LHX2 Is a Potential Biomarker and Associated with Immune Infiltration in Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Resources
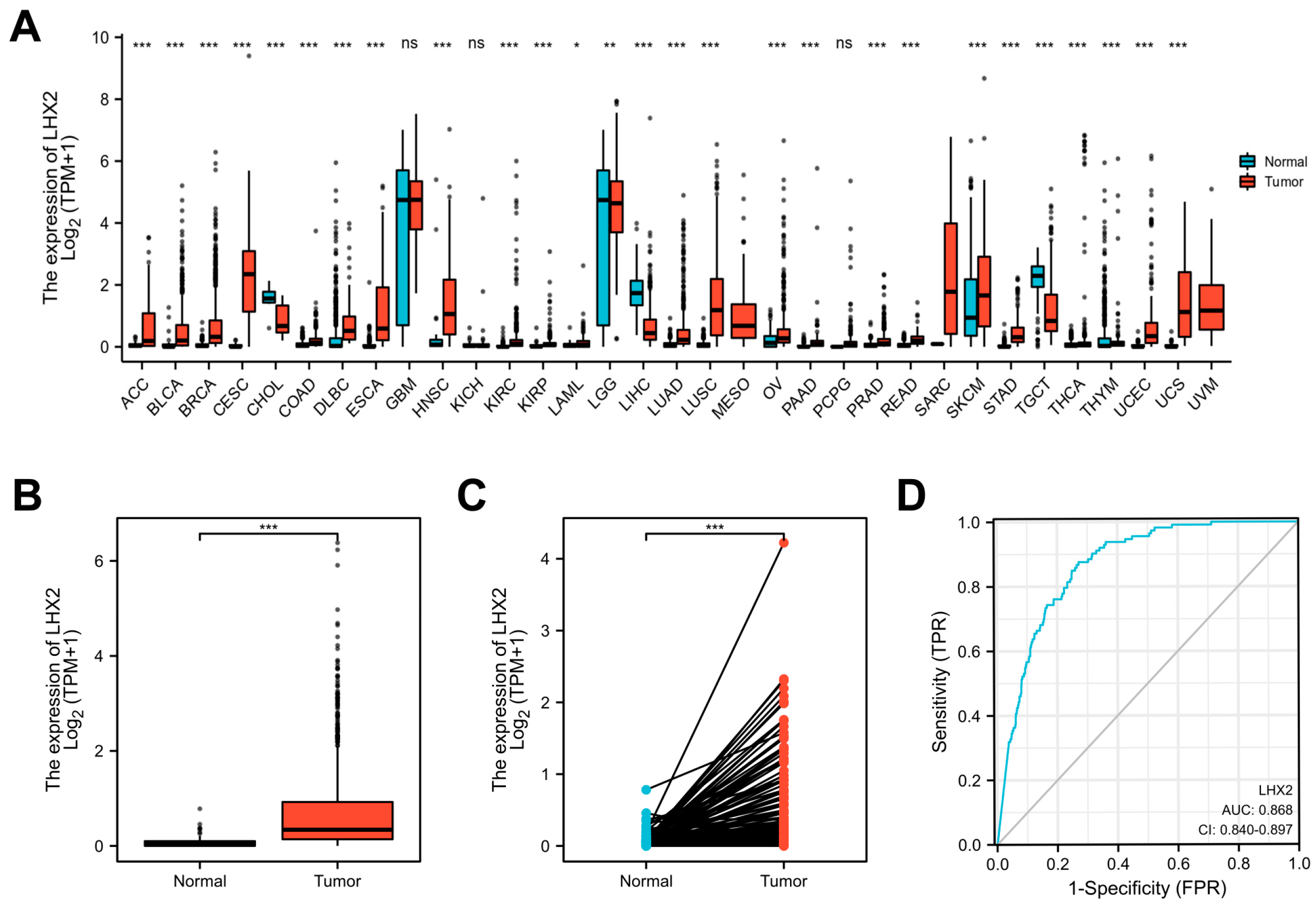
2.2. Differential Expression of LHX2
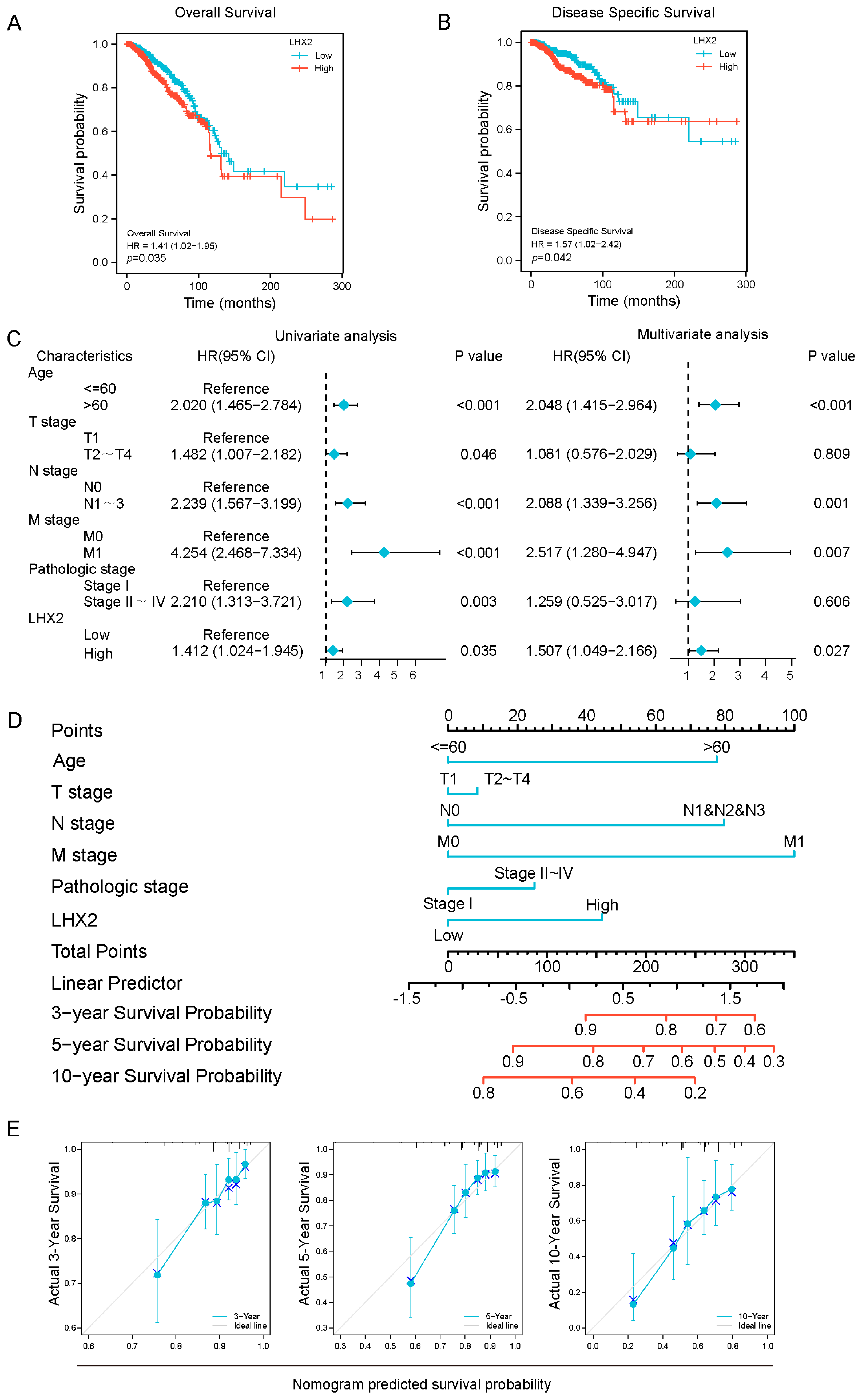
2.3. Prognostic Analyses of Different Malignancies
2.4. Analyses of Clinicopathological Features of BRCA
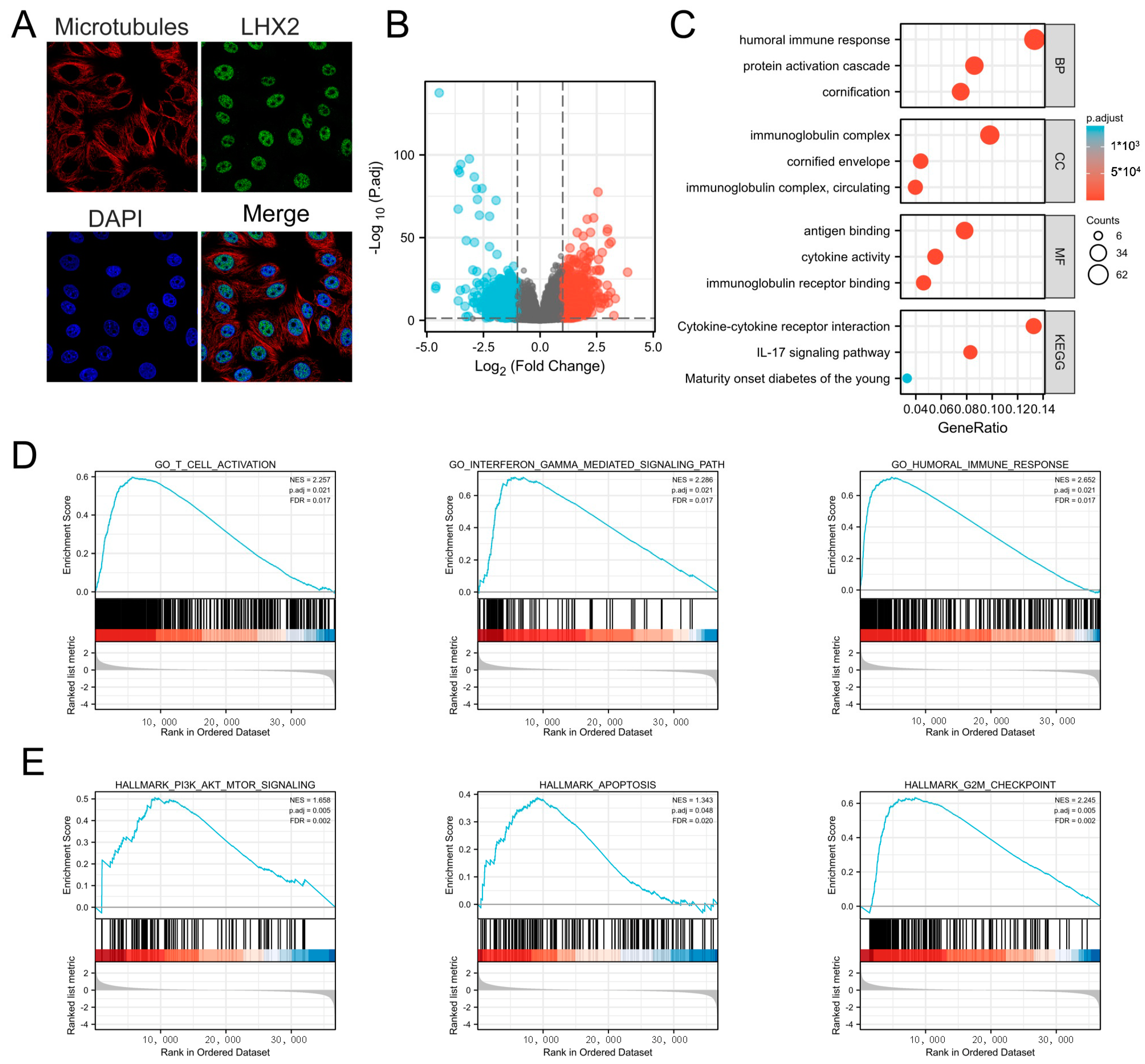
2.5. Detection of Differential Expressed Genes (DEGs) and Functional Enrichment Analyses
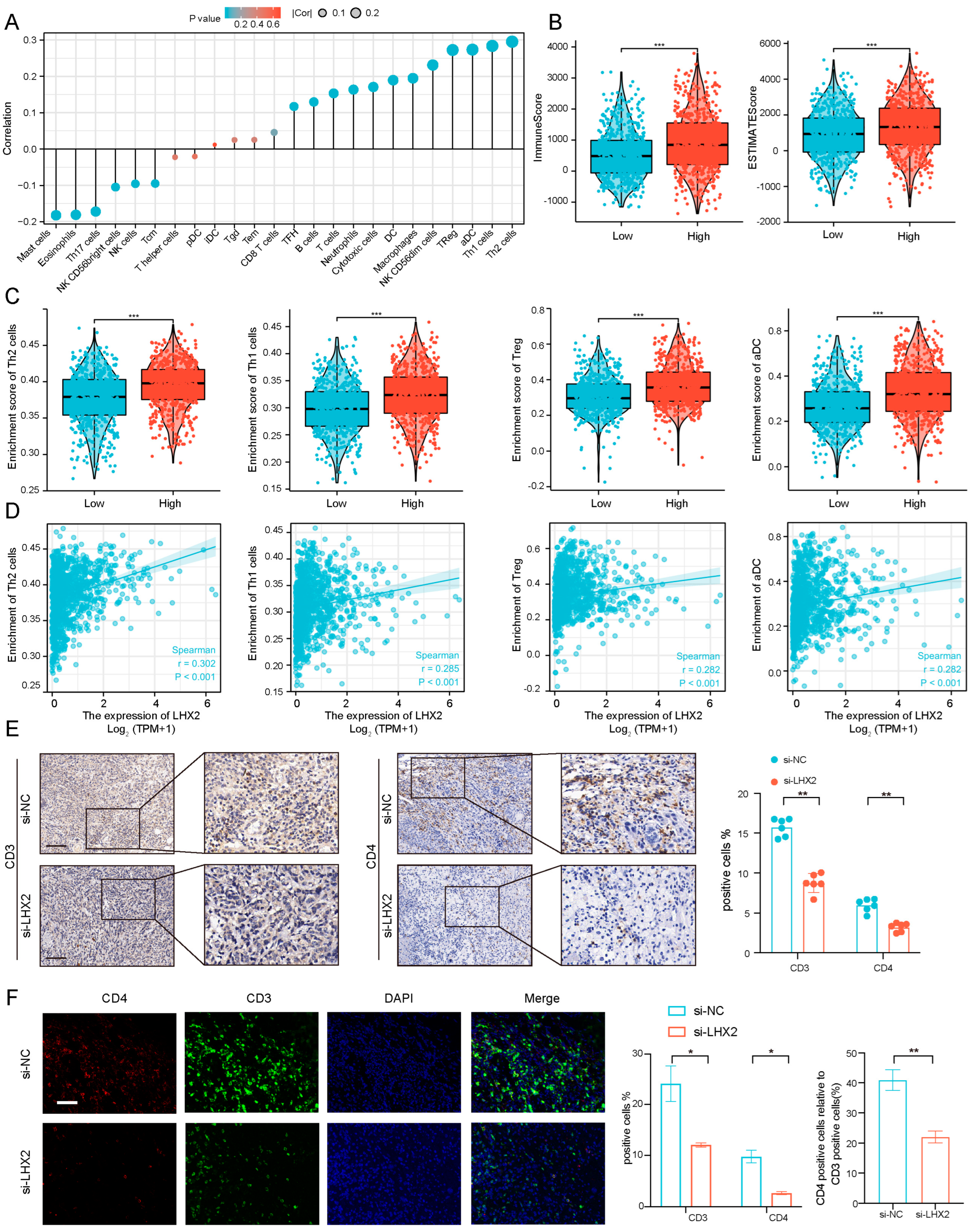
2.6. Immune Cell Infiltration
2.7. Cell Culture
2.8. Real-Time Quantitative PCR
2.9. Transfection
2.10. CCK-8 Assay
2.11. 5-Ethynyl-2′-Deoxyuridine (EdU) Assay
2.12. Colony Formation Assay
2.13. Transwell Assay
2.14. Wound Healing Assay
2.15. Flow Cytometry
2.16. In Vivo Assay
2.17. Western Blotting Assay
2.18. Immunohistochemistry (IHC)
2.19. Immunofluorescence
2.20. TdT-Mediated dUTP Nick-End Labeling (TUNEL) Assay
2.21. Statistical Analysis
3. Results
3.1. LHX2 Is Highly Expressed in BRCA
3.2. Prognostic Value of LHX2 in BRCA
3.3. Association between LHX2 Expression and Clinicopathological Features
3.4. Subcellular Location of LHX2 and Functional Enrichment Analyses of LHX2-Related Genes in BRCA
3.5. Immune Infiltration Correlated with LHX2 Expression in BRCA
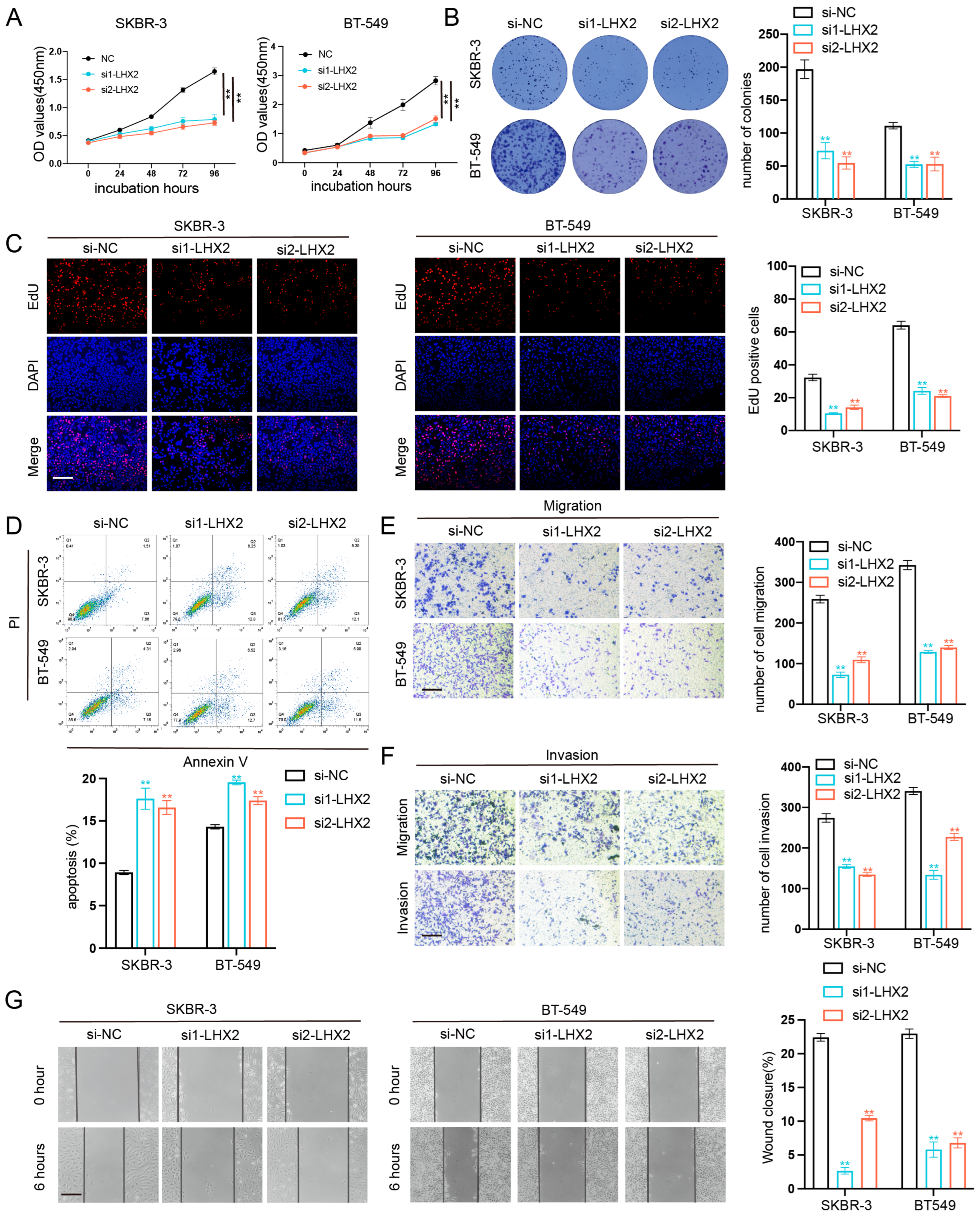
3.6. LHX2 Regulates BRCA Cell Proliferation Migration, Invasion Ability, and Apoptosis In Vitro
3.7. LHX2 Regulates BRCA Cell Proliferation In Vivo
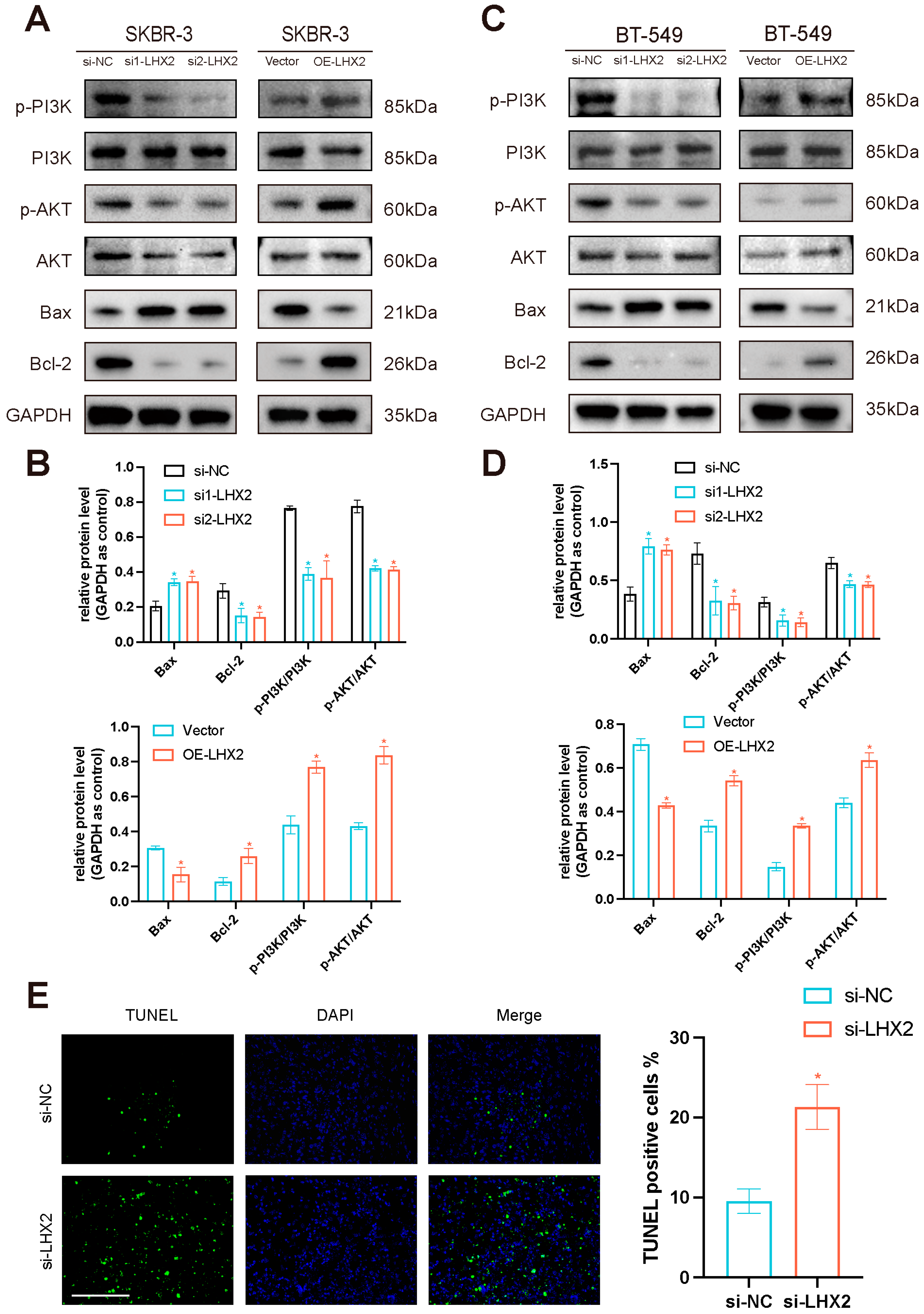
3.8. LHX2 Activates the PI3K/AKT/mTOR Pathway and Apoptosis Pathway in Breast Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Zhang, H.; Song, X.; Yang, Q. Metastatic heterogeneity of breast cancer: Molecular mechanism and potential therapeutic targets. Semin. Cancer Biol. 2020, 60, 14–27. [Google Scholar] [CrossRef] [PubMed]
- Porter, F.D.; Drago, J.; Xu, Y.; Cheema, S.S.; Wassif, C.; Huang, S.P.; Lee, E.; Grinberg, A.; Massalas, J.S.; Bodine, D.; et al. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development 1997, 124, 2935–2944. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wu, X.; Chen, H.; Liu, Z.; He, H.; Wang, L. LHX2 Enhances the Malignant Phenotype of Esophageal Squamous Cell Carcinoma by Upregulating the Expression of SERPINE2. Genes 2022, 13, 1457. [Google Scholar] [CrossRef]
- Mosca, N.; Khoubai, F.Z.; Fedou, S.; Carrillo-Reixach, J.; Caruso, S.; Del Rio-Alvarez, A.; Dubois, E.; Avignon, C.; Dugot-Senant, N.; Guettier, C.; et al. LIM Homeobox-2 Suppresses Hallmarks of Adult and Pediatric Liver Cancers by Inactivating MAPK/ERK and Wnt/Beta-Catenin Pathways. Liver Cancer 2022, 11, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Emens, L.A. Breast Cancer Immunotherapy: Facts and Hopes. Clin. Cancer Res. 2018, 24, 511–520. [Google Scholar] [CrossRef]
- Subramanian, L.; Sarkar, A.; Shetty, A.S.; Muralidharan, B.; Padmanabhan, H.; Piper, M.; Monuki, E.S.; Bach, I.; Gronostajski, R.M.; Richards, L.J.; et al. Transcription factor Lhx2 is necessary and sufficient to suppress astrogliogenesis and promote neurogenesis in the developing hippocampus. Proc. Natl. Acad. Sci. USA 2011, 108, E265–E274. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Wang, X.; Zhou, Y.; Wang, X.; Yu, Y. Autophagy, ferroptosis, pyroptosis, and necroptosis in tumor immunotherapy. Signal Transduct. Target. Ther. 2022, 7, 196. [Google Scholar] [CrossRef] [PubMed]
- Dieci, M.V.; Miglietta, F.; Guarneri, V. Immune Infiltrates in Breast Cancer: Recent Updates and Clinical Implications. Cells 2021, 10, 223. [Google Scholar] [CrossRef]
- Liu, J.; Li, X.; Yang, S.; Mou, J.; Lu, H. Whole exome sequencing and transcriptome-wide profiling identify potentially subtype-relevant genes of nasopharyngeal carcinoma. Pathol. Res. Pract. 2020, 216, 153244. [Google Scholar] [CrossRef] [PubMed]
- Alberti, G.; Vergilio, G.; Paladino, L.; Barone, R.; Cappello, F.; de Macario, E.C.; Macario, A.J.L.; Bucchieri, F.; Rappa, F. The Chaperone System in Breast Cancer: Roles and Therapeutic Prospects of the Molecular Chaperones Hsp27, Hsp60, Hsp70, and Hsp90. Int. J. Mol. Sci. 2022, 23, 7792. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Manning, B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Miricescu, D.; Totan, A.; Stanescu, S., II; Badoiu, S.C.; Stefani, C.; Greabu, M. PI3K/AKT/mTOR Signaling Pathway in Breast Cancer: From Molecular Landscape to Clinical Aspects. Int. J. Mol. Sci. 2020, 22, 173. [Google Scholar] [CrossRef] [PubMed]
- Shorning, B.Y.; Dass, M.S.; Smalley, M.J.; Pearson, H.B. The PI3K-AKT-mTOR Pathway and Prostate Cancer: At the Crossroads of AR, MAPK, and WNT Signaling. Int. J. Mol. Sci. 2020, 21, 4507. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Li, G.; Lu, J.; Li, L. Crosstalk between circRNAs and the PI3K/AKT signaling pathway in cancer progression. Signal Transduct. Target. Ther. 2021, 6, 400. [Google Scholar] [CrossRef] [PubMed]
- Fruman, D.A.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef]
- Song, H.; Liu, J.; Wu, X.; Zhou, Y.; Chen, X.; Chen, J.; Deng, K.; Mao, C.; Huang, S.; Liu, Z. LHX2 promotes malignancy and inhibits autophagy via mTOR in osteosarcoma and is negatively regulated by miR-129-5p. Aging 2019, 11, 9794–9810. [Google Scholar] [CrossRef]
- Yang, Q.; Guan, K.-L. Expanding mTOR signaling. Cell Res. 2007, 17, 666–681. [Google Scholar] [CrossRef]
- Manning, B.D.; Toker, A. AKT/PKB Signaling: Navigating the Network. Cell 2017, 169, 381–405. [Google Scholar] [CrossRef]
- Gaumer, S.; Guénal, I.; Brun, S.; Théodore, L.; Mignotte, B. Bcl-2 and Bax mammalian regulators of apoptosis are functional in Drosophila. Cell Death Differ. 2000, 7, 804–814. [Google Scholar] [CrossRef] [PubMed]
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast cancer. Lancet 2021, 397, 1750–1769. [Google Scholar] [CrossRef] [PubMed]
- Tiede, S.; Paus, R. Lhx2—Decisive role in epithelial stem cell maintenance, or just the “tip of the iceberg”? Bioessays 2006, 28, 1157–1160. [Google Scholar] [CrossRef]
- Muralidharan, B.; Khatri, Z.; Maheshwari, U.; Gupta, R.; Roy, B.; Pradhan, S.J.; Karmodiya, K.; Padmanabhan, H.; Shetty, A.S.; Balaji, C.; et al. LHX2 Interacts with the NuRD Complex and Regulates Cortical Neuron Subtype Determinants Fezf2 and Sox11. J. Neurosci. 2017, 37, 194–203. [Google Scholar] [CrossRef]
- Hou, P.-S.; Chuang, C.-Y.; Kao, C.-F.; Chou, S.-J.; Stone, L.; Ho, H.-N.; Chien, C.-L.; Kuo, H.-C. LHX2 regulates the neural differentiation of human embryonic stem cells via transcriptional modulation of PAX6 and CER1. Nucleic Acids Res. 2013, 41, 7753–7770. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.K.; Minden, M.D. Transcriptional activation of human LIM-HOX gene, hLH-2, in chronic myelogenous leukemia is due to a cis-acting effect of Bcr-Abl. Biochem. Biophys. Res. Commun. 1997, 233, 806–812. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Gou, S.; Xiong, J.; Wu, H.; Wang, C.; Liu, T. Oncogenicity of LHX2 in pancreatic ductal adenocarcinoma. Mol. Biol. Rep. 2014, 41, 8163–8167. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Du, K.; Liu, W.; Liu, C.; Wang, B.; Tian, Y.; Li, R.; Huang, X.; Lin, J.; Jian, H.; et al. LHX2 facilitates the progression of nasopharyngeal carcinoma via activation of the FGF1/FGFR axis. Br. J. Cancer 2022, 127, 1239–1253. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhan, L.; Xiao, C.; Lei, Z.; Yang, H.; Wang, L.; Zhao, J.; Zhang, H.-T. miR-1238 inhibits cell proliferation by targeting LHX2 in non-small cell lung cancer. Oncotarget 2015, 6, 19043–19054. [Google Scholar] [CrossRef] [PubMed]
- Kuzmanov, A.; Hopfer, U.; Marti, P.; Meyer-Schaller, N.; Yilmaz, M.; Christofori, G. LIM-homeobox gene 2 promotes tumor growth and metastasis by inducing autocrine and paracrine PDGF-B signaling. Mol. Oncol. 2014, 8, 401–416. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Sun, N.; Zhao, T.; Sun, Y.; Gu, J.; Ma, D.; Tian, H.; Peng, Z.; Zhang, Y.; Han, F.; et al. Identification of prognostic indicators, diagnostic markers, and possible therapeutic targets among LIM homeobox transcription factors in breast cancer. Cancer Innov. 2022, 1, 252–269. [Google Scholar] [CrossRef]
- Stienstra, R.; Netea-Maier, R.T.; Riksen, N.P.; Joosten, L.A.; Netea, M.G. Specific and Complex Reprogramming of Cellular Metabolism in Myeloid Cells during Innate Immune Responses. Cell Metab. 2017, 26, 142–156. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Yu, D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 2021, 221, 107753. [Google Scholar] [CrossRef]
- Basu, A.; Ramamoorthi, G.; Jia, Y.; Faughn, J.; Wiener, D.; Awshah, S.; Kodumudi, K.; Czerniecki, B.J. Immunotherapy in breast cancer: Current status and future directions. Adv. Cancer Res. 2019, 143, 295–349. [Google Scholar] [PubMed]
- Dieci, M.V.; Radosevic-Robin, N.; Fineberg, S.; van den Eynden, G.; Ternes, N.; Penault-Llorca, F.; Pruneri, G.; D’alfonso, T.M.; Demaria, S.; Castaneda, C.; et al. International Immuno-Oncology Biomarker Working Group on Breast, C., Update on tumor-infiltrating lymphocytes (TILs) in breast cancer, including recommendations to assess TILs in residual disease after neoadjuvant therapy and in carcinoma in situ: A report of the International Immuno-Oncology Biomarker Working Group on Breast Cancer. Semin. Cancer Biol. 2018, 52 Pt 2, 16–25. [Google Scholar] [PubMed]
- Sanders, V.M. Epigenetic regulation of Th1 and Th2 cell development. Brain Behav. Immun. 2006, 20, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, A.; Sakaguchi, S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017, 27, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Datta, J.; Fracol, M.; McMillan, M.T.; Berk, E.; Xu, S.; Goodman, N.; Lewis, D.A.; DeMichele, A.; Czerniecki, B.J. Association of Depressed Anti-HER2 T-Helper Type 1 Response with Recurrence in Patients with Completely Treated HER2-Positive Breast Cancer: Role for Immune Monitoring. JAMA Oncol. 2016, 2, 242–246. [Google Scholar] [CrossRef]
- Speiser, D.E.; Ho, P.C.; Verdeil, G. Regulatory circuits of T cell function in cancer. Nat. Rev. Immunol. 2016, 16, 599–611. [Google Scholar] [CrossRef] [PubMed]
- Zou, W. Regulatory T cells, tumour immunity and immunotherapy. Nat. Rev. Immunol. 2006, 6, 295–307. [Google Scholar] [CrossRef]
- Steinman, R.M. Decisions about dendritic cells: Past, present, and future. Annu. Rev. Immunol. 2012, 30, 1–22. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, J.S.; Massi, D.; Teng, M.W.L.; Mandala, M. PI3K-AKT-mTOR inhibition in cancer immunotherapy, redux. Semin. Cancer Biol. 2018, 48, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Zotano, A.; Mayer, I.A.; Arteaga, C.L. PI3K/AKT/mTOR: Role in breast cancer progression, drug resistance, and treatment. Cancer Metastasis Rev. 2016, 35, 515–524. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Gu, M.; He, G.; Yu, X.; Yang, J.; Wu, X.; Zhang, X.; Lu, K.; Qian, F.; Shi, X.; et al. LHX2 Is a Potential Biomarker and Associated with Immune Infiltration in Breast Cancer. Cancers 2023, 15, 2773. https://doi.org/10.3390/cancers15102773
Zhang Z, Gu M, He G, Yu X, Yang J, Wu X, Zhang X, Lu K, Qian F, Shi X, et al. LHX2 Is a Potential Biomarker and Associated with Immune Infiltration in Breast Cancer. Cancers. 2023; 15(10):2773. https://doi.org/10.3390/cancers15102773
Chicago/Turabian StyleZhang, Ziwei, Minghao Gu, Gao He, Xiafei Yu, Junzhe Yang, Xian Wu, Xiaoqiang Zhang, Kaining Lu, Fangze Qian, Xiaoyue Shi, and et al. 2023. "LHX2 Is a Potential Biomarker and Associated with Immune Infiltration in Breast Cancer" Cancers 15, no. 10: 2773. https://doi.org/10.3390/cancers15102773
APA StyleZhang, Z., Gu, M., He, G., Yu, X., Yang, J., Wu, X., Zhang, X., Lu, K., Qian, F., Shi, X., Xu, J., Zhuang, M., Liu, X., & Zhu, Y. (2023). LHX2 Is a Potential Biomarker and Associated with Immune Infiltration in Breast Cancer. Cancers, 15(10), 2773. https://doi.org/10.3390/cancers15102773





