Machine Learning Improves the Prediction of Responses to Immune Checkpoint Inhibitors in Metastatic Melanoma
Abstract
Simple Summary
Abstract
1. Introduction
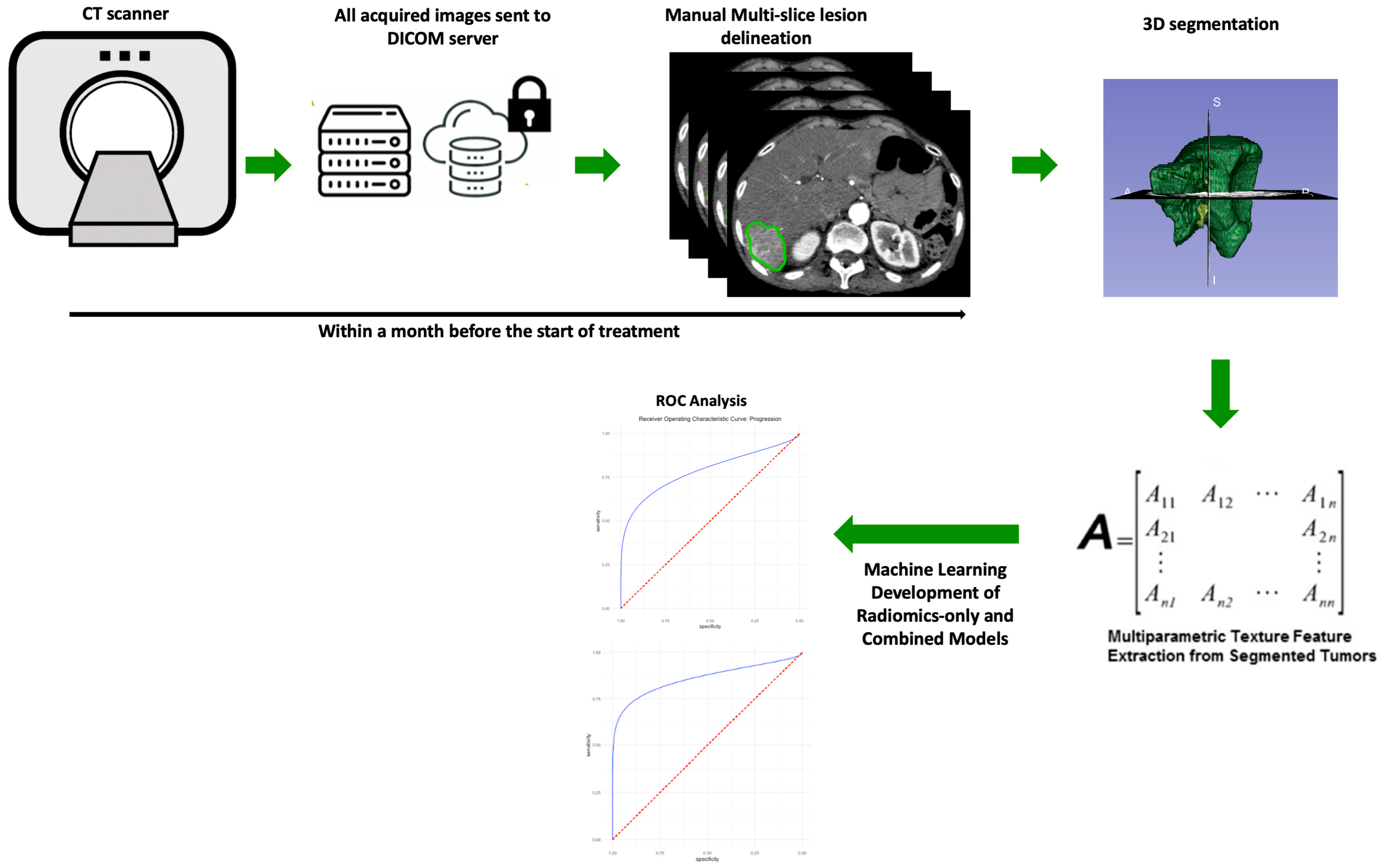
2. Materials and Methods
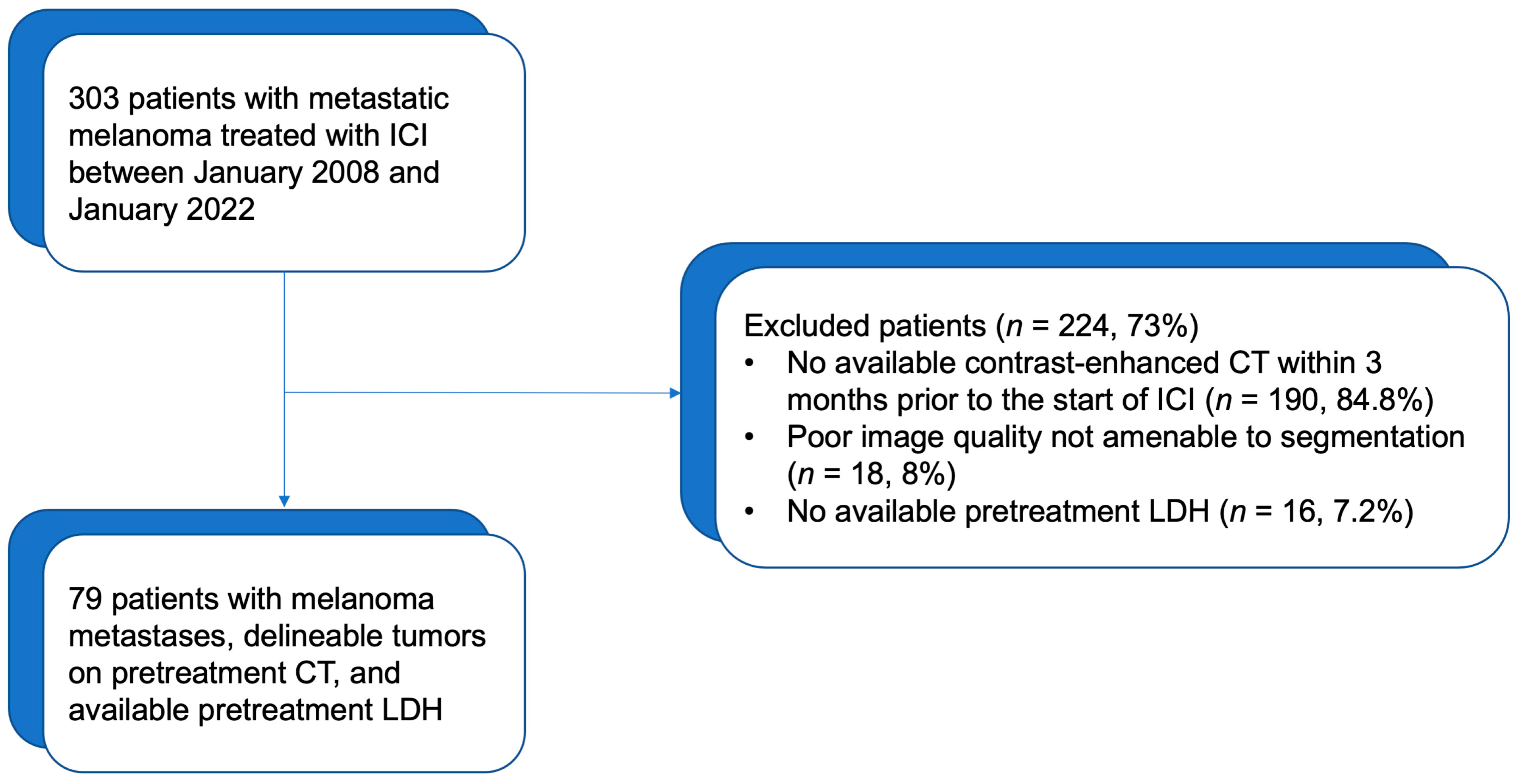
2.1. Patients and Clinical Data
2.2. CT Acquisition
2.3. Segmentation and Image Processing
2.4. Extraction of Radiomic Features and Machine Learning Model Training
3. Results
3.1. Patients and Tumor Characteristics
3.2. ICI Response Kinetics for the Whole Population
3.3. Comparison of Baseline Characteristics between Responding and Progressing Tumors
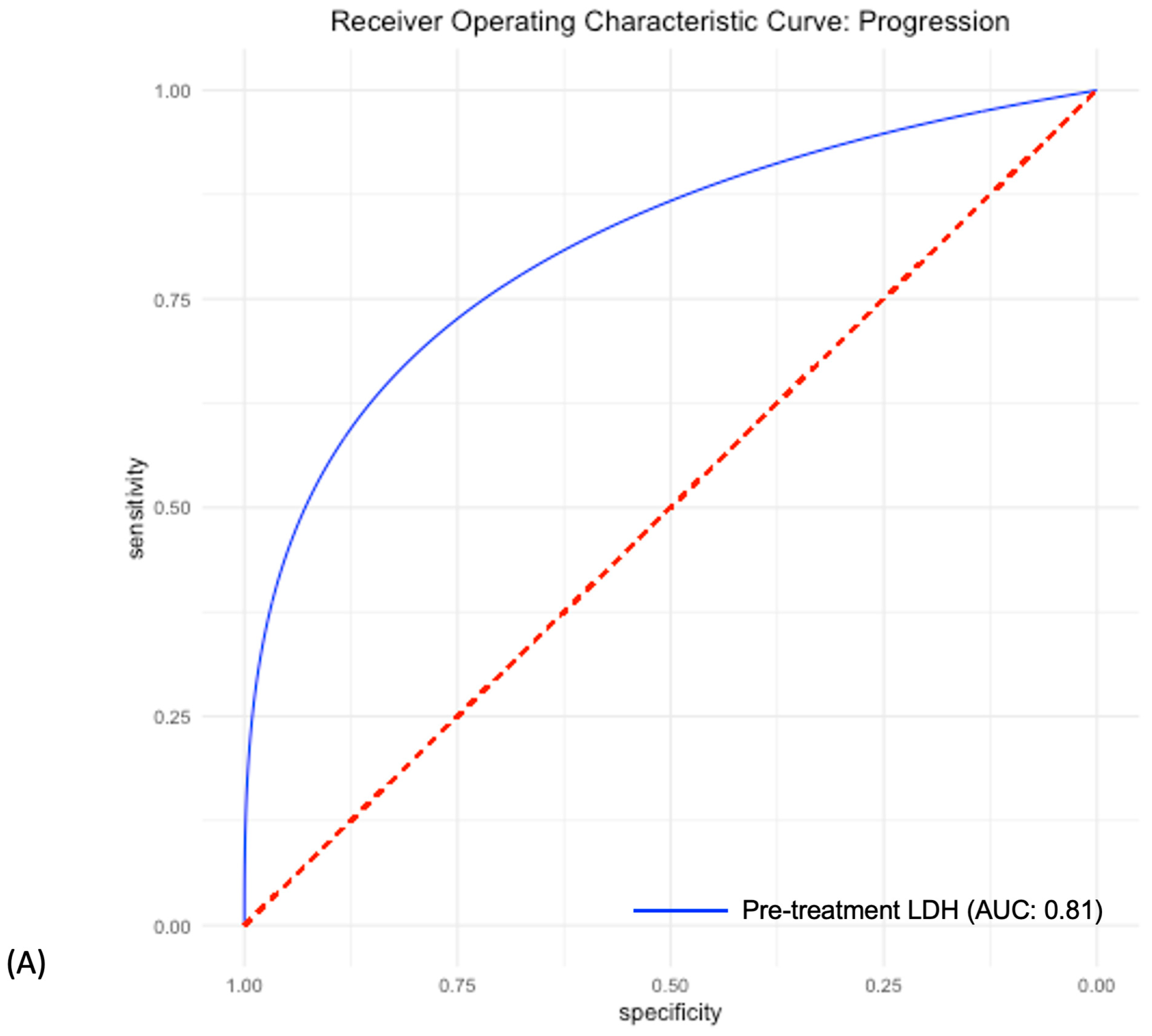
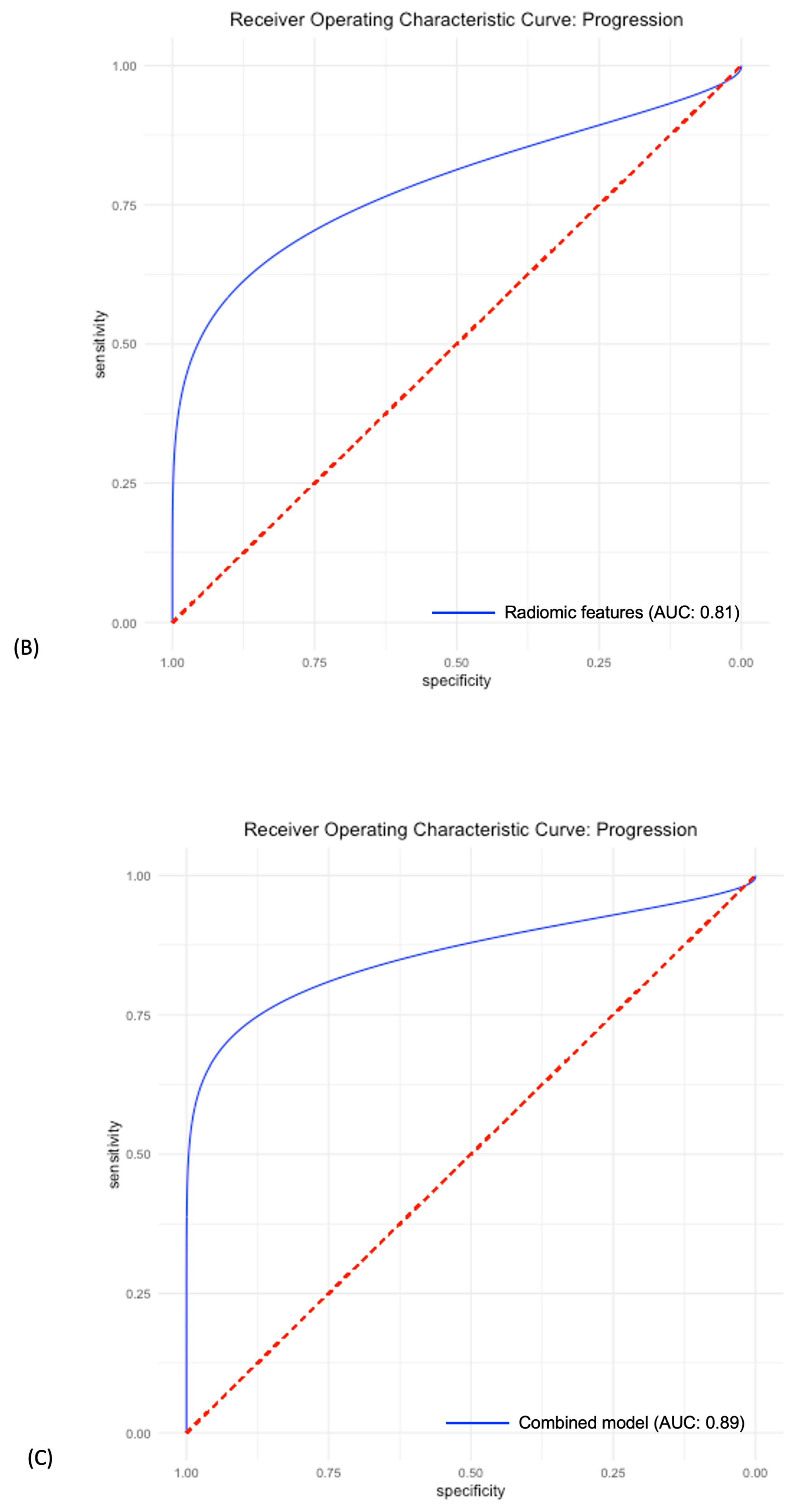
3.4. Predictive Power of Individual LDH, Radiomic and Combined Models
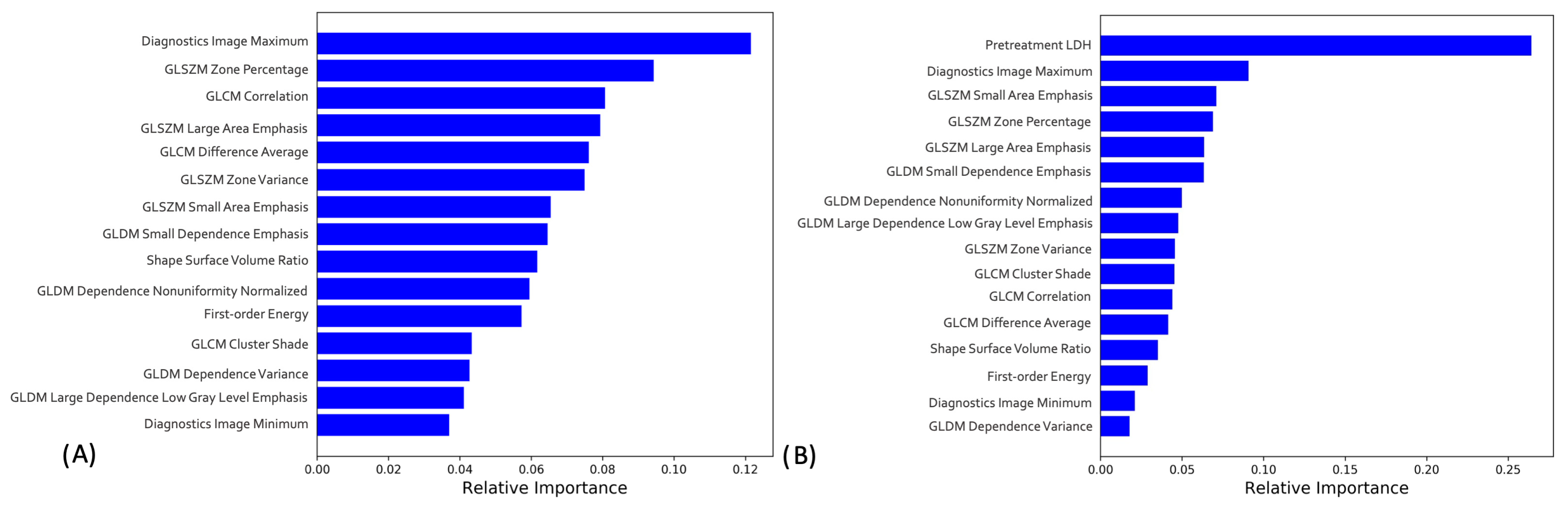
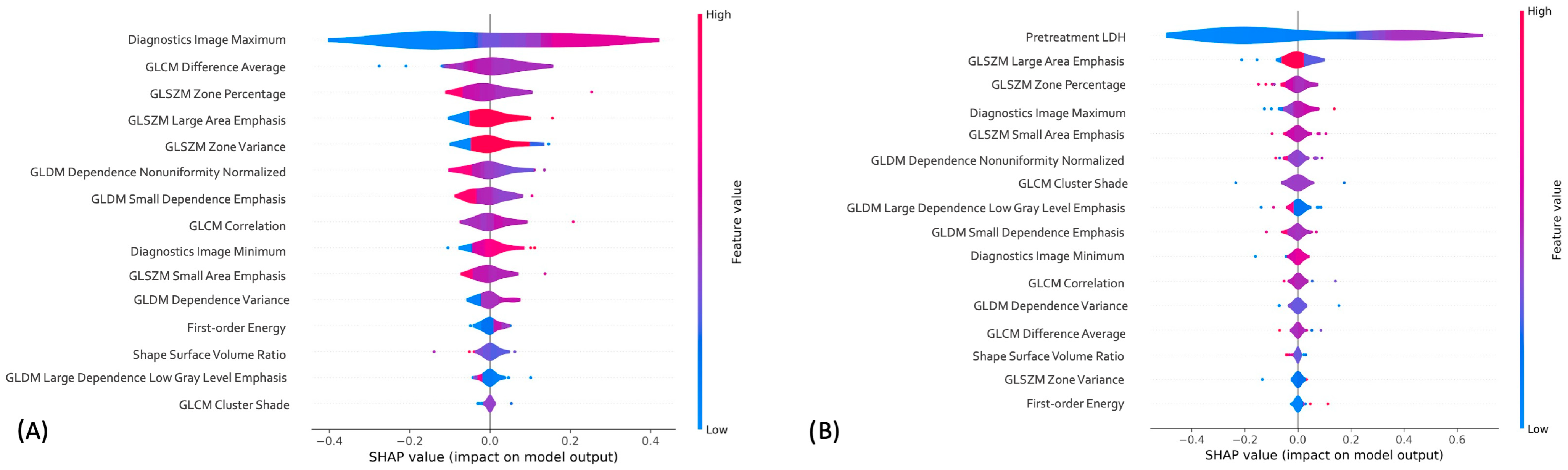
3.5. Radiomics-only and Combined Machine Learning Models Interpretation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- National Cancer Institute Surveillance Epidemiology and End Results Program. Cancer Statistics Cancer Stat Facts: Melanoma of the Skin. 2014. Available online: https://seer.cancer.gov/statfacts/html/melan.html (accessed on 11 March 2018).
- Available online: https://www.cancer.org/cancer/melanoma-skin-cancer/detection-diagnosis-staging/survival-rates-for-melanoma-skin-cancer-by-stage.html (accessed on 13 March 2023).
- Wolchok, J.D.; Chiarion-Sileni, V.; Gonzalez, R.; Rutkowski, P.; Grob, J.J.; Cowey, C.L.; Lao, C.D.; Wagstaff, J.; Schadendorf, D.; Ferrucci, P.F.; et al. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 2017, 377, 1345–1356. [Google Scholar] [PubMed]
- Pelizzari, G.; Basile, D.; Zago, S.; Lisanti, C.; Bartoletti, M.; Bortot, L.; Vitale, M.G.; Fanotto, V.; Barban, S.; Cinausero, M.; et al. Lactate Dehydrogenase (LDH) Response to First-Line Treatment Predicts Survival in Metastatic Breast Cancer: First Clues for a Cost-Effective and Dynamic Biomarker. Cancers 2019, 11, 1243. [Google Scholar] [CrossRef]
- Diem, S.; Kasenda, B.; Spain, L.; Martin-Liberal, J.; Marconcini, R.; Gore, M.; Larkin, J. Serum lactate dehydrogenase as an early marker for outcome in patients treated with anti-PD-1 therapy in metastatic melanoma. Br. J. Cancer 2016, 114, 256–261. [Google Scholar] [CrossRef]
- Van Wilpe, S.; Koornstra, R.; Den Brok, M.; De Groot, J.W.; Blank, C.; De Vries, J.; Gerritsen, W.; Mehra, N. Lactate dehydrogenase: A marker of diminished antitumor immunity. Oncoimmunology 2020, 9, 1731942. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, Q.; Dong, Q.; Zhan, L.; Zhang, J. How to differentiate pseudoprogression from true progression in cancer patients treated with immunotherapy. Am. J. Cancer Res. 2019, 9, 1546–1553. [Google Scholar] [PubMed]
- Jia, W.; Gao, Q.; Han, A.; Zhu, H.; Yu, J. The potential mechanism, recognition and clinical significance of tumor pseudoprogression after immunotherapy. Cancer Biol. Med. 2019, 16, 655–670. [Google Scholar] [CrossRef] [PubMed]
- Dromain, C.; Beigelman, C.; Pozzessere, C.; Duran, R.; Digklia, A. Imaging of tumour response to immunotherapy. Eur. Radiol. Exp. 2020, 4, 1–15. [Google Scholar] [CrossRef]
- Garutti, M.; Bonin, S.; Buriolla, S.; Bertoli, E.; Pizzichetta, M.A.; Zalaudek, I.; Puglisi, F. Find the Flame: Predictive Biomarkers for Im-munotherapy in Melanoma. Cancers 2021, 13, 1819. [Google Scholar] [CrossRef] [PubMed]
- Rizk, E.M.; Seffens, A.M.; Trager, M.H.; Moore, M.R.; Geskin, L.J.; Gartrell-Corrado, R.D.; Wong, W.; Saenger, Y.M. Biomarkers Predictive of Survival and Response to Immune Checkpoint Inhibitors in Melanoma. Am. J. Clin. Dermatol. 2019, 21, 1–11. [Google Scholar] [CrossRef]
- Bai, R.; Lv, Z.; Xu, D.; Cui, J. Predictive biomarkers for cancer immunotherapy with immune checkpoint inhibitors. Biomark. Res. 2020, 8, 1–17. [Google Scholar] [CrossRef]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef]
- Lin, Z.; Meng, X.; Wen, J.; Corral, J.M.; Andreev, D.; Kachler, K.; Schett, G.; Chen, X.; Bozec, A. Intratumor Heterogeneity Correlates with Reduced Immune Activity and Worse Survival in Melanoma Patients. Front. Oncol. 2020, 10, 596493. [Google Scholar] [CrossRef] [PubMed]
- Rauwerdink, D.J.W.; Molina, G.; Frederick, D.T.; Sharova, T.; van der Hage, J.; Cohen, S.; Boland, G.M. Mixed Response to Immunotherapy in Patients with Metastatic Melanoma. Ann. Surg. Oncol. 2020, 27, 3488–3497. [Google Scholar] [CrossRef] [PubMed]
- Nishino, M.; Jackman, D.M.; Hatabu, H.; Yeap, B.Y.; Cioffredi, L.-A.; Yap, J.T.; Jänne, P.A.; Johnson, B.E.; Abbeele, A.D.V.D. New Response Evaluation Criteria in Solid Tumors (RECIST) Guidelines for Advanced Non–Small Cell Lung Cancer: Comparison with Original RECIST and Impact on Assessment of Tumor Response to Targeted Therapy. Am. J. Roentgenol. 2010, 195, W221–W228. [Google Scholar] [CrossRef]
- Schwartz, L.H.; Litière, S.; de Vries, E.; Ford, R.; Gwyther, S.; Mandrekar, S.; Shankar, L.; Bogaerts, J.; Chen, A.; Dancey, J.; et al. RECIST 1.1-Update and clarification: From the RECIST committee. Eur. J. Cancer 2016, 62, 132–137. [Google Scholar] [CrossRef]
- Subbiah, V.; Chuang, H.H.; Gambhire, D.; Kairemo, K. Defining Clinical Response Criteria and Early Response Criteria for Precision Oncology: Current State-of-the-Art and Future Perspectives. Diagnostics 2017, 7, 10. [Google Scholar] [CrossRef] [PubMed]
- Aykan, N.F.; Özatlı, T. Objective response rate assessment in oncology: Current situation and future expectations. World J. Clin. Oncol. 2020, 11, 53–73. [Google Scholar] [CrossRef] [PubMed]
- Ko, C.-C.; Yeh, L.-R.; Kuo, Y.-T.; Chen, J.-H. Imaging biomarkers for evaluating tumor response: RECIST and beyond. Biomark. Res. 2021, 9, 1–20. [Google Scholar] [CrossRef]
- Radziuviene, G.; Rasmusson, A.; Augulis, R.; Grineviciute, R.B.; Zilenaite, D.; Laurinaviciene, A.; Ostapenko, V.; Laurinavicius, A. In-tratumoral Heterogeneity and Immune Response Indicators to Predict Overall Survival in a Retrospective Study of HER2-Borderline (IHC 2+) Breast Cancer Patients. Front Oncol. 2021, 11, 774088. [Google Scholar] [PubMed]
- Yoshimura, K.; Tsujikawa, T.; Mitsuda, J.; Ogi, H.; Saburi, S.; Ohmura, G.; Arai, A.; Shibata, S.; Thibault, G.; Chang, Y.H.; et al. Spatial Profiles of Intratumoral PD-1+ Helper T Cells Predict Prognosis in Head and Neck Squamous Cell Carcinoma. Front. Immunol. 2021, 12, 4554. [Google Scholar] [CrossRef] [PubMed]
- Laurinavicius, A.; Rasmusson, A.; Plancoulaine, B.; Shribak, M.; Levenson, R. Machine-Learning–Based Evaluation of Intratumoral Heterogeneity and Tumor-Stroma Interface for Clinical Guidance. Am. J. Pathol. 2021, 191, 1724–1731. [Google Scholar] [CrossRef] [PubMed]
- Seymour, L.; Bogaerts, J.; Perrone, A.; Ford, R.; Schwartz, L.H.; Mandrekar, S.; Lin, N.U.; Litière, S.; Dancey, J.; Chen, A.; et al. iRECIST: Guidelines for response criteria for use in trials testing immunotherapeutics. Lancet Oncol. 2017, 18, e143–e152, Erratum in Lancet Oncol. 2017, 18, e143–e152. [Google Scholar] [CrossRef]
- Weide, B.; Martens, A.; Hassel, J.C.; Berking, C.; Postow, M.A.; Bisschop, K.; Simeone, E.; Mangana, J.; Schilling, B.; Di Giacomo, A.M.; et al. Baseline Biomarkers for Outcome of Melanoma Patients Treated with Pembrolizumab. Clin. Cancer Res. 2016, 22, 5487–5496. [Google Scholar] [CrossRef] [PubMed]
- Walenta, S.; Mueller-Klieser, W.F. Lactate: Mirror and motor of tumor malignancy. Semin. Radiat. Oncol. 2004, 14, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Suh, S.-Y.; Ahn, H. Lactate dehydrogenase as a prognostic factor for survival time of terminally ill cancer patients: A preliminary study. Eur. J. Cancer 2007, 43, 1051–1059. [Google Scholar] [CrossRef] [PubMed]
- Burgermeister, S.; Gabryś, H.S.; Basler, L.; Hogan, S.A.; Pavic, M.; Bogowicz, M.; Martínez Gómez, J.M.; Vuong, D.; Tanadini-Lang, S.; Foerster, R.; et al. Improved Survival Prediction by Combining Radiological Imaging and S-100B Levels into a Multivariate Model in Metastatic Melanoma Patients Treated with Immune Checkpoint In-hibition. Front Oncol. 2022, 12, 830627. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, A.; Beichel, R.; Kalpathy-Cramer, J.; Finet, J.; Fillion-Robin, J.-C.; Pujol, S.; Bauer, C.; Jennings, D.; Fennessy, F.; Sonka, M.; et al. 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn. Reson. Imaging 2012, 30, 1323–1341. [Google Scholar] [CrossRef] [PubMed]
- Van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S. Aerts HJWL: Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef] [PubMed]
- Daye, D.; Tabari, A.; Kim, H.; Chang, K.; Kamran, S.C.; Hong, T.S.; Kalpathy-Cramer, J.; Gee, M.S. Quantitative tumor heterogeneity MRI profiling improves machine learning–based prognostication in patients with metastatic colon cancer. Eur. Radiol. 2021, 31, 5759–5767. [Google Scholar] [CrossRef] [PubMed]
- Bertsimas, D.; Pawlowski, C.; Zhuo, Y.D. From predictive methods to missing data imputation: An optimization approach. J. Mach. Learn. Res. 2017, 18, 7133–7171. [Google Scholar]
- He, H.; Bai, Y.; Garcia, E.A.; Li, S. ADASYN: Adaptive synthetic sampling approach for imbalanced learning. In Proceeding of the IEEE International Joint Conference on Neural Networks, Hong Kong, China, 1–8 June 2008; pp. 1322–1328. [Google Scholar] [CrossRef]
- Available online: https://statisticaloddsandends.wordpress.com/2019/12/29/what-is-a-brier-score/ (accessed on 13 March 2023).
- Schisterman, E.; Faraggi, D.; Reiser, B.; Hu, J. Youden Index and the optimal threshold for markers with mass at zero. Stat. Med. 2007, 27, 297–315. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Pérez, R.; Bajorath, J. Interpretation of machine learning models using shapley values: Application to compound potency and multi-target activity predictions. J. Comput. Mol. Des. 2020, 34, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Nosrati, A.; Tsai, K.K.; Goldinger, S.M.; Tumeh, P.; Grimes, B.; Loo, K.; Algazi, A.P.; Nguyen-Kim, T.D.L.; Levesque, M.; Dummer, R.; et al. Evaluation of clinicopathological factors in PD-1 response: Derivation and validation of a prediction scale for response to PD-1 monotherapy. Br. J. Cancer 2017, 116, 1141–1147. [Google Scholar] [CrossRef] [PubMed]
- Park, B.W.; Kim, J.K.; Heo, C.; Park, K.J. Reliability of CT radiomic features reflecting tumour heterogeneity according to image quality and image processing parameters. Sci. Rep. 2020, 10, 3852. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Gopal, Y.N.; Scott, A.; Chen, G.; Woodman, S.E.; Davies, M.A. Role and therapeutic potential of PI3K-mTOR sig-naling in de novo resistance to BRAF inhibition. Pigment. Cell Melanoma Res. 2012, 25, 248–258. [Google Scholar] [CrossRef] [PubMed]
- Khorrami, M.; Prasanna, P.; Gupta, A.; Patil, P.; Velu, P.D.; Thawani, R.; Corredor, G.; Alilou, M.; Bera, K.; Fu, P.; et al. Changes in CT Radiomic Features Associated with Lymphocyte Distribution Predict Overall Survival and Response to Immunotherapy in Non–Small Cell Lung Cancer. Cancer Immunol. Res. 2020, 8, 108–119. [Google Scholar] [CrossRef] [PubMed]
- Guerrisi, A.; Russillo, M.; Loi, E.; Ganeshan, B.; Ungania, S.; Desiderio, F.; Bruzzaniti, V.; Falcone, I.; Renna, D.; Ferraresi, V.; et al. Exploring CT Texture Parameters as Predictive and Response Imaging Biomarkers of Survival in Patients with Metastatic Melanoma Treated With PD-1 Inhibitor Nivolumab: A Pilot Study Using a Delta-Radiomics Approach. Front. Oncol. 2021, 11. [Google Scholar] [CrossRef]
- Peisen, F.; Hänsch, A.; Hering, A.; Brendlin, A.S.; Afat, S.; Nikolaou, K.; Gatidis, S.; Eigentler, T.; Amaral, T.; Moltz, J.H.; et al. Combination of Whole-Body Baseline CT Radiomics and Clinical Parameters to Predict Response and Survival in a Stage-IV Melanoma Cohort Undergoing Immunotherapy. Cancers 2022, 14, 2992. [Google Scholar] [CrossRef] [PubMed]
- Bonnin, A.; Durot, C.; Barat, M.; Djelouah, M.; Grange, F.; Mulé, S.; Soyer, P.; Hoeffel, C. CT texture analysis as a predictor of favorable response to anti-PD1 monoclonal antibodies in metastatic skin melanoma. Diagn. Interv. Imaging 2022, 103, 97–102. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-L.; Mao, L.-L.; Zhou, Z.-G.; Si, L.; Zhu, H.-T.; Chen, X.; Zhou, M.-J.; Sun, Y.-S.; Guo, J. Pilot Study of CT-Based Radiomics Model for Early Evaluation of Response to Immunotherapy in Patients with Metastatic Melanoma. Front. Oncol. 2020, 10. [Google Scholar] [CrossRef] [PubMed]
- Trebeschi, S.; Drago, S.G.; Birkbak, N.J.; Kurilova, I.; Cǎlin, A.M.; Pizzi, A.D.; Lalezari, F.; Lambregts, D.M.J.; Rohaan, M.W.; Parmar, C.; et al. Predicting response to cancer immunotherapy using non-invasive radiomic biomarkers. Ann. Oncol. 2019, 30, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Durot, C.; Mulé, S.; Soyer, P.; Marchal, A.; Grange, F.; Hoeffel, C. Metastatic melanoma: Pretreatment contrast-enhanced CT texture parameters as predictive biomarkers of survival in patients treated with pembrolizumab. Eur. Radiol. 2019, 29, 3183–3191. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Limkin, E.J.; Vakalopoulou, M.; Dercle, L.; Champiat, S.; Han, S.R.; Verlingue, L.; Brandao, D.; Lancia, A.; Ammari, S.; et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to an-ti-PD-1 or anti-PD-L1immunotherapy: An imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018, 19, 1180–1191. [Google Scholar] [CrossRef] [PubMed]
- Madonna, G.; Masucci, G.V.; Capone, M.; Mallardo, D.; Grimaldi, A.M.; Simeone, E.; Vanella, V.; Festino, L.; Palla, M.; Scarpato, L.; et al. Clinical Categorization Algorithm (CLICAL) and Machine Learning Approach (SRF-CLICAL) to Predict Clinical Benefit to Immunotherapy in Metastatic Melanoma Patients: Real-World Evidence from the Istituto Nazionale Tumori IRCCS Fondazione Pascale, Napoli, Italy. Cancers 2021, 13, 4164. [Google Scholar] [CrossRef] [PubMed]
| Variables | n (%) |
|---|---|
| Mean Age a (years ± SD) | 62.1 ± 17 |
| Gender | |
| Male | 51 (64.6%) |
| Female | 28 (35.4%) |
| NRAS mutation | |
| Positive | 8 (10%) |
| Negative | 71 (90%) |
| BRAF V600 mutation | |
| Positive | 23 (29%) |
| Negative | 56 (71%) |
| Primary Site of Disease | |
| Skin | 57 (72%) |
| Other | 22 (28%) |
| LDH > 248 (UI/L) | 36 (45.5%) |
| Cancer Stage | 4 |
| Number of Liver Lesions | |
| 1 | 41 (52%) |
| 2 | 19 (24%) |
| 3 | 9 (11.3%) |
| 4 | 3 (3.7%) |
| 5 | 3 (3.7%) |
| 7 | 3 (3.7%) |
| 9 | 1 (1.6%) |
| ICI Type | |
| Ipilimumab | 47 (59.4%) |
| Pembrozilumab | 14 (17.7%) |
| Nivolumab | 12 (15%) |
| Other ICI | 6 (7.9%) |
| RECIST status at three months | |
| Progression | 23 (29%) |
| Stable, partial or complete response | 56 (71%) |
| Variable | Responders | Non-Responders | p Value | ||
|---|---|---|---|---|---|
| Mean ± SD | Min–Max | Mean ± SD | Min–Max | ||
| Age (years) | 63 ± 15 | 22–89 | 60 ± 15 | 29–90 | 0.27 |
| Gender (M:F) | 28:28 | 18:5 | 0.01 | ||
| LDH (UI/L) | 318 ± 214 | 108–1242 | 914 ± 672 | 160–2751 | 0.00 |
| Maximum dimension (cm) | 22 ± 14 | 4.5–107 | 29 ± 26 | 4.5–126 | 0.027 |
| Tumor volume (mL) | 20.4 ± 13 | 4.1–92.7 | 37.3 ± 75 | 4.4–562 | 0.022 |
| Metric | LDH only | Radiomics Only | Combined |
|---|---|---|---|
| Sensitivity | 0.70 | 0.58 | 0.75 |
| Specificity | 0.87 | 0.91 | 0.95 |
| Accuracy | 0.76 | 0.76 | 0.85 |
| AUC | 0.81 CI: [0.72–0.91] | 0.81 CI: [0.65–0.94] | 0.89 CI: [0.76–0.99] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tabari, A.; Cox, M.; D’Amore, B.; Mansur, A.; Dabbara, H.; Boland, G.; Gee, M.S.; Daye, D. Machine Learning Improves the Prediction of Responses to Immune Checkpoint Inhibitors in Metastatic Melanoma. Cancers 2023, 15, 2700. https://doi.org/10.3390/cancers15102700
Tabari A, Cox M, D’Amore B, Mansur A, Dabbara H, Boland G, Gee MS, Daye D. Machine Learning Improves the Prediction of Responses to Immune Checkpoint Inhibitors in Metastatic Melanoma. Cancers. 2023; 15(10):2700. https://doi.org/10.3390/cancers15102700
Chicago/Turabian StyleTabari, Azadeh, Meredith Cox, Brian D’Amore, Arian Mansur, Harika Dabbara, Genevieve Boland, Michael S. Gee, and Dania Daye. 2023. "Machine Learning Improves the Prediction of Responses to Immune Checkpoint Inhibitors in Metastatic Melanoma" Cancers 15, no. 10: 2700. https://doi.org/10.3390/cancers15102700
APA StyleTabari, A., Cox, M., D’Amore, B., Mansur, A., Dabbara, H., Boland, G., Gee, M. S., & Daye, D. (2023). Machine Learning Improves the Prediction of Responses to Immune Checkpoint Inhibitors in Metastatic Melanoma. Cancers, 15(10), 2700. https://doi.org/10.3390/cancers15102700









