K.E. Stability Studies of Twenty-Four Analytes in Human Plasma and Serum. Clin. Chem. 2002, 48, 2242–2247. [Google Scholar] [CrossRef]
- Haymond, R.E.; Knight, J.A. Venous Serum, Capillary Serum, and Capillary Plasma Compared for Use in Determination of Lactate Dehydrogenase and Aspartate Aminotransferase Activities. Clin. Chem. 1975, 21, 896–897. [Google Scholar] [CrossRef] [PubMed]
- Hyötyläinen, T.; Orešič, M. Optimizing the Lipidomics Workflow for Clinical Studies—Practical Considerations. Anal. Bioanal. Chem. 2015, 407, 4973–4993. [Google Scholar] [CrossRef] [PubMed]
- Lima-Oliveira, G.; Monneret, D.; Guerber, F.; Guidi, G.C. Sample Management for Clinical Biochemistry Assays: Are Serum and Plasma Interchangeable Specimens? Crit. Rev. Clin. Lab. Sci. 2018, 55, 480–500. [Google Scholar] [CrossRef]
- Breier, M.; Wahl, S.; Prehn, C.; Fugmann, M.; Ferrari, U.; Weise, M.; Banning, F.; Seissler, J.; Grallert, H.; Adamski, J.; et al. Targeted Metabolomics Identifies Reliable and Stable Metabolites in Human Serum and Plasma Samples. PLoS ONE 2014, 9, e89728. [Google Scholar] [CrossRef]
- Paglia, G.; Del Greco, F.M.; Sigurdsson, B.B.; Rainer, J.; Volani, C.; Hicks, A.A.; Pramstaller, P.P.; Smarason, S.V. Influence of Collection Tubes during Quantitative Targeted Metabolomics Studies in Human Blood Samples. Clin. Chim. Acta 2018, 486, 320–328. [Google Scholar] [CrossRef]
- Yu, Z.; Kastenmüller, G.; He, Y.; Belcredi, P.; Möller, G.; Prehn, C.; Mendes, J.; Wahl, S.; Roemisch-Margl, W.; Ceglarek, U.; et al. Differences between Human Plasma and Serum Metabolite Profiles. PLoS ONE 2011, 6, e21230. [Google Scholar] [CrossRef]
- Ishikawa, M.; Maekawa, K.; Saito, K.; Senoo, Y.; Urata, M.; Murayama, M.; Tajima, Y.; Kumagai, Y.; Saito, Y. Plasma and Serum Lipidomics of Healthy White Adults Shows Characteristic Profiles by Subjects’ Gender and Age. PLoS ONE 2014, 9, e91806. [Google Scholar] [CrossRef] [PubMed]
- Ignjatovic, V.; Geyer, P.E.; Palaniappan, K.K.; Chaaban, J.E.; Omenn, G.S.; Baker, M.S.; Deutsch, E.W.; Schwenk, J.M. Mass Spectrometry-Based Plasma Proteomics: Considerations from Sample Collection to Achieving Translational Data. J. Proteome Res. 2019, 18, 4085–4097. [Google Scholar] [CrossRef] [PubMed]
- Geyer, P.E.; Voytik, E.; Treit, P.V.; Doll, S.; Kleinhempel, A.; Niu, L.; Müller, J.B.; Buchholtz, M.; Bader, J.M.; Teupser, D.; et al. Plasma Proteome Profiling to Detect and Avoid Sample-related Biases in Biomarker Studies. EMBO Mol. Med. 2019, 11, e10427. [Google Scholar] [CrossRef]
- Tammen, H.; Schulte, I.; Hess, R.; Menzel, C.; Kellmann, M.; Mohring, T.; Schulz-Knappe, P. Peptidomic Analysis of Human Blood Specimens: Comparison between Plasma Specimens and Serum by Differential Peptide Display. Proteomics 2005, 5, 3414–3422. [Google Scholar] [CrossRef] [PubMed]
- Omenn, G.S.; States, D.J.; Adamski, M.; Blackwell, T.W.; Menon, R.; Hermjakob, H.; Apweiler, R.; Haab, B.B.; Simpson, R.J.; Eddes, J.S.; et al. Overview of the HUPO Plasma Proteome Project: Results from the Pilot Phase with 35 Collaborating Laboratories and Multiple Analytical Groups, Generating a Core Dataset of 3020 Proteins and a Publicly-Available Database. Proteomics 2005, 5, 3226–3245. [Google Scholar] [CrossRef]
- Thongboonkerd, V. Urinary Proteomics: Towards Biomarker Discovery, Diagnostics and Prognostics. Mol. Biosyst. 2008, 4, 810–815. [Google Scholar] [CrossRef]
- Beretov, J.; Wasinger, V.C.; Graham, P.H.; Millar, E.K.; Kearsley, J.H.; Li, Y. Proteomics for Breast Cancer Urine Biomarkers. Adv. Clin. Chem. 2014, 63, 123–167. [Google Scholar] [PubMed]
- Lehmann, R. From Bedside to Bench—Practical Considerations to Avoid Pre-Analytical Pitfalls and Assess Sample Quality for High-Resolution Metabolomics and Lipidomics Analyses of Body Fluids. Anal. Bioanal. Chem. 2021, 413, 5567–5585. [Google Scholar] [CrossRef] [PubMed]
- Bauça, J.M.; Martínez-Morillo, E.; Diamandis, E.P. Peptidomics of Urine and Other Biofluids for Cancer Diagnostics. Clin. Chem. 2014, 60, 1052–1061. [Google Scholar] [CrossRef]
- Thongboonkerd, V.; Chutipongtanate, S.; Kanlaya, R. Systematic Evaluation of Sample Preparation Methods for Gel-Based Human Urinary Proteomics: Quantity, Quality, and Variability. J. Proteome Res. 2006, 5, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Eric Thomas, C.; Sexton, W.; Benson, K.; Sutphen, R.; Koomen, J. Urine Collection and Processing for Protein Biomarker Discovery and Quantification. Cancer Epidemiol. Biomark. Prev. 2010, 19, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yin, P.; Shao, Y.; Wang, Z.; Wang, B.; Lehmann, R.; Xu, G. Which Is the Urine Sample Material of Choice for Metabolomics-Driven Biomarker Studies? Anal. Chim. Acta 2020, 1105, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Slupsky, C.M.; Steed, H.; Wells, T.H.; Dabbs, K.; Schepansky, A.; Capstick, V.; Faught, W.; Sawyer, M.B. Urine Metabolite Analysis Offers Potential Early Diagnosis of Ovarian and Breast Cancers. Clin. Cancer Res. 2010, 16, 5835–5841. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Woo, H.M.; Kong, G.; Nam, S.J.; Chung, B.C. Discovery of Urinary Biomarkers in Patients with Breast Cancer Based on Metabolomics. Mass Spectrom. Lett. 2013, 4, 59–66. [Google Scholar] [CrossRef]
- More, T.H.; Taware, R.; Taunk, K.; Chanukuppa, V.; Naik, V.; Mane, A.; Rapole, S. Investigation of Altered Urinary Metabolomic Profiles of Invasive Ductal Carcinoma of Breast Using Targeted and Untargeted Approaches. Metabolomics 2018, 14, 107. [Google Scholar] [CrossRef]
- Silva, C.L.; Passos, M.; Câmara, J.S. Solid Phase Microextraction, Mass Spectrometry and Metabolomic Approaches for Detection of Potential Urinary Cancer Biomarkers—A Powerful Strategy for Breast Cancer Diagnosis. Talanta 2012, 89, 360–368. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, Y.; Zhang, X. Metabonomics Studies on Serum and Urine of Patients with Breast Cancer Using 1H-NMR Spectroscopy. Oncotarget 2017, 5. [Google Scholar] [CrossRef]
- Kim, H.; Min, H.K.; Kong, G.; Moon, M.H. Quantitative Analysis of Phosphatidylcholines and Phosphatidylethanolamines in Urine of Patients with Breast Cancer by Nanoflow Liquid Chromatography/Tandem Mass Spectrometry. Anal. Bioanal. Chem. 2009, 393, 1649–1656. [Google Scholar] [CrossRef]
- Min, H.K.; Kong, G.; Moon, M.H. Quantitative Analysis of Urinary Phospholipids Found in Patients with Breast Cancer by Nanoflow Liquid Chromatography-Tandem Mass Spectrometry: II. Negative Ion Mode Analysis of Four Phospholipid Classes. Anal. Bioanal. Chem. 2010, 396, 1273–1280. [Google Scholar] [CrossRef]
- Wang, H.; Altemus, J.; Niazi, F.; Green, H.; Calhoun, B.C.; Sturgis, C.; Grobmyer, S.R.; Eng, C. Breast Tissue, Oral and Urinary Microbiomes in Breast Cancer. Oncotarget 2017, 8, 88122–88138. [Google Scholar] [CrossRef]
- Rentka, A.; Koroskenyi, K.; Harsfalvi, J.; Szekanecz, Z.; Szucs, G.; Szodoray, P.; Kemeny-Beke, A. Evaluation of Commonly Used Tear Sampling Methods and Their Relevance in Subsequent Biochemical Analysis. Ann. Clin. Biochem. 2017, 54, 521–529. [Google Scholar] [CrossRef]
- Nättinen, J.; Aapola, U.; Jylhä, A.; Vaajanen, A.; Uusitalo, H. Comparison of Capillary and Schirmer Strip Tear Fluid Sampling Methods Using Swath-Ms Proteomics Approach. Transl. Vis. Sci. Technol. 2020, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Ponzini, E.; Santambrogio, C.; De Palma, A.; Mauri, P.; Tavazzi, S.; Grandori, R. Mass Spectrometry-Based Tear Proteomics for Noninvasive Biomarker Discovery. Mass Spectrom. Rev. 2022, 41, 842–860. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Beuerman, R.W. Tear Analysis in Ocular Surface Diseases. Prog. Retin. Eye Res. 2012, 31, 527–550. [Google Scholar] [CrossRef] [PubMed]
- Pieragostino, D.; D’Alessandro, M.; di Ioia, M.; Di Ilio, C.; Sacchetta, P.; Del Boccio, P. Unraveling the Molecular Repertoire of Tears as a Source of Biomarkers: Beyond Ocular Diseases. Proteom. Clin. Appl. 2015, 9, 169–186. [Google Scholar] [CrossRef] [PubMed]
- Lebrecht, A.; Boehm, D.; Schmidt, M.; Koelbl, H.; Grus, F.H. Surface-Enhanced Laser Desorption/Ionisation Time-of-Flight Mass Spectrometry to Detect Breast Cancer Markers in Tears and Serum. Cancer Genom. Proteom. 2009, 6, 75–84. [Google Scholar]
- Morimoto, Y.; Conroy, S.M.; Franke, A.A.; Maskarinec, G. Nipple Aspirate Fluid Producer Status among Premenopausal Women in Hawaii. Breast J. 2012, 18, 504–505. [Google Scholar] [CrossRef]
- Suijkerbuijk, K.P.M.; Van Der Wall, E.; Meijrink, H.; Pan, X.; Rinkes, I.H.M.B.; Ausems, M.G.E.M.; Van Diest, P.J. Successful Oxytocin-Assisted Nipple Aspiration in Women at Increased Risk for Breast Cancer. Fam. Cancer 2010, 9, 321–325. [Google Scholar] [CrossRef]
- Shaheed, S.U.; Tait, C.; Kyriacou, K.; Mullarkey, J.; Burrill, W.; Patterson, L.H.; Linforth, R.; Salhab, M.; Sutton, C.W. Nipple Aspirate Fluid—A Liquid Biopsy for Diagnosing Breast Health. Proteom. Clin. Appl. 2017, 11, 1700015. [Google Scholar] [CrossRef]
- Shaheed, S.U.; Tait, C.; Kyriacou, K.; Linforth, R.; Salhab, M.; Sutton, C. Evaluation of Nipple Aspirate Fluid as a Diagnostic Tool for Early Detection of Breast Cancer. Clin. Proteom. 2018, 15, 3. [Google Scholar] [CrossRef]
- Patuleia, S.I.S.; Suijkerbuijk, K.P.M.; van der Wall, E.; van Diest, P.J.; Moelans, C.B. Nipple Aspirate Fluid at a Glance. Cancers 2022, 14, 159. [Google Scholar] [CrossRef]
- Chan, A.A.; Bashir, M.; Rivas, M.N.; Duvall, K.; Sieling, P.A.; Pieber, T.R.; Vaishampayan, P.A.; Love, S.M.; Lee, D.J. Characterization of the Microbiome of Nipple Aspirate Fluid of Breast Cancer Survivors. Sci. Rep. 2016, 6, 28061. [Google Scholar] [CrossRef] [PubMed]
- Bel’Skaya, L.V.; Sarf, E.A.; Solomatin, D.V.; Kosenok, V.K. Metabolic Features of Saliva in Breast Cancer Patients. Metabolites 2022, 12, 166. [Google Scholar] [CrossRef]
- Zhang, L.; Xiao, H.; Karlan, S.; Zhou, H.; Gross, J.; Elashoff, D.; Akin, D.; Yan, X.; Chia, D.; Karlan, B.; et al. Discovery and Preclinical Validation of Salivary Transcriptomic and Proteomic Biomarkers for the Non- Invasive Detection of Breast Cancer. PLoS ONE 2010, 5, e15573. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Yang, M.; Zhu, J.; Zhang, H.; Duan, Z.; Wang, S.; Liao, Z.; Liu, W. Developments in Diagnostic Applications of Saliva in Human Organ Diseases. Med. Nov. Technol. Devices 2022, 13, 100115. [Google Scholar] [CrossRef]
- Assad, D.X.; Acevedo, A.C.; Mascarenhas, E.C.P.; Normando, A.G.C.; Pichon, V.; Chardin, H.; Guerra, E.N.S.; Combes, A. Using an Untargeted Metabolomics Approach to Identify Salivary Metabolites in Women with Breast Cancer. Metabolites 2020, 10, 506. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.Y.; Liao, Y.; Hosseinifard, H.; Imani, S.; Wen, Q.L. Diagnostic Role of Extracellular Vesicles in Cancer: A Comprehensive Systematic Review and Meta-Analysis. Front. Cell Dev. Biol. 2021, 9, 2749. [Google Scholar] [CrossRef] [PubMed]
- Lane, R.E.; Korbie, D.; Hill, M.M.; Trau, M. Extracellular Vesicles as Circulating Cancer Biomarkers: Opportunities and Challenges. Clin. Transl. Med. 2018, 7, 14. [Google Scholar] [CrossRef]
- Grölz, D.; Hauch, S.; Schlumpberger, M.; Guenther, K.; Voss, T.; Sprenger-Haussels, M.; Oelmüller, U. Liquid Biopsy Preservation Solutions for Standardized Pre-Analytical Workflows—Venous Whole Blood and Plasma. Curr. Pathobiol. Rep. 2018, 6, 275–286. [Google Scholar] [CrossRef]
- Salvianti, F.; Gelmini, S.; Costanza, F.; Mancini, I.; Sonnati, G.; Simi, L.; Pazzagli, M.; Pinzani, P. The Pre-Analytical Phase of the Liquid Biopsy. N. Biotechnol. 2020, 55, 19–29. [Google Scholar] [CrossRef]
- Lacroix, R.; Judicone, C.; Poncelet, P.; Robert, S.; Arnaud, L.; Sampol, J.; Dignat-George, F. Impact of Pre-Analytical Parameters on the Measurement of Circulating Microparticles: Towards Standardization of Protocol. J. Thromb. Haemost. 2012, 10, 437–446. [Google Scholar] [CrossRef]
- Abramowicz, A.; Widlak, P.; Pietrowska, M. Proteomic Analysis of Exosomal Cargo: The Challenge of High Purity Vesicle Isolation. Mol. Biosyst. 2016, 12, 1407–1419. [Google Scholar] [CrossRef]
- Van Deun, J.; Mestdagh, P.; Agostinis, P.; Akay, Ö.; Anand, S.; Anckaert, J.; Martinez, Z.A.; Baetens, T.; Beghein, E.; Bertier, L.; et al. EV-TRACK: Transparent Reporting and Centralizing Knowledge in Extracellular Vesicle Research. Nat. Methods 2017, 14, 228–232. [Google Scholar]
- Siwaponanan, P.; Keawvichit, R.; Lekmanee, K.; Chomanee, N.; Pattanapanyasat, K. Enumeration and Phenotyping of Circulating Microvesicles by Flow Cytometry and Nanoparticle Tracking Analysis: Plasma versus Serum. Int. J. Lab. Hematol. 2021, 43, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Palviainen, M.; Saraswat, M.; Varga, Z.; Kitka, D.; Neuvonen, M.; Puhka, M.; Joenväärä, S.; Renkonen, R.; Nieuwland, R.; Takatalo, M.; et al. Extracellular Vesicles from Human Plasma and Serum Are Carriers of Extravesicular Cargo—Implications for Biomarker Discovery. PLoS ONE 2020, 15, e0236439. [Google Scholar] [CrossRef] [PubMed]
- Yekula, A.; Muralidharan, K.; Kang, K.M.; Wang, L.; Balaj, L.; Carter, B.S. From Laboratory to Clinic: Translation of Extracellular Vesicle Based Cancer Biomarkers. Methods 2020, 177, 58–66. [Google Scholar] [CrossRef]
- Daly, R.; O’Driscoll, L. Extracellular Vesicles in Blood: Are They Viable as Diagnostic and Predictive Tools in Breast Cancer? Drug Discov. Today 2021, 26, 778–785. [Google Scholar] [CrossRef] [PubMed]
- Hassis, M.E.; Niles, R.K.; Braten, M.N.; Albertolle, M.E.; Ewa Witkowska, H.; Hubel, C.A.; Fisher, S.J.; Williams, K.E. Evaluating the Effects of Pre-analytical Variables on the Stability of the Human Plasma Proteome. Anal. Biochem. 2015, 478, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Bowen, R.A.R.; Adcock, D.M. Blood Collection Tubes as Medical Devices: The Potential to Affect Assays and Proposed Verification and Validation Processes for the Clinical Laboratory. Clin. Biochem. 2016, 49, 1321–1330. [Google Scholar] [CrossRef]
- Greco, V.; Piras, C.; Pieroni, L.; Urbani, A. Direct Assessment of Plasma/Serum Sample Quality for Proteomics Biomarker Investigation. Methods Mol. Biol. 2017, 1619, 3–21. [Google Scholar] [PubMed]
- Villanueva, J.; Philip, J.; Chaparro, C.A.; Li, Y.; Toledo-Crow, R.; DeNoyer, L.; Fleisher, M.; Robbins, R.J.; Tempst, P. Correcting Common Errors in Identifying Cancer-Specific Serum Peptide Signatures. J. Proteome Res. 2005, 4, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, S.Y.; Chen, R.K.; Pan, Y.H.; Lee, H.L. Systematical Evaluation of the Effects of Sample Collection Procedures on Low-Molecular-Weight Serum/Plasma Proteome Profiling. Proteomics 2006, 6, 3189–3198. [Google Scholar] [CrossRef] [PubMed]
- Bowen, R.A.R.; Hortin, G.L.; Csako, G.; Otañez, O.H.; Remaley, A.T. Impact of Blood Collection Devices on Clinical Chemistry Assays. Clin. Biochem. 2010, 43, 4–25. [Google Scholar] [CrossRef] [PubMed]
- Yin, P.; Peter, A.; Franken, H.; Zhao, X.; Neukamm, S.S.; Rosenbaum, L.; Lucio, M.; Zell, A.; Häring, H.U.; Xu, G.; et al. Pre-analytical Aspects and Sample Quality Assessment in Metabolomics Studies of Human Blood. Clin Chem 2013, 59, 833–845. [Google Scholar] [CrossRef]
- Barri, T.; Dragsted, L.O. UPLC-ESI-QTOF/MS and Multivariate Data Analysis for Blood Plasma and Serum Metabolomics: Effect of Experimental Artefacts and Anticoagulant. Anal. Chim. Acta 2013, 768, 118–128. [Google Scholar] [CrossRef]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-Mcintyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for Large-Scale Metabolic Profiling of Serum and Plasma Using Gas Chromatography and Liquid Chromatography Coupled to Mass Spectrometry. Nat. Protoc. 2011, 6, 1060–1083. [Google Scholar] [CrossRef]
- Kamlage, B.; Maldonado, S.G.; Bethan, B.; Peter, E.; Schmitz, O.; Liebenberg, V.; Schatz, P. Quality Markers Addressing Pre-analytical Variations of Blood and Plasma Processing Identified by Broad and Targeted Metabolite Profiling. Clin. Chem. 2014, 60, 399–412. [Google Scholar] [CrossRef]
- Liu, X.; Hoene, M.; Yin, P.; Fritsche, L.; Plomgaard, P.; Hansen, J.S.; Nakas, C.T.; Niess, A.M.; Hudemann, J.; Haap, M.; et al. Quality Control of Serum and Plasma by Quantification of (4E,14Z)-Sphingadienine-C18-1-Phosphate Uncovers Common Pre-analytical Errors during Handling of Whole Blood. Clin. Chem. 2018, 64, 810–819. [Google Scholar] [CrossRef]
- Teahan, O.; Gamble, S.; Holmes, E.; Waxman, J.; Nicholson, J.K.; Bevan, C.; Keun, H.C. Impact of Analytical Bias in Metabonomic Studies of Human Blood Serum and Plasma. Anal. Chem. 2006, 78, 4307–4318. [Google Scholar] [CrossRef]
- Halvey, P.; Farutin, V.; Koppes, L.; Gunay, N.S.; Pappas, D.A.; Manning, A.M.; Capila, I. Variable Blood Processing Procedures Contribute to Plasma Proteomic Variability. Clin. Proteom. 2021, 18, 5. [Google Scholar] [CrossRef]
- Lippi, G.; Blanckaert, N.; Bonini, P.; Green, S.; Kitchen, S.; Palicka, V.; Vassault, A.J.; Plebani, M. Haemolysis: An Overview of the Leading Cause of Unsuitable Specimens in Clinical Laboratories. Clin. Chem. Lab. Med. 2008, 46, 764–772. [Google Scholar] [CrossRef] [PubMed]
- Ammerlaan, W.; Trezzi, J.P.; Lescuyer, P.; Mathay, C.; Hiller, K.; Betsou, F. Method Validation for Preparing Serum and Plasma Samples from Human Blood for Downstream Proteomic, Metabolomic, and Circulating Nucleic Acid-Based Applications. Biopreserv. Biobank. 2014, 12, 269–280. [Google Scholar] [CrossRef]
- Stevens, V.L.; Hoover, E.; Wang, Y.; Zanetti, K.A. Pre-Analytical Factors That Affect Metabolite Stability in Human Urine, Plasma, and Serum: A Review. Metabolites 2019, 9, 156. [Google Scholar] [CrossRef] [PubMed]
- Züllig, T.; Trötzmüller, M.; Köfeler, H.C. Lipidomics from Sample Preparation to Data Analysis: A Primer. Anal Bioanal Chem 2020, 412, 2191–2209. [Google Scholar] [CrossRef]
- Scherer, M.; Schmitz, G.; Liebisch, G. High-Throughput Analysis of Sphingosine 1-Phosphate, Sphinganine 1-Phosphate, and Lysophosphatidic Acid in Plasma Samples by Liquid Chromatography—Tandem Mass Spectrometry. Clin. Chem. 2009, 55, 1218–1222. [Google Scholar] [CrossRef]
- Jain, M.; Kennedy, A.D.; Elsea, S.H.; Miller, M.J. Analytes Related to Erythrocyte Metabolism Are Reliable Biomarkers for Pre-analytical Error Due to Delayed Plasma Processing in Metabolomics Studies. Clin. Chim. Acta 2017, 466, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Barelli, S.; Crettaz, D.; Thadikkaran, L.; Rubin, O.; Tissot, J.D. Plasma/Serum Proteomics: Pre-Analytical Issues. Expert Rev. Proteom. 2007, 4, 363–370. [Google Scholar] [CrossRef]
- Lesche, D.; Geyer, R.; Lienhard, D.; Nakas, C.T.; Diserens, G.; Vermathen, P.; Leichtle, A.B. Does Centrifugation Matter? Centrifugal Force and Spinning Time Alter the Plasma Metabolome. Metabolomics 2016, 12, 159. [Google Scholar] [CrossRef]
- Rai, A.J.; Vitzthum, F. Effects of Pre-analytical Variables on Peptide and Protein Measurements in Human Serum and Plasma: Implications for Clinical Proteomics. Expert Rev. Proteom. 2006, 3, 409–426. [Google Scholar] [CrossRef]
- Valo, E.; Colombo, M.; Sandholm, N.; McGurnaghan, S.J.; Blackbourn, L.A.K.; Dunger, D.B.; McKeigue, P.M.; Forsblom, C.; Groop, P.H.; Colhoun, H.M.; et al. Effect of Serum Sample Storage Temperature on Metabolomic and Proteomic Biomarkers. Sci. Rep. 2022, 12, 4571. [Google Scholar] [CrossRef]
- Ferguson, R.E.; Hochstrasser, D.F.; Banks, R.E. Impact of Pre-analytical Variables on the Analysis of Biological Fluids in Proteomic Studies. Proteom. Clin. Appl. 2007, 1, 739–746. [Google Scholar] [CrossRef] [PubMed]
- Zander, J.; Bruegel, M.; Kleinhempel, A.; Becker, S.; Petros, S.; Kortz, L.; Dorow, J.; Kratzsch, J.; Baber, R.; Ceglarek, U.; et al. Effect of Biobanking Conditions on Short-Term Stability of Biomarkers in Human Serum and Plasma. Clin. Chem. Lab. Med. 2014, 52, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Kim, J.W.; Jeon, S.Y.; Park, B.K.; Han, B.G. Proteomic Analysis of the Effect of Storage Temperature on Human Serum. Ann. Clin. Lab. Sci. 2010, 40, 61–70. [Google Scholar] [PubMed]
- Mitchell, B.L.; Yasui, Y.; Li, C.I.; Fitzpatrick, A.L.; Lampe, P.D. Impact of Freeze-Thaw Cycles and Storage Time on Plasma Samples Used in Mass Spectrometry Based Biomarker Discovery Projects. Cancer Inform. 2005, 1, 98–104. [Google Scholar] [CrossRef]
- Fliniaux, O.; Gaillard, G.; Lion, A.; Cailleu, D.; Mesnard, F.; Betsou, F. Influence of Common Pre-analytical Variations on the Metabolic Profile of Serum Samples in Biobanks. J. Biomol. NMR 2011, 51, 457–465. [Google Scholar] [CrossRef]
- Gardner, A.; Carpenter, G.; So, P.W. Salivary Metabolomics: From Diagnostic Biomarker Discovery to Investigating Biological Function. Metabolites 2020, 10, 47. [Google Scholar] [CrossRef]
- Li, Y.; Xun, D.; Li, L.; Wang, B.; Lv, J.; Liu, H.; Zhu, L.; Ma, F.; Chen, X.; Tian, S.; et al. Deep Dive on the Proteome of Human Body Fluids: A Valuable Data Resource for Biomarker Discovery. Cancer Genom. Proteom. 2021, 18, 549–568. [Google Scholar] [CrossRef]
- Dayon, L.; Cominetti, O.; Affolter, M. Proteomics of Human Biological Fluids for Biomarker Discoveries: Technical Advances and Recent Applications. Expert Rev. Proteom. 2022, 19, 131–151. [Google Scholar] [CrossRef]
- Panneerselvam, K.; Ishikawa, S.; Krishnan, R.; Sugimoto, M. Salivary Metabolomics for Oral Cancer Detection: A Narrative Review. Metabolites 2022, 12, 436. [Google Scholar] [CrossRef]
- Srivastava, A.; Creek, D.J. Discovery and Validation of Clinical Biomarkers of Cancer: A Review Combining Metabolomics and Proteomics. Proteomics 2019, 19, e1700448. [Google Scholar] [CrossRef]
- Pusch, W.; Kostrzewa, M. Application of MALDI-TOF Mass Spectrometry in Screening and Diagnostic Research. Curr. Pharm. Des. 2005, 11, 2577–2591. [Google Scholar] [CrossRef] [PubMed]
- Hosnedlova, B.; Kepinska, M.; Ruttkay-Nedecky, B.; Fernandez, C.; Parak, T.; Milnerowicz, H.; Sochor, J.; Bjørklund, G.; Kizek, R. Matrix Assisted Laser Desorption/Ionization as a New Cancer Diagnostic Tool. In Encyclopedia of Biomedical Engineering; Elsevier: Cambridge, MA, USA, 2019; Volume 1–3, pp. 400–414. ISBN 9780128051443. [Google Scholar]
- Gutierrez, J.A.; Dorocke, J.A.; Knierman, M.D.; Gelfanova, V.; Higgs, R.E.; Koh, N.L.; Hale, J.E. Quantitative Determination of Peptides Using Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry. Biotechniques 2005, 38, S13–S17. [Google Scholar] [CrossRef] [PubMed]
- Albrethsen, J. Reproducibility in Protein Profiling by MALDI-TOF Mass Spectrometry. Clin. Chem. 2007, 53, 852–858. [Google Scholar] [CrossRef] [PubMed]
- Kiehntopf, M.; Siegmund, R.; Deufel, T. Use of SELDI-TOF Mass Spectrometry for Identification of New Biomarkers: Potential and Limitations. Clin. Chem. Lab. Med. 2007, 45, 1435–1449. [Google Scholar] [CrossRef]
- Hu, T.; Zhang, J.L. Mass-Spectrometry-Based Lipidomics. J. Sep. Sci. 2018, 41, 351–372. [Google Scholar] [CrossRef]
- Xie, F.; Liu, T.; Qian, W.J.; Petyuk, V.A.; Smith, R.D. Liquid Chromatography-Mass Spectrometry-Based Quantitative Proteomics. J. Biol. Chem. 2011, 286, 25443–25449. [Google Scholar] [CrossRef]
- Grebe, S.K.G.; Singh, R.J. LC-MS/MS in the Clinical Laboratory—Where to from Here? Clin. Biochem. Rev. 2011, 32, 5–31. [Google Scholar]
- Lubes, G.; Goodarzi, M. GC–MS Based Metabolomics Used for the Identification of Cancer Volatile Organic Compounds as Biomarkers. J. Pharm. Biomed. Anal. 2018, 147, 313–322. [Google Scholar] [CrossRef]
- Emwas, A.H.M. The Strengths and Weaknesses of NMR Spectroscopy and Mass Spectrometry with Particular Focus on Metabolomics Research. Methods Mol. Biol. 2015, 1277, 161–193. [Google Scholar] [CrossRef]
- Meftahi, G.H.; Bahari, Z.; Zarei Mahmoudabadi, A.; Iman, M.; Jangravi, Z. Applications of Western Blot Technique: From Bench to Bedside. Biochem. Mol. Biol. Educ. 2021, 49, 509–517. [Google Scholar] [CrossRef]
- Monteoliva, L.; Albar, J.P. Differential Proteomics: An Overview of Gel and Non-Gel Based Approaches. Brief. Funct. Genom. Proteom. 2004, 3, 220–239. [Google Scholar] [CrossRef] [PubMed]
- Baggerman, G.; Vierstraete, E.; De Loof, A.; Schoofs, L. Gel-Based Versus Gel-Free Proteomics: A Review. Comb. Chem. High Throughput Screen. 2005, 8, 669–677. [Google Scholar] [CrossRef] [PubMed]
- Meleady, P. Two-Dimensional Gel Electrophoresis and 2D-DIGE. Methods Mol. Biol. 2018, 1664, 3–14. [Google Scholar] [PubMed]
- Beckett, P. The Basics of 2D DIGE. Methods Mol. Biol. 2012, 854, 9–19. [Google Scholar] [CrossRef]
- Hosseini, S.; Vázquez-Villegas, P.; Rito-Palomares, M.; Martinez-Chapa, S.O. Advantages, Disadvantages and Modifications of Conventional ELISA. Springer Briefs in Applied Sciences and Technology; Springer: Singapore, 2018; pp. 67–115. [Google Scholar]
- Sakamoto, S.; Putalun, W.; Vimolmangkang, S.; Phoolcharoen, W.; Shoyama, Y.; Tanaka, H.; Morimoto, S. Enzyme-Linked Immunosorbent Assay for the Quantitative/Qualitative Analysis of Plant Secondary Metabolites. J. Nat. Med. 2018, 72, 32–42. [Google Scholar] [CrossRef]
- Wang, K.; Huang, C.; Nice, E.C. Proteomics, Genomics and Transcriptomics: Their Emerging Roles in the Discovery and Validation of Colorectal Cancer Biomarkers. Expert Rev. Proteom. 2014, 11, 179–205. [Google Scholar] [CrossRef]
- Bratulic, S.; Gatto, F.; Nielsen, J. The Translational Status of Cancer Liquid Biopsies. Regen. Eng. Transl. Med. 2021, 7, 312–352. [Google Scholar] [CrossRef]
- Hajduk, J.; Matysiak, J.; Kokot, Z.J. Challenges in Biomarker Discovery with MALDI-TOF MS. Clin. Chim. Acta 2016, 458, 84–98. [Google Scholar] [CrossRef]
- Chandramouli, K.; Qian, P.-Y. Proteomics: Challenges, Techniques and Possibilities to Overcome Biological Sample Complexity. Hum. Genom. Proteom. 2009, 1, 239204. [Google Scholar] [CrossRef]
- Jiang, M.; Gu, G.; Ni, B.; Wang, W.; Shi, J.; Liao, P.; Hu, H. Detection of Serum Protein Biomarkers by Surface Enhanced Laser Desorption/Ionization in Patients with Adenocarcinoma of the Lung. Asia Pac. J. Clin. Oncol. 2014, 10, e7–e12. [Google Scholar] [CrossRef]
- Muthu, M.; Vimala, A.; Mendoza, O.H.; Gopal, J. Tracing the Voyage of SELDI-TOF MS in Cancer Biomarker Discovery and Its Current Depreciation Trend—Need for Resurrection? TrAC Trends Anal. Chem. 2016, 76, 95–101. [Google Scholar] [CrossRef]
- Van, Q.N.; Veenstra, T.D. How Close Is the Bench to the Bedside? Metabolic Profiling in Cancer Research. Genome Med. 2009, 1, 5. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.; Zhang, X.; He, Y.; Chen, H.; Liu, M.; Wang, H.; Tang, L.; Tu, G.; Ding, M. A Pseudo-Targeted Metabolomics Study Based on Serum Bile Acids Profiling for the Differential Diagnosis of Benign and Malignant Breast Lesions. Steroids 2021, 175, 108914. [Google Scholar] [CrossRef] [PubMed]
- Marian, C.; Varghese, R.S.; Ahn, J.; Da Cunha, P.A.; Willey, S.; Sidawy, M.; Rone, J.D.; Cheema, A.K.; Luta, G.; Nezami Ranjbar, M.R.; et al. Metabolomic Profiling of Breast Tumors Using Ductal Fluid. Int. J. Oncol. 2016, 49, 2245–2254. [Google Scholar] [CrossRef]
- Oktay, K.; Santaliz-Casiano, A.; Patel, M.; Marino, N.; Storniolo, A.M.V.; Torun, H.; Acar, B.; Madak Erdogan, Z. A Computational Statistics Approach to Evaluate Blood Biomarkers for Breast Cancer Risk Stratification. Horm. Cancer 2020, 11, 17–33. [Google Scholar] [CrossRef]
- Hadi, N.I.; Jamal, Q.; Iqbal, A.; Shaikh, F.; Somroo, S.; Musharraf, S.G. Serum Metabolomic Profiles for Breast Cancer Diagnosis, Grading and Staging by Gas Chromatography-Mass Spectrometry. Sci. Rep. 2017, 7, 1715. [Google Scholar] [CrossRef]
- Rashed, R.; Darwish, H.; Omran, M.; Belal, A.; Zahran, F. A Novel Serum Metabolome Score for Breast Cancer Diagnosis. Br. J. Biomed. Sci. 2020, 77, 196–201. [Google Scholar] [CrossRef]
- Jové, M.; Collado, R.; Quiles, J.L.; Ramírez-Tortosa, M.C.; Sol, J.; Ruiz-Sanjuan, M.; Fernandez, M.; Cabrera, C.d.l.T.; Ramírez-Tortosa, C.; Granados-Principal, S.; et al. A Plasma Metabolomic Signature Discloses Human Breast Cancer. Oncotarget 2017, 8, 19522–19533. [Google Scholar] [CrossRef]
- Huang, J.; Sun, J.; Chen, Y.; Song, Y.; Dong, L.; Zhan, Q.; Zhang, R.; Abliz, Z. Analysis of Multiplex Endogenous Estrogen Metabolites in Human Urine Using Ultra-Fast Liquid Chromatography-Tandem Mass Spectrometry: A Case Study for Breast Cancer. Anal. Chim. Acta 2012, 711, 60–68. [Google Scholar] [CrossRef]
- Hadi, N.I.; Jamal, Q. “OMIC” Tumor Markers for Breast Cancer: A Review. Pak. J. Med. Sci. 2015, 31, 1256. [Google Scholar] [CrossRef]
- Huang, Q.; Tan, Y.; Yin, P.; Ye, G.; Gao, P.; Lu, X.; Wang, H.; Xu, G. Metabolic Characterization of Hepatocellular Carcinoma Using Nontargeted Tissue Metabolomics. Cancer Res. 2013, 73, 4992–5002. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xie, G.; Wang, X.; Fan, J.; Qiu, Y.; Zheng, X.; Qi, X.; Cao, Y.; Su, M.; Wang, X.; et al. Serum and Urine Metabolite Profiling Reveals Potential Biomarkers of Human Hepatocellular Carcinoma. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef]
- Ghosh, A.; Nishtala, K. Biofluid Lipidome: A Source for Potential Diagnostic Biomarkers. Clin. Transl. Med. 2017, 6, 22. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Han, X. Lipidomics: Techniques, Applications, and Outcomes Related to Biomedical Sciences. Trends Biochem. Sci. 2016, 41, 954–969. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, C.; Han, X. Tutorial on Lipidomics. Anal. Chim. Acta 2019, 1061, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Vosegaard, T.; Guo, Z. Applications of Nuclear Magnetic Resonance in Lipid Analyses: An Emerging Powerful Tool for Lipidomics Studies. Prog. Lipid Res. 2017, 68, 37–56. [Google Scholar] [CrossRef]
- Zhang, F.; Ge, W.; Ruan, G.; Cai, X.; Guo, T. Data-Independent Acquisition Mass Spectrometry-Based Proteomics and Software Tools: A Glimpse in 2020. Proteomics 2020, 20, e1900276. [Google Scholar] [CrossRef]
- Ràfols, P.; Vilalta, D.; Brezmes, J.; Cañellas, N.; del Castillo, E.; Yanes, O.; Ramírez, N.; Correig, X. Signal Preprocessing, Multivariate Analysis and Software Tools for MA(LDI)-TOF Mass Spectrometry Imaging for Biological Applications. Mass Spectrom. Rev. 2018, 37, 281–306. [Google Scholar] [CrossRef]
- Teleman, J.; Röst, H.L.; Rosenberger, G.; Schmitt, U.; Malmström, L.; Malmström, J.; Levander, F. DIANA-Algorithmic Improvements for Analysis of Data-Independent Acquisition MS Data. Bioinformatics 2015, 31, 555–562. [Google Scholar] [CrossRef]
- Mertens, B.J.A. Transformation, Normalization, and Batch Effect in the Analysis of Mass Spectrometry Data for Omics Studies. In Statistical Analysis of Proteomics, Metabolomics, and Lipidomics Data Using Mass Spectrometry; Springer: Cham, Switzerland, 2017; pp. 1–21. [Google Scholar]
- Välikangas, T.; Suomi, T.; Elo, L.L. A Systematic Evaluation of Normalization Methods in Quantitative Label-Free Proteomics. Brief. Bioinform. 2018, 19, 1–11. [Google Scholar] [CrossRef]
- Vafaee, F.; Diakos, C.; Kirschner, M.B.; Reid, G.; Michael, M.Z.; Horvath, L.G.; Alinejad-Rokny, H.; Cheng, Z.J.; Kuncic, Z.; Clarke, S. A Data-Driven, Knowledge-Based Approach to Biomarker Discovery: Application to Circulating MicroRNA Markers of Colorectal Cancer Prognosis. NPJ Syst. Biol. Appl. 2018, 4, 20. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, A.; Fatima, S.; Sowmya, A.; Vafaee, F. Blood-Based Transcriptomic Signature Panel Identification for Cancer Diagnosis: Benchmarking of Feature Extraction Methods. Brief. Bioinform. 2022, 23, bbac315. [Google Scholar] [CrossRef]
- Liu, L.; Chen, X.; Petinrin, O.O.; Zhang, W.; Rahaman, S.; Tang, Z.R.; Wong, K.C. Machine Learning Protocols in Early Cancer Detection Based on Liquid Biopsy: A Survey. Life 2021, 11, 638. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Jonassen, I.; Goksøyr, A. Machine Learning Approaches for Biomarker Discovery Using Gene Expression Data. In Bioinformatics; Exon Publications: Brisbane, QLD, Australia, 2021; pp. 53–64. [Google Scholar]
- Moons, K.G.M.; de Groot, J.A.H.; Bouwmeester, W.; Vergouwe, Y.; Mallett, S.; Altman, D.G.; Reitsma, J.B.; Collins, G.S. Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modelling Studies: The CHARMS Checklist. PLoS Med. 2014, 11, e1001744. [Google Scholar] [CrossRef] [PubMed]
- Darrow, J.J.; Avorn, J.; Kesselheim, A.S. FDA Regulation and Approval of Medical Devices: 1976–2020. JAMA J. Am. Med. Assoc. 2021, 326, 420–432. [Google Scholar] [CrossRef]
- Rathi, V.; Wright, G.; Constantin, D.; Chang, S.; Pham, H.; Jones, K.; Palios, A.; Mclachlan, S.A.; Conron, M.; McKelvie, P.; et al. Clinical Validation of the 50 Gene AmpliSeq Cancer Panel V2 for Use on a next Generation Sequencing Platform Using Formalin Fixed, Paraffin Embedded and Fine Needle Aspiration Tumour Specimens. Pathology 2017, 49, 75–82. [Google Scholar] [CrossRef]
- Wang, P.; Kricka, L.J. Current and Emerging Trends in Point-of-Care Technology and Strategies for Clinical Validation and Implementation. Clin. Chem. 2018, 64, 1439–1452. [Google Scholar] [CrossRef]
- Klein, E.A.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical Validation of a Targeted Methylation-Based Multi-Cancer Early Detection Test Using an Independent Validation Set. Ann. Oncol. 2021, 32, 1167–1177. [Google Scholar] [CrossRef]
- Nadauld, L.D.; McDonnell, C.H.; Beer, T.M.; Liu, M.C.; Klein, E.A.; Hudnut, A.; Whittington, R.A.; Taylor, B.; Oxnard, G.R.; Lipson, J.; et al. The Pathfinder Study: Assessment of the Implementation of an Investigational Multi-Cancer Early Detection Test into Clinical Practice. Cancers 2021, 13, 3501. [Google Scholar] [CrossRef]
- Chen, G.; Zhang, J.; Fu, Q.; Taly, V.; Tan, F. Integrative Analysis of Multi-Omics Data for Liquid Biopsy. Br. J. Cancer 2023, 128, 505–518. [Google Scholar] [CrossRef]
| Aim | Pre-Analytical Phase | Analytical Phase | Post-Analytical Phase | Ref | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BioSource | Collection Tube | Time to Sample Processing | Centrifugation | Storage | Tumour Grade | Technique | Validation Method | Hypothesis Test Performed | ||
| Proteomic | Serum | NA | 4 °C for 1–2 h | 3000 rpm for 5 min + 12,000 rpm for 5 min | −80 °C | NA | SELDI-TOF-MS | SDS-PAGE MALDI-TOF/TOF |
| [26] |
| Serum | Plastic tube with clot activator | 15 min | 3280× g for 5 min, 4 °C | −80 °C | NA | SELDI-TOF MALDI-TOF-TOF | NA |
| [27] | |
| Plasma | K2EDTA tube | 2 h | 1300× g for 10 min | −80 °C | NA | 1D gel electrophoresis 2D gel electrophoresis LC-MS/MS | WB | Unpaired t-test | [28] | |
| Plasma | EDTA tube | 30 min | 4000× g for 30 min | −80 °C | NA | LC-MS/MS | WB | t-test | [29] | |
| Plasma | Sodium EDTA tube | NA | 1400× g for 5 min, 4 °C | ND | Low and high grade | Label-free nano-LC/MSMS | WB | Mann–Whitney | [30] | |
| NAF | Graduated micropipette | Immediately | 1500 rpm for 10 min | −80 °C | I/II | SELDI-TOF-MS | ELISA | Supervised and unsupervised cluster analysis | [14] | |
| NAF | Tube pre-treated with cocktail mixture of protease inhibitor | <30 min | NA | ST: −20 °C LT: −80 °C | I–III | 1D LC-MS/MS | NA |
| [31] | |
| Urine | Sterile tube | Immediately | 2000× g for 10 min, 4 °C | ST: −20 °C LT: −80 °C | II–III | Label-free LC-MS/MS | WB | ANOVA | [9] | |
| First Morning Urine | Tube containing 0.02% w/v Sodium Azide) | NA | NA | ND | I/II | Standardisation phase: 2D gel electrophoresis Discovery phase: 2D-DIGE, MALDI-TOF-TOF, SWATH-MS, iTRAQ, LC-QTOF | WB MRM |
| [10] | |
| Metabolomic | Plasma | EDTA tube | <2 h | 3000× g for 10 min, 4 °C | −80 °C | I–III | LC-MS | NA |
| [32] |
| Plasma | K2EDTA tube | Immediately | 1500× g for 10 min, RT | −80 °C | I–III | LC-QTOF-MS LC-QQQ-MS | NA |
| [33] | |
| Serum | Vacutainer tube | 30 min | 3000 rpm for 10 min, 4 °C | −80 °C | I–III | UHPLC-QTOF-(ESIþ)-MS | NA |
| [34] | |
| First Morning Urine | NA | NA | 3000× g for 10 min, RT | −80 °C | I/III | GC–MS LC-QTOF/MS | NA |
| [35] | |
| Saliva | Polypropylene tube | NA | NA | −80 °C | 0–IV | CE-TOF-MS | LC-QQQ-MS |
| [36] | |
| Saliva | NA | 10 min | 13,500 rpm for 20 min, 4 °C | −40 °C | I–IV | HILIC-ESI-MS RPLC-ESI-MS | NA |
| [37] | |
| Lipidomic | Plasma | Heparin tube | NA | 1500× g for 15 min | −80 °C | I/II | UPLC-QTOF/MS | NA |
| [38] |
| Plasma | EDTA tube | <2 h | 2600× g for 10 min, 4 °C | −80 °C | 0- II | LC-ESI-MS/MS | NA |
| [39] | |
| Serum | NA | NA | NA | −80 °C | NA | NMR spectroscopy | NA |
| [40] | |
| First Morning Urine | NA | NA | 3000× g for 10 min, RT | −80 °C | I/III | LC–MS | NA |
| [35] | |
| Saliva | Polypropylene tube | NA | 10,000× g for 10 min | Without freezing and storage | I–III | IR spectroscopy | NA |
| [41] | |
| Pre-Analytical Variable | Literature Findings |
|---|---|
| Collection Tubes |
|
| Anti-Coagulant |
|
| Hemolysis | |
| Incubation Time | |
| Centrifugation Force | |
| Storage Conditions | |
| Freeze–Thaw Cycles |
|
| Techniques | Advantages | Limitations | Biomarker Type |
|---|---|---|---|
| MALDI-TOF-MS [142,143,144,145] |
|
|
|
| SELDI-TOF-MS [146] |
|
|
|
| LC-MS [68,147,148,149,150] |
|
|
|
| GC-MS [147,150] |
|
|
|
| NMR [150,151] |
|
|
|
| 1DGE [68,152,153,154] |
|
|
|
| 2DGE [68,152,153,154] |
|
|
|
| 2D-DIGE [153,155,156] |
|
|
|
| Immunoassay techniques (ELISA, Western Blot) [152,157,158] |
|
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Safari, F.; Kehelpannala, C.; Safarchi, A.; Batarseh, A.M.; Vafaee, F. Biomarker Reproducibility Challenge: A Review of Non-Nucleotide Biomarker Discovery Protocols from Body Fluids in Breast Cancer Diagnosis. Cancers 2023, 15, 2780. https://doi.org/10.3390/cancers15102780
Safari F, Kehelpannala C, Safarchi A, Batarseh AM, Vafaee F. Biomarker Reproducibility Challenge: A Review of Non-Nucleotide Biomarker Discovery Protocols from Body Fluids in Breast Cancer Diagnosis. Cancers. 2023; 15(10):2780. https://doi.org/10.3390/cancers15102780
Chicago/Turabian StyleSafari, Fatemeh, Cheka Kehelpannala, Azadeh Safarchi, Amani M. Batarseh, and Fatemeh Vafaee. 2023. "Biomarker Reproducibility Challenge: A Review of Non-Nucleotide Biomarker Discovery Protocols from Body Fluids in Breast Cancer Diagnosis" Cancers 15, no. 10: 2780. https://doi.org/10.3390/cancers15102780
APA StyleSafari, F., Kehelpannala, C., Safarchi, A., Batarseh, A. M., & Vafaee, F. (2023). Biomarker Reproducibility Challenge: A Review of Non-Nucleotide Biomarker Discovery Protocols from Body Fluids in Breast Cancer Diagnosis. Cancers, 15(10), 2780. https://doi.org/10.3390/cancers15102780






