Evaluation of Gliomas with Magnetic Resonance Fingerprinting with PET Correlation—A Comparative Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. MR Fingerprinting Protocol
2.2. PET
2.3. Co-Registration
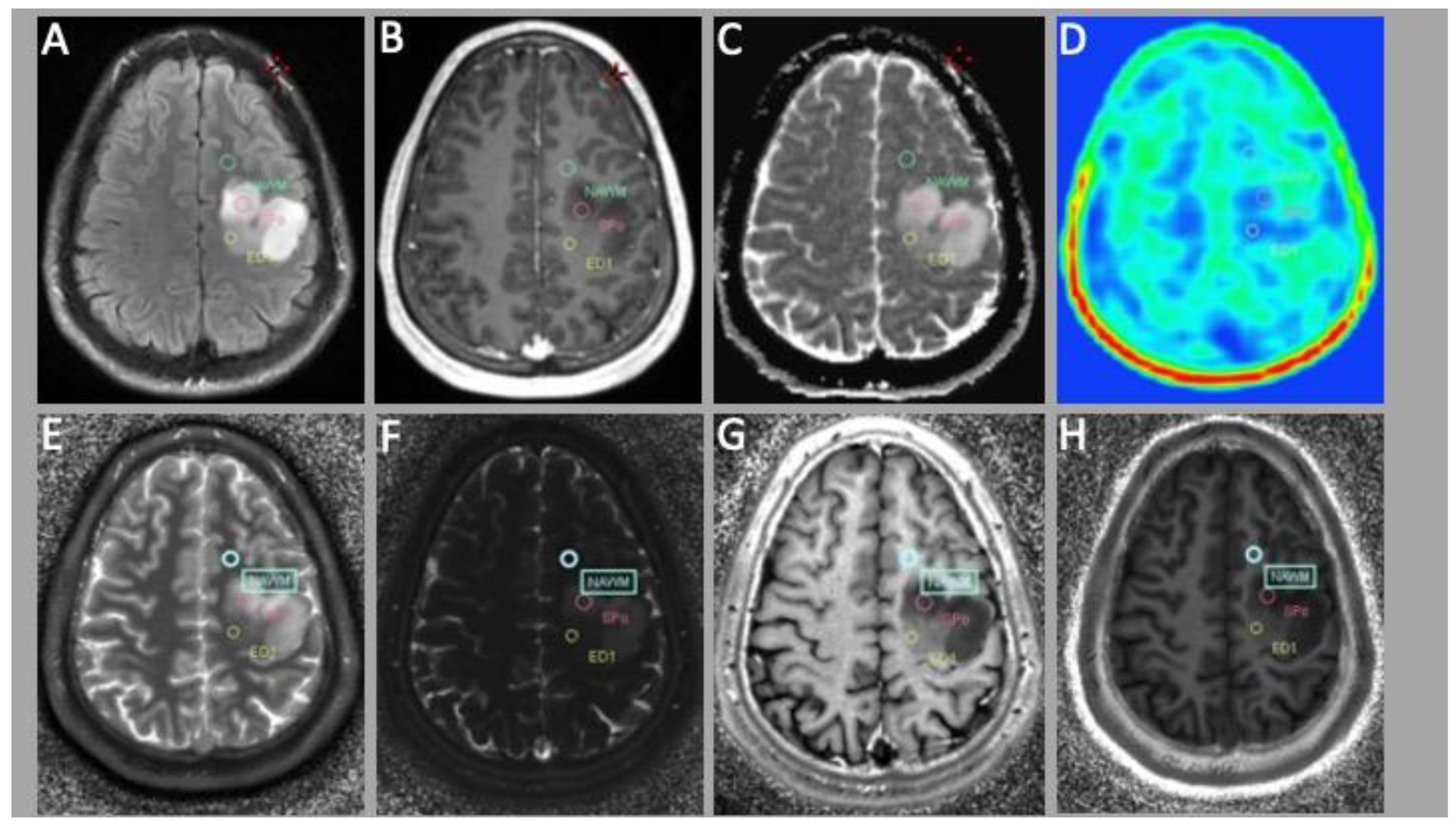
2.4. Region-of-Interest (ROI) Evaluation
2.5. Statistical Analysis
3. Results
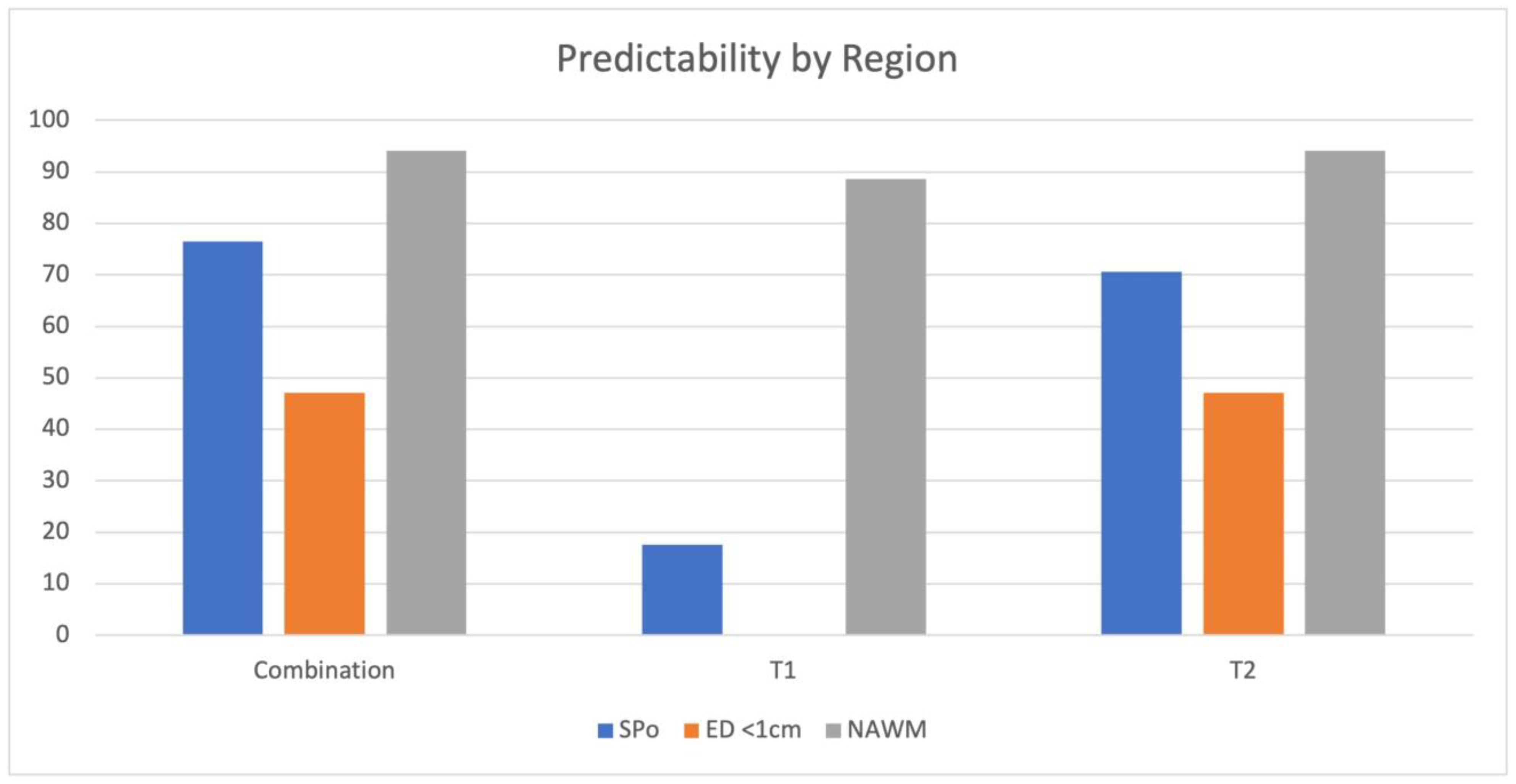
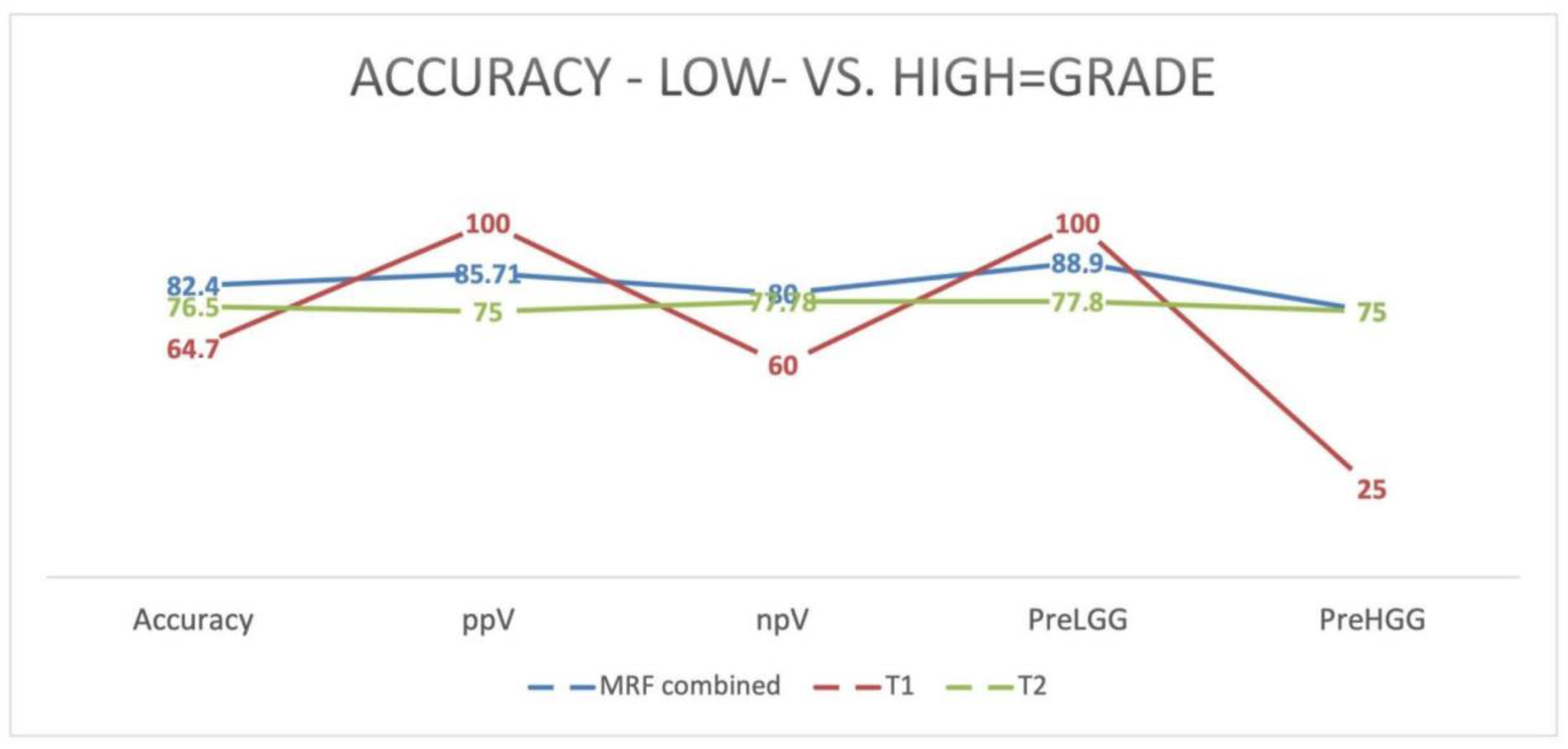
3.1. MRF
3.2. PET Evaluation
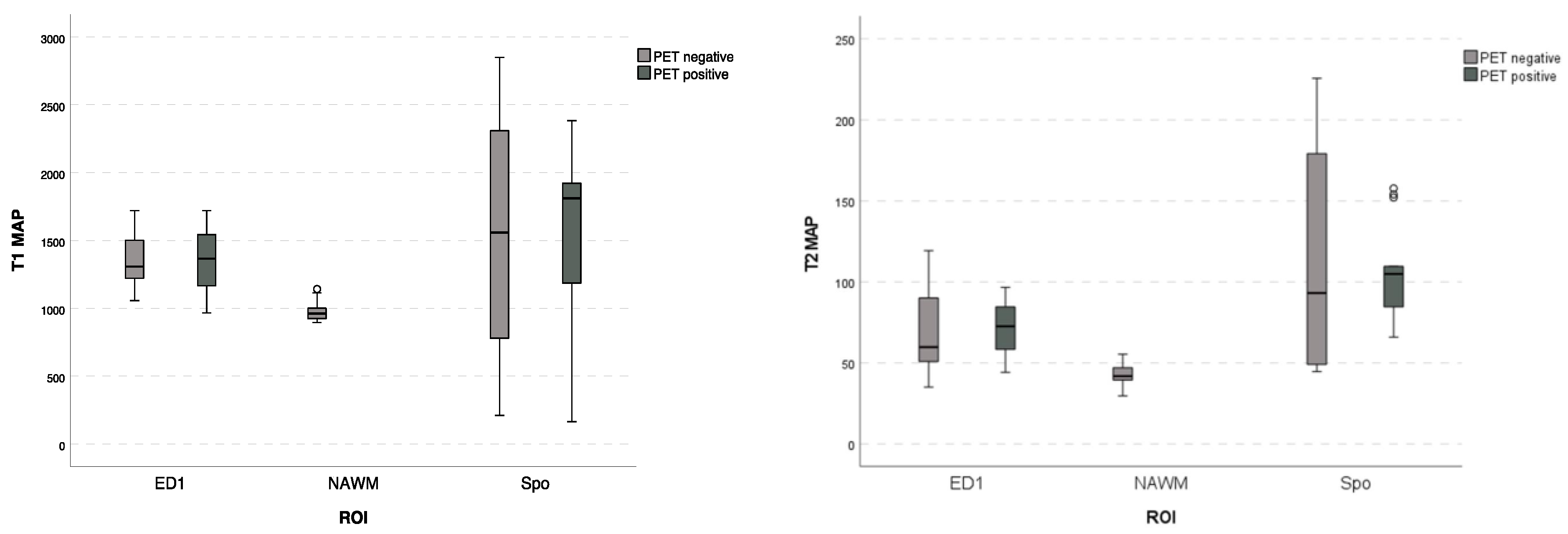
3.3. Correlation of MRF and PET
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADC | Apparent diffusion coefficient |
| CNS | Central nervous system |
| CVR | Cerebrovascular reactivity |
| ED1 | Peritumoral edema |
| FET | Fluorethyl-L-Tyrosine MET ([11C]-methionine) |
| FLAIR | Fluid attenuated inversion recovery |
| FWHM | Full width at half maximum |
| GBM | Glioblastoma |
| HGG | High grade glioma |
| IDH | Isocitratdehydrogenase |
| LGG | Low-grade glioma |
| MET | Methionine |
| MGMT | Methylguanine-DNA-methyltransferase |
| MPRAGE | Magnetization prepared—rapid gradient echo |
| MRI | Magnetic resonance imaging |
| MRF | Magnetic resonance finger printing |
| NAWM | Normal appearing white matter |
| npV | Negative predictive value |
| OSEM | Ordered subset expectation maximization algorithm |
| PET | Positron emission tomography |
| ppV | Positive predictive value |
| ROI | Region of interest |
| Sd | Standard deviation |
| SPo | Solid tumor part |
| TE | Echo time |
| TI | Inversion time |
| TR | Repetition time |
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Smits, M. Update on neuroimaging in brain tumours. Curr. Opin. Neurol. 2021, 34, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, V.G.; Korzdorfer, G.; Gall, P. Toward Quantification: Microstructure and Magnetic Resonance Fingerprinting. Investig. Radiol. 2021, 56, 1–9. [Google Scholar] [CrossRef]
- Ma, D.; Gulani, V.; Seiberlich, N.; Liu, K.; Sunshine, J.L.; Duerk, J.L.; Griswold, M.A. Magnetic resonance fingerprinting. Nature 2013, 495, 187–192. [Google Scholar] [CrossRef] [PubMed]
- Springer, E.; Cardoso, P.L.; Strasser, B.; Bogner, W.; Preusser, M.; Widhalm, G.; Nittka, M.; Koerzdoerfer, G.; Szomolanyi, P.; Hangel, G.; et al. MR Fingerprinting-A Radiogenomic Marker for Diffuse Gliomas. Cancers 2022, 14, 723. [Google Scholar] [CrossRef] [PubMed]
- Palanichamy, K.; Chakravarti, A. Diagnostic and Prognostic Significance of Methionine Uptake and Methionine Positron Emission Tomography Imaging in Gliomas. Front. Oncol. 2017, 7, 257. [Google Scholar] [CrossRef] [PubMed]
- Badve, C.; Yu, A.; Dastmalchian, S.; Rogers, M.; Ma, D.; Jiang, Y.; Margevicius, S.; Pahwa, S.; Lu, Z.; Schluchter, M.; et al. MR Fingerprinting of Adult Brain Tumors: Initial Experience. AJNR Am. J. Neuroradiol. 2017, 38, 492–499. [Google Scholar] [CrossRef]
- Tippareddy, C.; Zhao, W.; Sunshine, J.L.; Griswold, M.; Ma, D.; Badve, C. Magnetic resonance fingerprinting: An overview. Eur. J. Nucl. Med. Mol. Imaging 2021, 48, 4189–4200. [Google Scholar] [CrossRef]
- Haubold, J.; Demircioglu, A.; Gratz, M.; Glas, M.; Wrede, K.; Sure, U.; Antoch, G.; Keyvani, K.; Nittka, M.; Kannengiesser, S.; et al. Non-invasive tumor decoding and phenotyping of cerebral gliomas utilizing multiparametric (18)F-FET PET-MRI and MR Fingerprinting. Eur. J. Nucl. Med. Mol. Imaging 2020, 47, 1435–1445. [Google Scholar] [CrossRef] [PubMed]
- Korzdorfer, G.; Kirsch, R.; Liu, K.; Pfeuffer, J.; Hensel, B.; Jiang, Y.; Ma, D.; Gratz, M.; Bar, P.; Bogner, W.; et al. Reproducibility and Repeatability of MR Fingerprinting Relaxometry in the Human Brain. Radiology 2019, 292, 429–437. [Google Scholar] [CrossRef]
- Körzdörfer, G.; Cardoso, P.L.; Bär, P.; Kitzer, S.; Bogner, W.; Trattnig, S.; Nittka, M. Data-Driven Motion Detection for MR Fingerprinting. In Proceedings of the ISMRM & SMRT Virtual Conference & Exhibition, 8–14 August 2020. Available online: https://www.ismrm.org/20/program_files/PP25.htm (accessed on 10 December 2021).
- Chung, S.; Kim, D.; Breton, E.; Axel, L. Rapid B1+ mapping using a preconditioning RF pulse with TurboFLASH readout. Magn. Reason. Med. 2010, 64, 439–446. [Google Scholar] [CrossRef]
- Ding, H.; Velasco, C.; Ye, H.; Lindner, T.; Grech-Sollars, M.; O’Callaghan, J.; Hiley, C.; Chouhan, M.D.; Niendorf, T.; Koh, D.M.; et al. Current Applications and Future Development of Magnetic Resonance Fingerprinting in Diagnosis, Characterization, and Response Monitoring in Cancer. Cancers 2021, 13, 4742. [Google Scholar] [CrossRef]
- Kern, M.; Auer, T.A.; Picht, T.; Misch, M.; Wiener, E. T2 mapping of molecular subtypes of WHO grade II/III gliomas. BMC Neurol. 2020, 20, 8. [Google Scholar] [CrossRef]
- Price, S.J.; Jena, R.; Burnet, N.G.; Hutchinson, P.J.; Dean, A.F.; Pena, A.; Pickard, J.D.; Carpenter, T.A.; Gillard, J.H. Improved delineation of glioma margins and regions of infiltration with the use of diffusion tensor imaging: An image-guided biopsy study. AJNR Am. J. Neuroradiol. 2006, 27, 1969–1974. [Google Scholar] [PubMed]
- Fatouros, P.P.; Marmarou, A. Use of magnetic resonance imaging for in vivo measurements of water content in human brain: Method and normal values. J. Neurosurg. 1999, 90, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Pirkl, C.M.; Nunez-Gonzalez, L.; Kofler, F.; Endt, S.; Grundl, L.; Golbabaee, M.; Gomez, P.A.; Cencini, M.; Buonincontri, G.; Schulte, R.F.; et al. Accelerated 3D whole-brain T1, T2, and proton density mapping: Feasibility for clinical glioma MR imaging. Neuroradiology 2021, 63, 1831–1851. [Google Scholar] [CrossRef]
- Albert, N.L.; Weller, M.; Suchorska, B.; Galldiks, N.; Soffietti, R.; Kim, M.M.; la Fougere, C.; Pope, W.; Law, I.; Arbizu, J.; et al. Response Assessment in Neuro-Oncology working group and European Association for Neuro-Oncology recommendations for the clinical use of PET imaging in gliomas. Neuro Oncol. 2016, 18, 1199–1208. [Google Scholar] [CrossRef] [PubMed]
- Stopa, B.M.; Juhasz, C.; Mittal, S. Comparison of Amino Acid PET to Advanced and Emerging MRI Techniques for Neurooncology Imaging: A Systematic Review of the Recent Studies. Mol. Imaging 2021, 2021, 8874078. [Google Scholar] [CrossRef]
- Lopci, E.; Riva, M.; Olivari, L.; Raneri, F.; Soffietti, R.; Piccardo, A.; Bizzi, A.; Navarria, P.; Ascolese, A.M.; Ruda, R.; et al. Prognostic value of molecular and imaging biomarkers in patients with supratentorial glioma. Eur. J. Nucl. Med. Mol. Imaging 2017, 44, 1155–1164. [Google Scholar] [CrossRef]
- Papp, L.; Potsch, N.; Grahovac, M.; Schmidbauer, V.; Woehrer, A.; Preusser, M.; Mitterhauser, M.; Kiesel, B.; Wadsak, W.; Beyer, T.; et al. Glioma Survival Prediction with Combined Analysis of In Vivo (11)C-MET PET Features, Ex Vivo Features, and Patient Features by Supervised Machine Learning. J. Nucl. Med. 2018, 59, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Sebok, M.; van Niftrik, C.H.B.; Muscas, G.; Pangalu, A.; Seystahl, K.; Weller, M.; Regli, L.; Fierstra, J. Hypermetabolism and impaired cerebrovascular reactivity beyond the standard MRI-identified tumor border indicate diffuse glioma extended tissue infiltration. Neurooncol. Adv. 2021, 3, vdab048. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Shan, Y.; Wang, L.; Cheng, Y.; Yang, H.; Zhao, G.; Wang, Z.; Lu, J. MGMT promoter methylation status shows no effect on [(18)F]FET uptake and CBF in gliomas: A stereotactic image-based histological validation study. Eur. Radiol. 2022, 32, 5577–5587. [Google Scholar] [CrossRef] [PubMed]
- Zhang-Yin, J.T.; Girard, A.; Bertaux, M. What Does PET Imaging Bring to Neuro-Oncology in 2022? A Review. Cancers 2022, 14, 879. [Google Scholar] [CrossRef] [PubMed]

| Nr | Age | PET | Gender | Classification (WHO 2021) | Classification (WHO 2016) | MGMT-Status (Methyliert = 1 Unmethyliert = 0) | 1p/19q (1 = Codel, O = n.a.) | IDH (Mutant = 1 Wildtyp = 0) |
|---|---|---|---|---|---|---|---|---|
| 1 | 23 | MET | m | Astrocytoma, IDH-mutant (CNS WHO grade 2) | Diffuse astrocytoma, IDH-mutant (WHO Gr. II) | 1 | 0 | 1 |
| 2 | 58 | MET | m | Astrocytoma, IDH-mutant (CNS WHO grade 3) | Anaplastic astrocytoma, IDH-mutant (WHO Gr. III) | 0 | 0 | 1 |
| 3 | 46 | MET | f | Astrocytoma, IDH-mutant (CNS WHO grade 4) | Glioblastoma, IDH-mutant (WHO Gr. IV) | 1 | 0 | 1 |
| 4 | 52 | FET | m | Glioblastoma, IDH-wildtype (CNS WHO grade 4) | Glioblastoma, IDH-wildtype (WHO Gr. IV) | 1 | 0 | 0 |
| 5 | 29 | MET | m | Astrocytoma, IDH-mutant (CNS WHO grade 3) | Anaplastic astrocytoma, IDH-mutant (WHO Gr. III) | 1 | 0 | 1 |
| 6 | 33 | FET | m | Astrocytoma, IDH-mutant (CNS WHO grade 2) | Diffuse astrocytoma, IDH-mutant (WHO Gr. II) | 1 | 0 | 1 |
| 7 | 54 | MET | f | Astrocytoma, IDH-mutant (CNS WHO grade 2) | Diffuse astrocytoma, IDH-mutant (WHO Gr. II) | 0 | 0 | 1 |
| 8 | 77 | FET | f | Astrocytoma, IDH-mutant (CNS WHO grade 2) | Diffuse astrocytoma, IDH-mutant (WHO Gr. II) | 1 | 0 | 1 |
| 9 | 46 | MET | f | Astrocytoma, IDH-mutant (CNS WHO grade 2) | Diffuse astrocytoma, IDH-mutant (WHO Gr. II) | 1 | 0 | 1 |
| 10 | 52 | FET | m | Oligodendroglioma, IDH-mutant and 1p/19q codeleted (CNS WHO grade 2) | Oligoendroglioma, IDH-mutant and 1p/19q codeleted (WHO Gr. II) | 1 | 1 | 1 |
| 11 | 57 | FET | m | Astrocytoma, IDH-mutant (CNS WHO grade 2) | Diffuse astrocytoma, IDH-mutant (WHO Gr. II) | 0 | 0 | 1 |
| 12 | 65 | MET | f | Glioblastoma, IDH-wildtype (CNS WHO grade 4) | Anaplastic astrocytoma, IDH-wildtype (WHO Gr. III) | 0 | 0 | 0 |
| 13 | 51 | FET | m | Oligodendroglioma, IDH-mutant and 1p/19q codeleted (CNS WHO grade 3) | Anaplastic oligodendroglioma, IDH-mutant and 1p/19q codeleted (WHO Gr. III) | 1 | 1 | 1 |
| 14 | 27 | FET | m | Glioblastoma, IDH-wildtype (CNS WHO grade 4) | Diffuse astrocytoma, IDH-wildtype (WHO Gr. II) | 0 | 0 | 0 |
| 15 | 28 | MET | f | Astrocytoma, IDH-mutant (CNS WHO grade 3) | Anaplastic astrocytoma, IDH-mutant (WHO Gr. III) | 1 | 0 | 1 |
| 16 | 39 | FET | f | Oligodendroglioma, IDH-mutant and 1p/19q codeleted (CNS WHO grade 2) | Oligodendroglioma, IDH-mutant and 1p/19q codeleted (WHO Gr. II) | 1 | 1 | 1 |
| 17 | 61 | FET | m | Oligodendroglioma, IDH-mutant and 1p/19q codeleted (CNS WHO grade 2) | Oligodendroglioma, IDH-mutant and 1p/19q codeleted (WHO Gr. II) | 1 | 1 | 1 |
| 2D ax T2 FLAIR | 2D T2 ax | DWI ax | 3D SWI ax | 3D T1 ax pre | 2D T2 cor | PWI ax | 3D T1 ax post | 2D CSI | DTI ax | |
|---|---|---|---|---|---|---|---|---|---|---|
| TSE + IR | TSE | EPI-SE | GRE | MPRAGE | TSE | SS-EPI | MPRAGE | S-LASER | EPI-SE | |
| Voxel dimensions | 0.9 × 0.9 | 0.8 × 0.6 | 1.8 × 1.8 | 0.9 × 0.9 | 1 × 1 | 0.4 × 0.4 | 1.8 × 1.8 | 1 × 1 | 10 × 10 | 2 × 2 |
| Matrix size | 256 × 256 | 250 × 384 | 128 × 128 | 256 × 192 | 256 × 256 | 531 × 640 | 128 × 128 | 256 × 256 | 16 × 16 | 128 × 128 |
| No. slices | 36 | 40 | 30 | 80 | 192 | 56 | 19 | 192 | 1 | 65 |
| Field of view (mm2) | 230 | 210 | 230 | 230 | 220 | 230 | 230 | 220 | 160 | 256 |
| Slice thickness, mm | 4 | 3 | 5 | 1.75 | 1 | 3 | 5 | 1 | 10 | 2 |
| TE (ms) | 100 | 88 | 78 | 20 | 3.79 | 115 | 32 | 3.79 | 135 | 83 |
| TI (ms) | 2500 | - | - | - | 1100 | - | - | - | - | - |
| TR (ms) | 9220 | 3490 | 4000 | 28 | 1800 | 4290 | 1400 | 1800 | 1510 | 8000 |
| TA (min:s) | 4:38 | 1:25 | 1:38 | 3:52 | 5:44 | 3:40 | 1:17 | 5:44 | 6:07 | 4:38 |
| GRAPPA factor | - | - | 2 | 2 | - | 2 | 2 | - | - | 2 |
| RB/pixel, Hz/pixel | 170 | 199 | 1502 | 120 | 200 | 176 | 1346 | 200 | 1200 | 1562 |
| FA (°) | 150 | 120 | - | 15 | 12 | 120 | 90 | 12 | 90 | - |
| Fat saturation | yes | no | yes | No | no | No | yes | no | yes | yes |
| 2D ax T2 FLAIR | 3D T1 Sag | Multi-Echo Spin Echo | MRF | |
|---|---|---|---|---|
| TSE + IR | MP2RAGE | (T2 Map) | ||
| Voxel dimensions (mm2) | 0.6 × 0.6 | 1.0 × 1.0 | 0.7 × 0.7 | 1.0 × 1.0 |
| Matrix size | 384 × 276 | 256 × 216 | 320 × 257 | 256 × 256 |
| No. slices | 10 | 160 | 10 | 10–13 |
| Field of view (mm2) | 230 × 166 | 256 × 216 | 230 × 180 | 256 × 256 |
| Slice thickness (mm) | 5.0 | 1.0 | 5.0 | 5.0 |
| TE (ms) | 126 | 2.98 | 12.6, 25.2,... 201.6 | 2 |
| TI (ms) | 2500 | 700, 2500 | – | 21 |
| TR (ms) | 8500 | 5000 | 2100 | 12.1–15.0 (varied by sequence) |
| TA (min:sec) | 3:43 | 8:02 | 3:38 | 3:51–4:51 |
| Acceleration factor | 1 (turbo factor: 19) | 2 | 3 | 24 (inner k-space), 48 (outer k-space) |
| RB/pixel (Hz/pixel) | 140 | 240 | 150 | RX-Bandwidth: 400 kHz |
| FA (°) | 180 | 4, 5 | 180 | 0–74 (varied by sequence) |
| Fat saturation | yes | no | no | no |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marik, W.; Cardoso, P.L.; Springer, E.; Bogner, W.; Preusser, M.; Widhalm, G.; Hangel, G.; Hainfellner, J.A.; Rausch, I.; Weber, M.; et al. Evaluation of Gliomas with Magnetic Resonance Fingerprinting with PET Correlation—A Comparative Study. Cancers 2023, 15, 2740. https://doi.org/10.3390/cancers15102740
Marik W, Cardoso PL, Springer E, Bogner W, Preusser M, Widhalm G, Hangel G, Hainfellner JA, Rausch I, Weber M, et al. Evaluation of Gliomas with Magnetic Resonance Fingerprinting with PET Correlation—A Comparative Study. Cancers. 2023; 15(10):2740. https://doi.org/10.3390/cancers15102740
Chicago/Turabian StyleMarik, Wolfgang, Pedro Lima Cardoso, Elisabeth Springer, Wolfgang Bogner, Matthias Preusser, Georg Widhalm, Gilbert Hangel, Johannes A. Hainfellner, Ivo Rausch, Michael Weber, and et al. 2023. "Evaluation of Gliomas with Magnetic Resonance Fingerprinting with PET Correlation—A Comparative Study" Cancers 15, no. 10: 2740. https://doi.org/10.3390/cancers15102740
APA StyleMarik, W., Cardoso, P. L., Springer, E., Bogner, W., Preusser, M., Widhalm, G., Hangel, G., Hainfellner, J. A., Rausch, I., Weber, M., Schmidbauer, V., Traub-Weidinger, T., & Trattnig, S. (2023). Evaluation of Gliomas with Magnetic Resonance Fingerprinting with PET Correlation—A Comparative Study. Cancers, 15(10), 2740. https://doi.org/10.3390/cancers15102740







