Circulating and Endometrial Tissue microRNA Markers Associated with Endometrial Cancer Diagnosis, Prognosis, and Response to Treatment
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
3. Results
3.1. Diagnostic and Classificatory Capacity of miRs for EC
3.1.1. miRs Studied in Plasma
3.1.2. Serum miRs
3.1.3. miRs Studied in Endometrial Tissue
Tissue miRs with Diagnostic Usefulness for EC
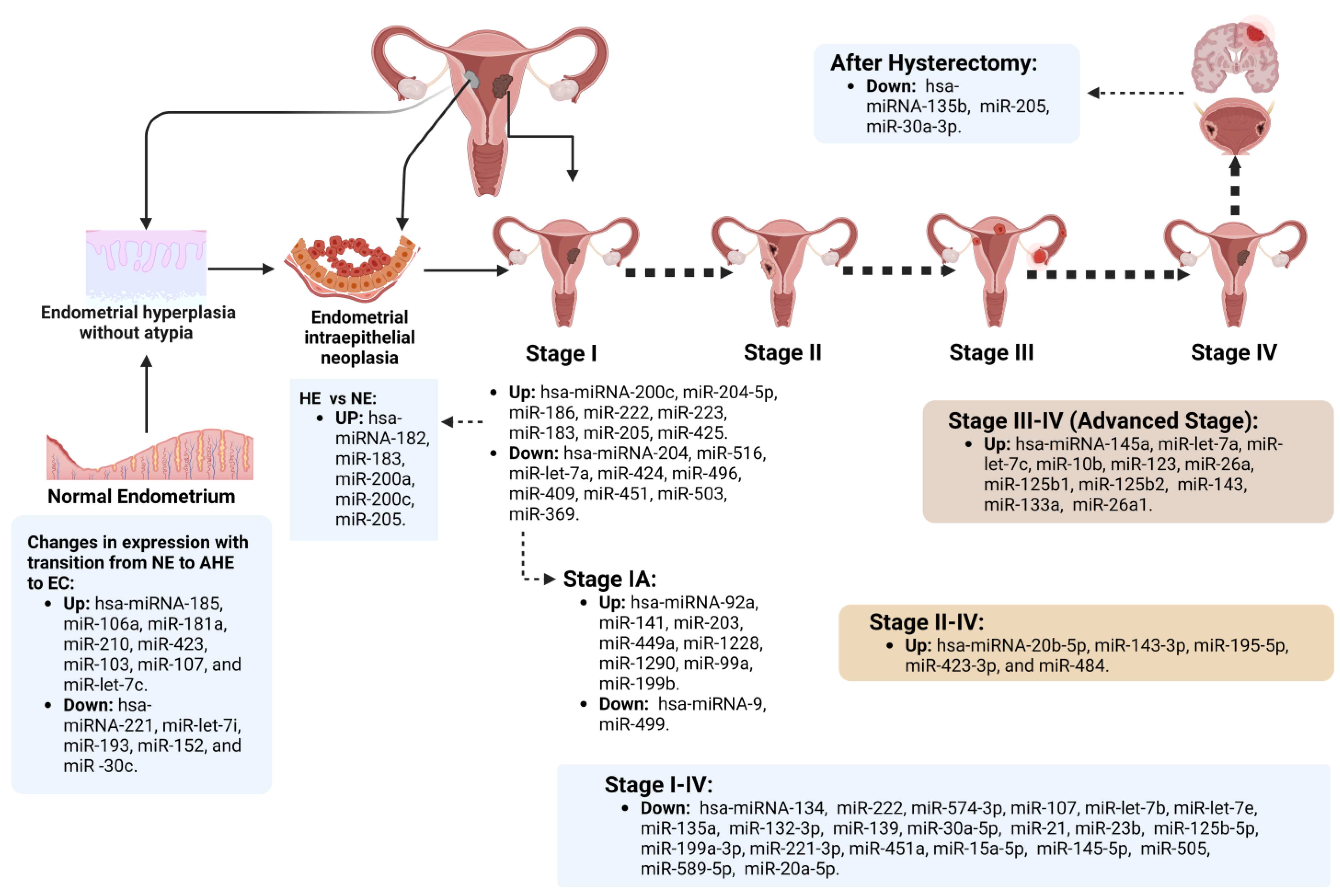
Endometrial miRs Expressed According to Specific EC Chronological Stage
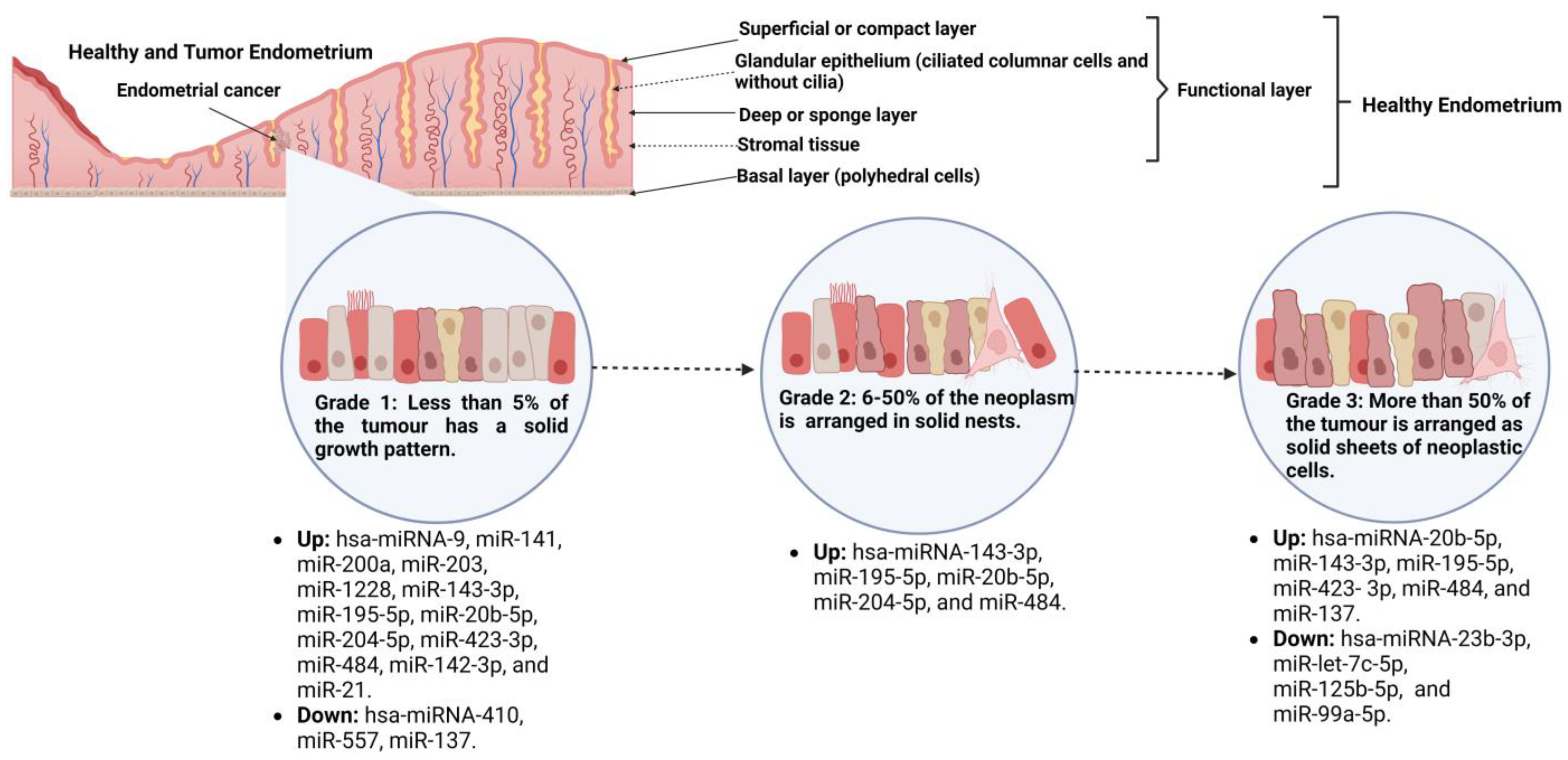
Tissue miRs Associated with EC, Tumor Grade, and Their Transcriptional Targets
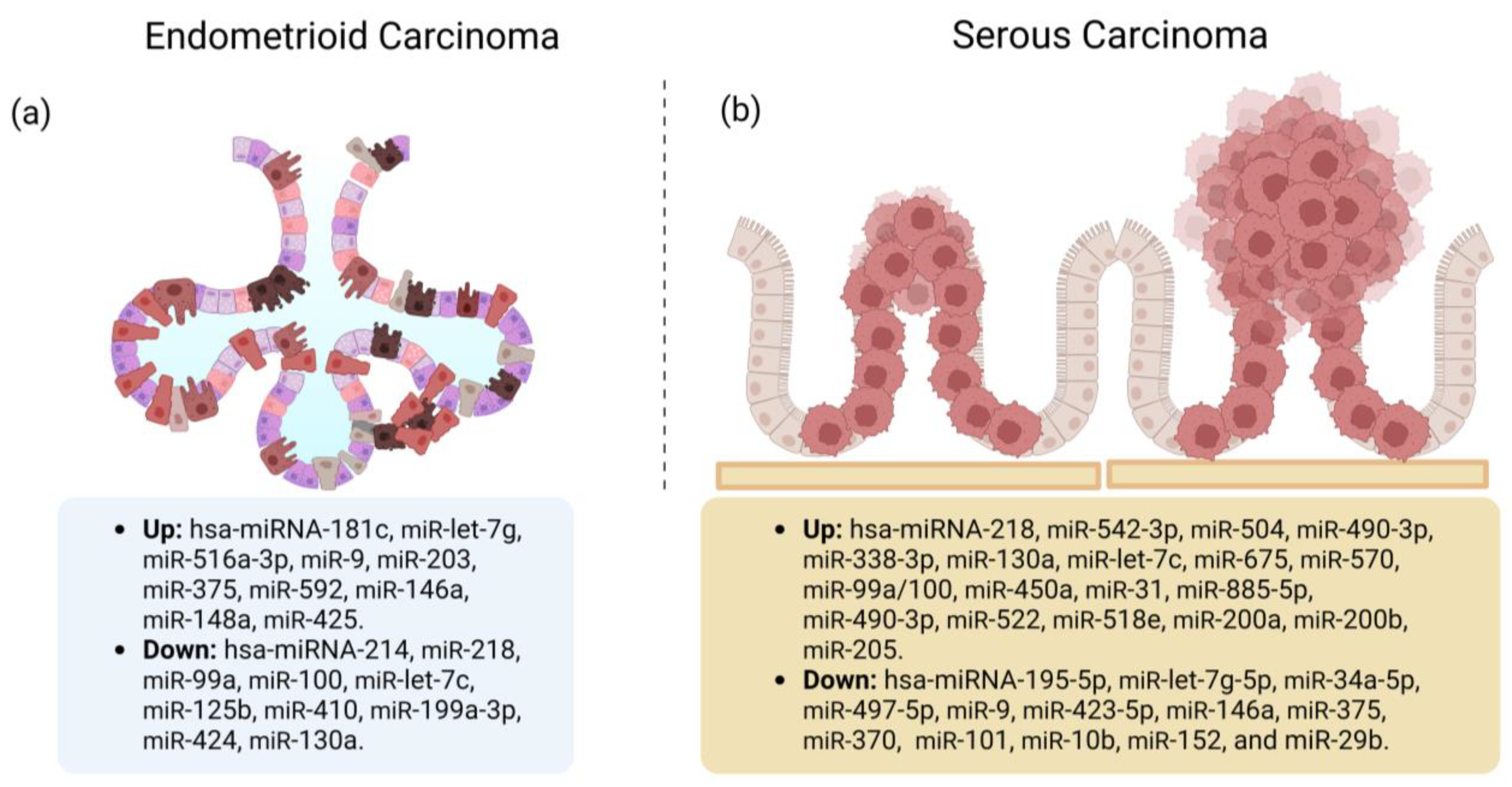
Tissue miRs Associated with the EC Histological Types
3.2. miRs and EC Prognosis
3.3. Treatment Response
3.3.1. Mechanisms of Chemoresistance in EC
3.3.2. miRs Involved with Treatment Response in EC
4. Conclusions
Gaps and Opportunity Areas for the Use of miRs as Markers with Clinical Value in EC
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Makker, V.; MacKay, H.; Ray-Coquard, I.; Levine, D.A.; Westin, S.N.; Aoki, D.; Oaknin, A. Endometrial cancer. Nat. Rev. Dis. Prim. 2021, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Gilks, C.B.; Oliva, E.; Soslow, R.A. Poor interobserver reproducibility in the diagnosis of high-grade endometrial carcinoma. Am. J. Surg. Pathol. 2013, 37, 874–881. [Google Scholar] [CrossRef] [PubMed]
- McAlpine, J.; Leon-Castillo, A.; Bosse, T. The rise of a novel classification system for endometrial carcinoma; integration of molecular subclasses. J. Pathol. 2018, 244, 538–549. [Google Scholar] [CrossRef]
- Talhouk, A.; McConechy, M.K.; Leung, S.; Li-Chang, H.H.; Kwon, J.S.; Melnyk, N.; Yang, W.; Senz, J.; Boyd, N.; Karnezis, A.N.; et al. A clinically applicable molecular-based classification for endometrial cancers. Br. J. Cancer 2015, 113, 299–310. [Google Scholar] [CrossRef]
- Stelloo, E.; Nout, R.A.; Osse, E.M.; Jurgenliemk-Schulz, I.J.; Jobsen, J.J.; Lutgens, L.C.; van der Steen-Banasik, E.M.; Nijman, H.W.; Putter, H.; Bosse, T.; et al. Improved Risk Assessment by Integrating Molecular and Clinicopathological Factors in Early-stage Endometrial Cancer-Combined Analysis of the PORTEC Cohorts. Clin. Cancer Res. 2016, 22, 4215–4224. [Google Scholar] [CrossRef]
- Levine, D.A. The Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar] [CrossRef]
- Concin, N.; Matias-Guiu, X.; Vergote, I.; Cibula, D.; Mirza, M.R.; Marnitz, S.; Ledermann, J.; Bosse, T.; Chargari, C.; Fagotti, A.; et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int. J. Gynecol. Cancer 2021, 31, 12–39. [Google Scholar] [CrossRef]
- Vermij, L.; Smit, V.; Nout, R.; Bosse, T. Incorporation of molecular characteristics into endometrial cancer management. Histopathology 2020, 76, 52–63. [Google Scholar] [CrossRef]
- Board, W. Female Genital Tumours. In WHO Classification of Tumours; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Passarello, K.; Kurian, S.; Villanueva, V. Endometrial Cancer: An Overview of Pathophysiology, Management, and Care. Semin. Oncol. Nurs. 2019, 35, 157–165. [Google Scholar] [CrossRef]
- Bokhman, J.V. Two pathogenetic types of endometrial carcinoma. Gynecol. Oncol. 1983, 15, 10–17. [Google Scholar] [CrossRef]
- Fidler, I.J. The pathogenesis of cancer metastasis: The ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer 2003, 3, 453–458. [Google Scholar] [CrossRef]
- Torres, A.; Torres, K.; Pesci, A.; Ceccaroni, M.; Paszkowski, T.; Cassandrini, P.; Zamboni, G.; Maciejewski, R. Diagnostic and prognostic significance of miRNA signatures in tissues and plasma of endometrioid endometrial carcinoma patients. Int. J. Cancer 2013, 132, 1633–1645. [Google Scholar] [CrossRef]
- Talhouk, A.; McAlpine, J.N. New classification of endometrial cancers: The development and potential applications of genomic-based classification in research and clinical care. Gynecol. Oncol. Res. Pr. 2016, 3, 14. [Google Scholar] [CrossRef]
- Wang, Q.; Xu, K.; Tong, Y.; Dai, X.; Xu, T.; He, D.; Ying, J. Novel miRNA markers for the diagnosis and prognosis of endometrial cancer. J. Cell Mol. Med. 2020, 24, 4533–4546. [Google Scholar] [CrossRef]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef]
- Banno, K.; Yanokura, M.; Kisu, I.; Yamagami, W.; Susumu, N.; Aoki, D. MicroRNAs in endometrial cancer. Int. J. Clin. Oncol. 2013, 18, 186–192. [Google Scholar] [CrossRef]
- Tsukamoto, O.; Miura, K.; Mishima, H.; Abe, S.; Kaneuchi, M.; Higashijima, A.; Miura, S.; Kinoshita, A.; Yoshiura, K.; Masuzaki, H. Identification of endometrioid endometrial carcinoma-associated microRNAs in tissue and plasma. Gynecol. Oncol. 2014, 132, 715–721. [Google Scholar] [CrossRef]
- Widodo; Djati, M.S.; Rifa’i, M. Role of MicroRNAs in carcinogenesis that potential for biomarker of endometrial cancer. Ann. Med. Surg. 2016, 7, 9–13. [Google Scholar] [CrossRef]
- Fan, X.; Cao, M.; Liu, C.; Zhang, C.; Li, C.; Cheng, W.; Zhang, S.; Zhang, H.; Zhu, W. Three plasma-based microRNAs as potent diagnostic biomarkers for endometrial cancer. Cancer Biomark. 2021, 31, 127–138. [Google Scholar] [CrossRef]
- Li, Z.; Li, N.; Sun, X.; Wang, J. FAM98A promotes cancer progression in endometrial carcinoma. Mol. Cell. Biochem. 2019, 459, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.X.; Zhao, W.; Mao, L.W.; Wang, Y.L.; Xia, L.Q.; Cao, M.; Shen, J.; Chen, J. Long non-coding RNA NIFK-AS1 inhibits M2 polarization of macrophages in endometrial cancer through targeting miR-146a. Int. J. Biochem. Cell Biol. 2018, 104, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zou, X.; Liu, C.; Cheng, W.; Zhang, S.; Geng, X.; Zhu, W. MicroRNA expression profile in serum reveals novel diagnostic biomarkers for endometrial cancer. Biosci. Rep. 2021, 41, BSR20210111. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Xu, J.L.; Chen, W.Q.; Xu, W.X.; Song, Y.X.; Tang, W.J.; Xu, D.; Jiang, M.P.; Tang, J. Roles and mechanisms of miR-195-5p in human solid cancers. Biomed. Pharm. 2022, 150, 112885. [Google Scholar] [CrossRef]
- Kong, F.; Ma, J.; Yang, H.; Yang, D.; Wang, C.; Ma, X. Long non-coding RNA PVT1 promotes malignancy in human endometrial carcinoma cells through negative regulation of miR-195-5p. Biochim. Biophys. Acta Mol. Cell Res. 2018. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Saini, H.K.; van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008, 36, D154–D158. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S. The microRNA Registry. Nucleic Acids Res. 2004, 32, D109–D111. [Google Scholar] [CrossRef]
- Li, M.; Zhou, Y.; Xia, T.; Zhou, X.; Huang, Z.; Zhang, H.; Zhu, W.; Ding, Q.; Wang, S. Circulating microRNAs from the miR-106a-363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res. Treat. 2018, 170, 257–270. [Google Scholar] [CrossRef]
- Zhou, W.; Shi, G.; Zhang, Q.; Wu, Q.; Li, B.; Zhang, Z. MicroRNA-20b promotes cell growth of breast cancer cells partly via targeting phosphatase and tensin homologue (PTEN). Cell Biosci. 2014, 4, 62. [Google Scholar] [CrossRef]
- Cascio, S.; D’Andrea, A.; Ferla, R.; Surmacz, E.; Gulotta, E.; Amodeo, V.; Bazan, V.; Gebbia, N.; Russo, A. miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. J. Cell. Physiol. 2010, 224, 242–249. [Google Scholar] [CrossRef]
- Karagkouni, D.; Paraskevopoulou, M.D.; Chatzopoulos, S.; Vlachos, I.S.; Tastsoglou, S.; Kanellos, I.; Papadimitriou, D.; Kavakiotis, I.; Maniou, S.; Skoufos, G.; et al. DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Res. 2018, 46, D239–D245. [Google Scholar] [CrossRef]
- Jia, W.; Wu, Y.; Zhang, Q.; Gao, G.; Zhang, C.; Xiang, Y. Identification of four serum microRNAs from a genome-wide serum microRNA expression profile as potential non-invasive biomarkers for endometrioid endometrial cancer. Oncol. Lett. 2013, 6, 261–267. [Google Scholar] [CrossRef]
- Chung, T.K.; Lau, T.S.; Cheung, T.H.; Yim, S.F.; Lo, K.W.; Siu, N.S.; Chan, L.K.; Yu, M.Y.; Kwong, J.; Doran, G.; et al. Dysregulation of microRNA-204 mediates migration and invasion of endometrial cancer by regulating FOXC1. Int. J. Cancer 2012, 130, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Choi, H.J.; Kang, C.S.; Lee, H.J.; Lee, W.S.; Park, C.S. Expression of miRNAs and PTEN in endometrial specimens ranging from histologically normal to hyperplasia and endometrial adenocarcinoma. Mod. Pathol. 2012, 25, 1508–1515. [Google Scholar] [CrossRef] [PubMed]
- Boren, T.; Xiong, Y.; Hakam, A.; Wenham, R.; Apte, S.; Wei, Z.; Kamath, S.; Chen, D.T.; Dressman, H.; Lancaster, J.M. MicroRNAs and their target messenger RNAs associated with endometrial carcinogenesis. Gynecol. Oncol. 2008, 110, 206–215. [Google Scholar] [CrossRef]
- Cohn, D.E.; Fabbri, M.; Valeri, N.; Alder, H.; Ivanov, I.; Liu, C.G.; Croce, C.M.; Resnick, K.E. Comprehensive miRNA profiling of surgically staged endometrial cancer. Am. J. Obstet. Gynecol. 2010, 202, 656.e1–656.e8. [Google Scholar] [CrossRef]
- Montagnana, M.; Benati, M.; Danese, E.; Giudici, S.; Perfranceschi, M.; Ruzzenenete, O.; Salvagno, G.L.; Bassi, A.; Gelati, M.; Paviati, E.; et al. Aberrant MicroRNA Expression in Patients With Endometrial Cancer. Int. J. Gynecol. Cancer 2017, 27, 459–466. [Google Scholar] [CrossRef]
- Torres, A.; Torres, K.; Pesci, A.; Ceccaroni, M.; Paszkowski, T.; Cassandrini, P.; Zamboni, G.; Maciejewski, R. Deregulation of miR-100, miR-99a and miR-199b in tissues and plasma coexists with increased expression of mTOR kinase in endometrioid endometrial carcinoma. BMC Cancer 2012, 12, 369. [Google Scholar] [CrossRef]
- Donkers, H.; Hirschfeld, M.; Weiss, D.; Erbes, T.; Jaeger, M.; Pijnenborg, J.M.A.; Bekkers, R.; Galaal, K.; Consortium, E. Usefulness of microRNA detection in the diagnostics of endometrial cancer. Acta Obstet. Gynecol. Scand. 2021, 100, 1148–1154. [Google Scholar] [CrossRef]
- Klicka, K.; Grzywa, T.M.; Klinke, A.; Mielniczuk, A.; Wejman, J.; Ostrowska, J.; Gondek, A.; Wlodarski, P.K. Decreased expression of miR-23b is associated with poor survival of endometrial cancer patients. Sci. Rep. 2022, 12, 18824. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.M.; Wan, X.H.; Sang, G.Y.; Zhao, J.D.; Zhu, Q.Y.; Wang, D.M. miR-15a-5p suppresses endometrial cancer cell growth via Wnt/beta-catenin signaling pathway by inhibiting WNT3A. Eur. Rev. Med. Pharm. Sci. 2017, 21, 4810–4818. [Google Scholar]
- Wu, X.; Han, Y.; Liu, F.; Ruan, L. Downregulations of miR-449a and miR-145-5p Act as Prognostic Biomarkers for Endometrial Cancer. J. Comput. Biol. 2020, 27, 834–844. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Sun, K.X.; Liu, B.L.; Zong, Z.H.; Zhao, Y. MicroRNA-505 functions as a tumor suppressor in endometrial cancer by targeting TGF-alpha. Mol. Cancer 2016, 15, 11. [Google Scholar] [CrossRef]
- Wang, Y.; Dong, L.; Liu, Y. Targeting Thyroid Receptor Interacting Protein 6 by MicroRNA-589-5p Inhibits Cell Proliferation, Migration, and Invasion in Endometrial Carcinoma. Cancer Biother. Radiopharm. 2019, 34, 529–536. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, N. MicroRNA-20a-5p inhibits epithelial to mesenchymal transition and invasion of endometrial cancer cells by targeting STAT3. Int. J. Clin. Exp. Pathol. 2018, 11, 5715–5724. [Google Scholar] [PubMed]
- He, Y.; Ma, H.; Wang, J.; Kang, Y.; Xue, Q. miR-20a-5p inhibits endometrial cancer progression by targeting janus kinase 1. Oncol. Lett. 2021, 21, 427. [Google Scholar] [CrossRef]
- Sun, R.; Liu, J.; Nie, S.; Li, S.; Yang, J.; Jiang, Y.; Cheng, W. Construction of miRNA-mRNA Regulatory Network and Prognostic Signature in Endometrial Cancer. Oncol. Targets 2021, 14, 2363–2378. [Google Scholar] [CrossRef]
- Park, Y.A.; Lee, J.W.; Choi, J.J.; Jeon, H.K.; Cho, Y.; Choi, C.; Kim, T.J.; Lee, N.W.; Kim, B.G.; Bae, D.S. The interactions between MicroRNA-200c and BRD7 in endometrial carcinoma. Gynecol. Oncol. 2012, 124, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, K.; Ishibashi, O.; Kawase, R.; Kurose, K.; Takeshita, T. miR-200a, miR-200b and miR-429 are onco-miRs that target the PTEN gene in endometrioid endometrial carcinoma. Anticancer Res. 2015, 35, 1401–1410. [Google Scholar]
- Esteva, F.J.; Guo, H.; Zhang, S.; Santa-Maria, C.; Stone, S.; Lanchbury, J.S.; Sahin, A.A.; Hortobagyi, G.N.; Yu, D. PTEN, PIK3CA, p-AKT, and p-p70S6K status: Association with trastuzumab response and survival in patients with HER2-positive metastatic breast cancer. Am. J. Pathol. 2010, 177, 1647–1656. [Google Scholar] [CrossRef]
- Dong, Y.; Si, J.-W.; Li, W.-T.; Liang, L.; Zhao, J.; Zhou, M.; Li, D.; Li, T. miR-200a/miR-141 and miR-205 upregulation might be associated with hormone receptor status and prognosis in endometrial carcinomas. Int. J. Clin. Exp. Pathol. 2015, 8, 2864. [Google Scholar] [PubMed]
- Gregory, P.A.; Bert, A.G.; Paterson, E.L.; Barry, S.C.; Tsykin, A.; Farshid, G.; Vadas, M.A.; Khew-Goodall, Y.; Goodall, G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008, 10, 593–601. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, D.R.; Howe, E.N.; Spoelstra, N.S.; Richer, J.K. Loss of miR-200c: A Marker of Aggressiveness and Chemoresistance in Female Reproductive Cancers. J. Oncol. 2010, 2010, 821717. [Google Scholar] [CrossRef]
- Zhang, X.; Dong, Y.; Ti, H.; Zhao, J.; Wang, Y.; Li, T.; Zhang, B. Down-regulation of miR-145 and miR-143 might be associated with DNA methyltransferase 3B overexpression and worse prognosis in endometrioid carcinomas. Hum. Pathol. 2013, 44, 2571–2580. [Google Scholar] [CrossRef]
- Jin, B.; Robertson, K.D. DNA methyltransferases, DNA damage repair, and cancer. Adv. Exp. Med. Biol. 2013, 754, 3–29. [Google Scholar] [CrossRef] [PubMed]
- Jin, F.; Dowdy, S.C.; Xiong, Y.; Eberhardt, N.L.; Podratz, K.C.; Jiang, S.W. Up-regulation of DNA methyltransferase 3B expression in endometrial cancers. Gynecol. Oncol. 2005, 96, 531–538. [Google Scholar] [CrossRef]
- Kalinkova, L.; Kajo, K.; Karhanek, M.; Wachsmannova, L.; Suran, P.; Zmetakova, I.; Fridrichova, I. Discriminating miRNA Profiles between Endometrioid Well- and Poorly-Differentiated Tumours and Endometrioid and Serous Subtypes of Endometrial Cancers. Int. J. Mol. Sci. 2020, 21, 6071. [Google Scholar] [CrossRef]
- Devor, E.J.; Hovey, A.M.; Goodheart, M.J.; Ramachandran, S.; Leslie, K.K. microRNA expression profiling of endometrial endometrioid adenocarcinomas and serous adenocarcinomas reveals profiles containing shared, unique and differentiating groups of microRNAs. Oncol. Rep. 2011, 26, 995–1002. [Google Scholar] [CrossRef]
- Fridrichova, I.; Kalinkova, L.; Karhanek, M.; Smolkova, B.; Machalekova, K.; Wachsmannova, L.; Nikolaieva, N.; Kajo, K. miR-497-5p Decreased Expression Associated with High-Risk Endometrial Cancer. Int. J. Mol. Sci. 2020, 22, 127. [Google Scholar] [CrossRef] [PubMed]
- Hiroki, E.; Akahira, J.; Suzuki, F.; Nagase, S.; Ito, K.; Suzuki, T.; Sasano, H.; Yaegashi, N. Changes in microRNA expression levels correlate with clinicopathological features and prognoses in endometrial serous adenocarcinomas. Cancer Sci. 2010, 101, 241–249. [Google Scholar] [CrossRef]
- Bernegger, G.; Musalek, M.; Rehmann-Sutter, C. An alternative view on the task of prognosis. Crit. Rev. Oncol. Hematol. 2012, 84 (Suppl. 2), S17–S24. [Google Scholar] [CrossRef] [PubMed]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef]
- Sorosky, J.I. Endometrial cancer. Obstet. Gynecol. 2012, 120, 383–397. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Su, N.; Qiu, H.; Chen, Y.; Yang, T.; Yan, Q.; Wan, X. miR-205 promotes tumor proliferation and invasion through targeting ESRRG in endometrial carcinoma. Oncol. Rep. 2013, 29, 2297–2302. [Google Scholar] [CrossRef]
- Zhang, G.; Hou, X.; Li, Y.; Zhao, M. MiR-205 inhibits cell apoptosis by targeting phosphatase and tensin homolog deleted on chromosome ten in endometrial cancer Ishikawa cells. BMC Cancer 2014, 14, 440. [Google Scholar] [CrossRef] [PubMed]
- Wilczynski, M.; Danielska, J.; Dzieniecka, M.; Szymanska, B.; Wojciechowski, M.; Malinowski, A. Prognostic and Clinical Significance of miRNA-205 in Endometrioid Endometrial Cancer. PLoS ONE 2016, 11, e0164687. [Google Scholar] [CrossRef]
- Hendrickson, M.; Ross, J.; Eifel, P.; Martinez, A.; Kempson, R. Uterine papillary serous carcinoma: A highly malignant form of endometrial adenocarcinoma. Am. J. Surg. Pathol. 1982, 6, 93–108. [Google Scholar] [CrossRef]
- Bancher-Todesca, D.; Neunteufel, W.; Williams, K.E.; Prainsack, D.; Breitenecker, G.; Friedlander, M.L.; Hacker, N.F. Influence of postoperative treatment on survival in patients with uterine papillary serous carcinoma. Gynecol. Oncol. 1998, 71, 344–347. [Google Scholar] [CrossRef]
- Wang, Y.; Yin, L.; Sun, X. CircRNA hsa_circ_0002577 accelerates endometrial cancer progression through activating IGF1R/PI3K/Akt pathway. J. Exp. Clin. Cancer Res. 2020, 39, 169. [Google Scholar] [CrossRef]
- Yuan, J.; Yin, Z.; Tao, K.; Wang, G.; Gao, J. Function of insulin-like growth factor 1 receptor in cancer resistance to chemotherapy. Oncol. Lett. 2018, 15, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Peng, X.; Du, W.; Huang, Y.; Zhang, C.; Zhang, X. circSLC6A6 Sponges miR-497-5p to Promote Endometrial Cancer Progression via the PI4KB/Hedgehog Axis. J. Immunol. Res. 2021, 2021, 5512391. [Google Scholar] [CrossRef]
- Xia, Y.; Hu, C.; Lian, L.; Hui, K.; Wang, L.; Qiao, Y.; Liu, L.; Liang, L.; Jiang, X. miR497 suppresses malignant phenotype in nonsmall cell lung cancer via targeting KDR. Oncol. Rep. 2019, 42, 443–452. [Google Scholar] [CrossRef]
- Pengcheng, S.; Ziqi, W.; Luyao, Y.; Xiangwei, Z.; Liang, L.; Yuwei, L.; Lechen, L.; Wanhai, X. MicroRNA-497 suppresses renal cell carcinoma by targeting VEGFR-2 in ACHN cells. Biosci. Rep. 2017, 37, BSR20170270. [Google Scholar] [CrossRef] [PubMed]
- Tao, L.; Zhang, C.Y.; Guo, L.; Li, X.; Han, N.N.; Zhou, Q.; Liu, Z.L. MicroRNA-497 accelerates apoptosis while inhibiting proliferation, migration, and invasion through negative regulation of the MAPK/ERK signaling pathway via RAF-1. J. Cell. Physiol. 2018, 233, 6578–6588. [Google Scholar] [CrossRef]
- Xu, X.; Liu, T.; Wang, Y.; Fu, J.; Yang, Q.; Wu, J.; Zhou, H. miRNA-mRNA Associated With Survival in Endometrial Cancer. Front. Genet 2019, 10, 743. [Google Scholar] [CrossRef] [PubMed]
- de Foucher, T.; Sbeih, M.; Uzan, J.; Bendifallah, S.; Lefevre, M.; Chabbert-Buffet, N.; Aractingi, S.; Uzan, C.; Abd Alsalam, I.; Mitri, R.; et al. Identification of micro-RNA expression profile related to recurrence in women with ESMO low-risk endometrial cancer. J. Transl. Med. 2018, 16, 131. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Bi, X.; Yang, Y. Circular RNA hsa_circ_0011324 is involved in endometrial cancer progression and the evolution of its mechanism. Bioengineered 2022, 13, 7485–7499. [Google Scholar] [CrossRef]
- Liu, Y.; Yuan, H.; He, T. Downregulated circular RNA hsa_circ_0005797 inhibits endometrial cancer by modulating microRNA-298/Catenin delta 1 signaling. Bioengineered 2022, 13, 4634–4645. [Google Scholar] [CrossRef]
- Yang, J.; Bassuk, A.G.; Merl-Pham, J.; Hsu, C.W.; Colgan, D.F.; Li, X.; Au, K.S.; Zhang, L.; Smemo, S.; Justus, S.; et al. Catenin delta-1 (CTNND1) phosphorylation controls the mesenchymal to epithelial transition in astrocytic tumors. Hum. Mol. Genet. 2016, 25, 4201–4210. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Wang, N.; Chen, H.; Zhang, M.; Lin, Q. MicroRNA-199a/b-5p inhibits endometrial cancer cell metastasis and invasion by targeting FAM83B in the epithelial-to-mesenchymal transition signaling pathway. Mol. Med. Rep. 2021, 23, 304. [Google Scholar] [CrossRef]
- Chang, L.; Zhang, D.; Shi, H.; Bian, Y.; Guo, R. MiR-143 inhibits endometrial cancer cell proliferation and metastasis by targeting MAPK1. Oncotarget 2017, 8, 84384–84395. [Google Scholar] [CrossRef]
- Li, J.; Sun, H.; Liu, T.; Kong, J. MicroRNA-423 promotes proliferation, migration and invasion and induces chemoresistance of endometrial cancer cells. Exp. Med. 2018, 16, 4213–4224. [Google Scholar] [CrossRef]
- Guo, S.; Zhang, J.; Zhao, Y.Y.; Zhou, L.Y.; Xie, Y.; Wu, X.Y.; Bian, X.; Yu, X.Y. The expressions of miR-151a-5p and miR-23b in lung cancer tissues and their effects on the biological functions of lung cancer A549 cells. Eur. Rev. Med. Pharm. Sci. 2020, 24, 6779–6785. [Google Scholar] [CrossRef]
- Wei, Y.; Li, H.; Qu, Q. miR-484 suppresses endocrine therapy-resistant cells by inhibiting KLF4-induced cancer stem cells in estrogen receptor-positive cancers. Breast Cancer 2021, 28, 175–186. [Google Scholar] [CrossRef]
- Jing, L.; Hua, X.; Yuanna, D.; Rukun, Z.; Junjun, M. Exosomal miR-499a-5p Inhibits Endometrial Cancer Growth and Metastasis via Targeting VAV3. Cancer Manag. Res. 2020, 12, 13541–13552. [Google Scholar] [CrossRef]
- Yue, Z.; Shen, J.J.; Huang, Q.T.; Qin, Y.F.; Li, X.N.; Liu, G.B. [MiR-135b promotes proliferation of endometrial carcinoma cells by targeting FOXO1]. Nan Fang Yi Ke Da Xue Xue Bao 2016, 36, 675–680. [Google Scholar] [PubMed]
- Greene, S.B.; Gunaratne, P.H.; Hammond, S.M.; Rosen, J.M. A putative role for microRNA-205 in mammary epithelial cell progenitors. J. Cell Sci. 2010, 123, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Karaayvaz, M.; Zhang, C.; Liang, S.; Shroyer, K.R.; Ju, J. Prognostic significance of miR-205 in endometrial cancer. PLoS ONE 2012, 7, e35158. [Google Scholar] [CrossRef]
- Chen, H.; Fan, Y.; Xu, W.; Chen, J.; Xu, C.; Wei, X.; Fang, D.; Feng, Y. miR-10b Inhibits Apoptosis and Promotes Proliferation and Invasion of Endometrial Cancer Cells via Targeting HOXB3. Cancer Biother. Radiopharm. 2016, 31, 225–231. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Dai, L.; Yue, Q.; Wang, H.; Wang, X.U.; Li, Y.; Chen, R. MiR-195 inhibits migration, invasion and epithelial-mesenchymal transition (EMT) of endometrial carcinoma cells by targeting SOX4. J. Biosci. 2019, 44, 146. [Google Scholar] [CrossRef]
- Deng, J.; Wang, W.; Yu, G.; Ma, X. MicroRNA-195 inhibits epithelial-mesenchymal transition by targeting G protein-coupled estrogen receptor 1 in endometrial carcinoma. Mol. Med. Rep. 2019, 20, 4023–4032. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Wei, B.; Ye, Q.; Liu, W. MiR-30a-5p/UBE3C axis regulates breast cancer cell proliferation and migration. Biochem. Biophys. Res. Commun. 2019, 516, 1013–1018. [Google Scholar] [CrossRef]
- Lin, C.Y.; Wu, R.C.; Yang, L.Y.; Jung, S.M.; Ueng, S.H.; Tang, Y.H.; Huang, H.J.; Tung, H.J.; Lin, C.T.; Chen, H.Y.; et al. MicroRNAs as Predictors of Future Uterine Malignancy in Endometrial Hyperplasia without Atypia. J. Pers. Med. 2022, 12, 311. [Google Scholar] [CrossRef]
- Liu, P.; Qi, M.; Ma, C.; Lao, G.; Liu, Y.; Liu, Y.; Liu, Y. Let7a inhibits the growth of endometrial carcinoma cells by targeting Aurora-B. FEBS Lett. 2013, 587, 2523–2529. [Google Scholar] [CrossRef]
- Penolazzi, L.; Bonaccorsi, G.; Gafa, R.; Ravaioli, N.; Gabriele, D.; Bosi, C.; Lanza, G.; Greco, P.; Piva, R. SLUG/HIF1-alpha/miR-221 regulatory circuit in endometrial cancer. Gene 2019, 711, 143938. [Google Scholar] [CrossRef]
- Xu, J.H.; Zhao, J.X.; Jiang, M.Y.; Yang, L.P.; Sun, M.L.; Wang, H.W. MiR-193 promotes cell proliferation and invasion by ING5/PI3K/AKT pathway of triple-negative breast cancer. Eur. Rev. Med. Pharm. Sci. 2020, 24, 3122–3129. [Google Scholar] [CrossRef]
- Xie, D.; Liang, Y.; Su, Y.; An, Y.; Qu, P. miR-152 inhibits proliferation of human endometrial cancer cells via inducing G2/M phase arrest by suppressing CDC25B expression. Biomed. Pharm. 2018, 99, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Kong, X.; Liu, T.; Zhou, L.; Wu, J.; Fu, J.; Wang, Y.; Zhu, M.; Yao, S.; Ding, Y.; et al. Metastasis-associated protein 1, modulated by miR-30c, promotes endometrial cancer progression through AKT/mTOR/4E-BP1 pathway. Gynecol. Oncol. 2019, 154, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Shahabi, A.; Naghili, B.; Ansarin, K.; Montazeri, M.; Dadashpour, M.; Zarghami, N. Let-7d and miR-185 Impede Epithelial-Mesenchymal Transition by Downregulating Rab25 in Breast Cancer. Asian Pac. J. Cancer Prev. 2021, 22, 305–313. [Google Scholar] [CrossRef]
- Tang, W.; Li, J.; Liu, H.; Zhou, F.; Liu, M. MiR-106a promotes tumor growth, migration, and invasion by targeting BCL2L11 in human endometrial adenocarcinoma. Am. J. Transl. Res. 2017, 9, 4984–4993. [Google Scholar]
- Geletina, N.S.; Kobelev, V.S.; Babayants, E.V.; Feng, L.; Pustylnyak, V.O.; Gulyaeva, L.F. PTEN negative correlates with miR-181a in tumour tissues of non-obese endometrial cancer patients. Gene 2018, 655, 20–24. [Google Scholar] [CrossRef]
- Yang, L.; Yang, Z.; Yao, R.; Li, Y.; Liu, Z.; Chen, X.; Zhang, G. miR-210 promotes progression of endometrial carcinoma by regulating the expression of NFIX. Int. J. Clin. Exp. Pathol. 2018, 11, 5213–5222. [Google Scholar]
- Du, J.; Zhang, F.; Zhang, L.; Jia, Y.; Chen, H. MicroRNA-103 regulates the progression in endometrial carcinoma through ZO-1. Int. J. Immunopathol. Pharm. 2019, 33, 2058738419872621. [Google Scholar] [CrossRef]
- Yu, D.; Zhou, H.; Xun, Q.; Xu, X.; Ling, J.; Hu, Y. microRNA-103 regulates the growth and invasion of endometrial cancer cells through the downregulation of tissue inhibitor of metalloproteinase 3. Oncol. Lett. 2012, 3, 1221–1226. [Google Scholar] [CrossRef]
- Sato, I.; Ishibashi, M.; Tokunaga, H.; Shigeta, S.; Sakurada, S.; Shimada, M.; Nagase, S.; Watanabe, Y.; Yaegashi, N. MicroRNA Let-7c Contributes to Paclitaxel Resistance via Aurora-B in Endometrial Serous Carcinoma. Tohoku J. Exp. Med. 2020, 251, 263–272. [Google Scholar] [CrossRef]
- Panda, H.; Pelakh, L.; Chuang, T.D.; Luo, X.; Bukulmez, O.; Chegini, N. Endometrial miR-200c is altered during transformation into cancerous states and targets the expression of ZEBs, VEGFA, FLT1, IKKbeta, KLF9, and FBLN5. Reprod. Sci. 2012, 19, 786–796. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.; Liang, X.; Zhao, W.; Ma, L.; Zhao, Y. The effects of microRNA-183 promots cell proliferation and invasion by targeting MMP-9 in endometrial cancer. Biomed. Pharm. 2017, 89, 812–818. [Google Scholar] [CrossRef]
- Myatt, S.S.; Wang, J.; Monteiro, L.J.; Christian, M.; Ho, K.K.; Fusi, L.; Dina, R.E.; Brosens, J.J.; Ghaem-Maghami, S.; Lam, E.W. Definition of microRNAs that repress expression of the tumor suppressor gene FOXO1 in endometrial cancer. Cancer.Res. 2010, 70, 367–377. [Google Scholar] [CrossRef]
- Cui, Z.; An, X.; Li, J.; Liu, Q.; Liu, W. LncRNA MIR22HG negatively regulates miR-141-3p to enhance DAPK1 expression and inhibits endometrial carcinoma cells proliferation. Biomed. Pharm. 2018, 104, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.J.; Gao, L.; Guo, Y.N.; Liang, Z.Q.; Li, D.M.; Tang, Y.L.; Liu, Y.H.; Gao, W.J.; Zeng, J.J.; Shi, L.; et al. Upregulation of microRNA miR-141-3p and its prospective targets in endometrial carcinoma: A comprehensive study. Bioengineered 2021, 12, 2941–2956. [Google Scholar] [CrossRef]
- Shi, W.; Wang, X.; Ruan, L.; Fu, J.; Liu, F.; Qu, J. MiR-200a promotes epithelial-mesenchymal transition of endometrial cancer cells by negatively regulating FOXA2 expression. Pharmazie 2017, 72, 694–699. [Google Scholar] [CrossRef]
- Yan, J.; Jiang, J.Y.; Meng, X.N.; Xiu, Y.L.; Zong, Z.H. MiR-23b targets cyclin G1 and suppresses ovarian cancer tumorigenesis and progression. J. Exp. Clin. Cancer Res. 2016, 35, 31. [Google Scholar] [CrossRef] [PubMed]
- Un, F. G1 arrest induction represents a critical determinant for cisplatin cytotoxicity in G1 checkpoint-retaining human cancers. Anticancer Drugs 2007, 18, 411–417. [Google Scholar] [CrossRef]
- Li, J.; Feng, Q.; Kim, J.M.; Schneiderman, D.; Liston, P.; Li, M.; Vanderhyden, B.; Faught, W.; Fung, M.F.; Senterman, M.; et al. Human ovarian cancer and cisplatin resistance: Possible role of inhibitor of apoptosis proteins. Endocrinology 2001, 142, 370–380. [Google Scholar] [CrossRef]
- Mizutani, H.; Tada-Oikawa, S.; Hiraku, Y.; Kojima, M.; Kawanishi, S. Mechanism of apoptosis induced by doxorubicin through the generation of hydrogen peroxide. Life Sci. 2005, 76, 1439–1453. [Google Scholar] [CrossRef] [PubMed]
- Schiff, P.B.; Horwitz, S.B. Taxol stabilizes microtubules in mouse fibroblast cells. Proc. Natl. Acad. Sci. USA 1980, 77, 1561–1565. [Google Scholar] [CrossRef]
- Ramos, A.; Sadeghi, S.; Tabatabaeian, H. Battling Chemoresistance in Cancer: Root Causes and Strategies to Uproot Them. Int. J. Mol. Sci. 2021, 22, 9451. [Google Scholar] [CrossRef]
- Burks, R.T.; Kessis, T.D.; Cho, K.R.; Hedrick, L. Microsatellite instability in endometrial carcinoma. Oncogene 1994, 9, 1163–1166. [Google Scholar] [PubMed]
- Catasus, L.; Matias-Guiu, X.; Machin, P.; Munoz, J.; Prat, J. BAX somatic frameshift mutations in endometrioid adenocarcinomas of the endometrium: Evidence for a tumor progression role in endometrial carcinomas with microsatellite instability. Lab. Investig. 1998, 78, 1439–1444. [Google Scholar] [PubMed]
- Wahl, A.F.; Donaldson, K.L.; Fairchild, C.; Lee, F.Y.; Foster, S.A.; Demers, G.W.; Galloway, D.A. Loss of normal p53 function confers sensitization to Taxol by increasing G2/M arrest and apoptosis. Nat. Med. 1996, 2, 72–79. [Google Scholar] [CrossRef]
- Perego, P.; Righetti, S.C.; Supino, R.; Delia, D.; Caserini, C.; Carenini, N.; Bedogne, B.; Broome, E.; Krajewski, S.; Reed, J.C.; et al. Role of apoptosis and apoptosis-related proteins in the cisplatin-resistant phenotype of human tumor cell lines. Apoptosis 1997, 2, 540–548. [Google Scholar] [CrossRef]
- Yang, X.; Fraser, M.; Moll, U.M.; Basak, A.; Tsang, B.K. Akt-mediated cisplatin resistance in ovarian cancer: Modulation of p53 action on caspase-dependent mitochondrial death pathway. Cancer Res. 2006, 66, 3126–3136. [Google Scholar] [CrossRef] [PubMed]
- Rouette, A.; Parent, S.; Girouard, J.; Leblanc, V.; Asselin, E. Cisplatin increases B-cell-lymphoma-2 expression via activation of protein kinase C and Akt2 in endometrial cancer cells. Int. J. Cancer 2012, 130, 1755–1767. [Google Scholar] [CrossRef]
- Haggblad Sahlberg, S.; Mortensen, A.C.; Haglof, J.; Engskog, M.K.; Arvidsson, T.; Pettersson, C.; Glimelius, B.; Stenerlow, B.; Nestor, M. Different functions of AKT1 and AKT2 in molecular pathways, cell migration and metabolism in colon cancer cells. Int. J. Oncol. 2017, 50, 5–14. [Google Scholar] [CrossRef]
- Girouard, J.; Lafleur, M.J.; Parent, S.; Leblanc, V.; Asselin, E. Involvement of Akt isoforms in chemoresistance of endometrial carcinoma cells. Gynecol. Oncol. 2013, 128, 335–343. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Stokoe, D.; Taketani, Y.; McCormick, F. High frequency of coexistent mutations of PIK3CA and PTEN genes in endometrial carcinoma. Cancer Res. 2005, 65, 10669–10673. [Google Scholar] [CrossRef]
- Yan, X.; Fraser, M.; Qiu, Q.; Tsang, B.K. Over-expression of PTEN sensitizes human ovarian cancer cells to cisplatin-induced apoptosis in a p53-dependent manner. Gynecol. Oncol. 2006, 102, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Choi, E.J.; Jin, C.; Kim, D.H. Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA amplification contributes to cisplatin resistance in an ovarian cancer cell line. Gynecol. Oncol. 2005, 97, 26–34. [Google Scholar] [CrossRef]
- Bhattacharya, R.; Cabral, F. Molecular basis for class V beta-tubulin effects on microtubule assembly and paclitaxel resistance. J. Biol. Chem. 2009, 284, 13023–13032. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Luduena, R.F. Removal of beta III isotype enhances taxol induced microtubule assembly. Cell Struct. Funct. 1993, 18, 173–182. [Google Scholar] [CrossRef]
- Umezu, T.; Shibata, K.; Kajiyama, H.; Terauchi, M.; Ino, K.; Nawa, A.; Kikkawa, F. Taxol resistance among the different histological subtypes of ovarian cancer may be associated with the expression of class III beta-tubulin. Int. J. Gynecol. Pathol. 2008, 27, 207–212. [Google Scholar] [CrossRef]
- Lee, J.W.; Park, Y.A.; Choi, J.J.; Lee, Y.Y.; Kim, C.J.; Choi, C.; Kim, T.J.; Lee, N.W.; Kim, B.G.; Bae, D.S. The expression of the miRNA-200 family in endometrial endometrioid carcinoma. Gynecol. Oncol. 2011, 120, 56–62. [Google Scholar] [CrossRef]
- Wu, Y.; Xiao, Y.; Ding, X.; Zhuo, Y.; Ren, P.; Zhou, C.; Zhou, J. A miR-200b/200c/429-binding site polymorphism in the 3′ untranslated region of the AP-2alpha gene is associated with cisplatin resistance. PLoS ONE 2011, 6, e29043. [Google Scholar] [CrossRef]
- Schwartz, B.; Melnikova, V.O.; Tellez, C.; Mourad-Zeidan, A.; Blehm, K.; Zhao, Y.J.; McCarty, M.; Adam, L.; Bar-Eli, M. Loss of AP-2alpha results in deregulation of E-cadherin and MMP-9 and an increase in tumorigenicity of colon cancer cells in vivo. Oncogene 2007, 26, 4049–4058. [Google Scholar] [CrossRef]
- Choi, H.J.; Chung, T.W.; Kim, S.J.; Cho, S.Y.; Lee, Y.S.; Lee, Y.C.; Ko, J.H.; Kim, C.H. The AP-2alpha transcription factor is required for the ganglioside GM3-stimulated transcriptional regulation of a PTEN gene. Glycobiology 2008, 18, 395–407. [Google Scholar] [CrossRef]
- Wajapeyee, N.; Britto, R.; Ravishankar, H.M.; Somasundaram, K. Apoptosis induction by activator protein 2alpha involves transcriptional repression of Bcl-2. J. Biol. Chem. 2006, 281, 16207–16219. [Google Scholar] [CrossRef]
- Cochrane, D.R.; Spoelstra, N.S.; Howe, E.N.; Nordeen, S.K.; Richer, J.K. MicroRNA-200c mitigates invasiveness and restores sensitivity to microtubule-targeting chemotherapeutic agents. Mol. Cancer 2009, 8, 1055–1066. [Google Scholar] [CrossRef]
- Kong, J.; He, X.; Wang, Y.; Li, J. Effect of microRNA-29b on proliferation, migration, and invasion of endometrial cancer cells. J. Int. Med. Res. 2019, 47, 3803–3817. [Google Scholar] [CrossRef]
- Djordjevic, B.; Hennessy, B.T.; Li, J.; Barkoh, B.A.; Luthra, R.; Mills, G.B.; Broaddus, R.R. Clinical assessment of PTEN loss in endometrial carcinoma: Immunohistochemistry outperforms gene sequencing. Mod. Pathol. 2012, 25, 699–708. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, L.; Jiang, W.; Zhang, R.; Zhang, B.; Silayiding, A.; Duan, X. MicroRNA-135a promotes proliferation, migration, invasion and induces chemoresistance of endometrial cancer cells. Eur. J. Obstet. Gynecol. Reprod. Biol. X 2020, 5, 100103. [Google Scholar] [CrossRef]
- Yi, H.; Han, Y.; Li, S. Oncogenic circular RNA circ_0007534 contributes to paclitaxel resistance in endometrial cancer by sponging miR-625 and promoting ZEB2 expression. Front. Oncol. 2022, 12, 985470. [Google Scholar] [CrossRef]
- Fardi, M.; Alivand, M.; Baradaran, B.; Farshdousti Hagh, M.; Solali, S. The crucial role of ZEB2: From development to epithelial-to-mesenchymal transition and cancer complexity. J. Cell. Physiol. 2019, 234, 14783–14799. [Google Scholar] [CrossRef]
- Ran, X.; Yang, J.; Liu, C.; Zhou, P.; Xiao, L.; Zhang, K. MiR-218 inhibits HMGB1-mediated autophagy in endometrial carcinoma cells during chemotherapy. Int. J. Clin. Exp. Pathol. 2015, 8, 6617–6626. [Google Scholar] [PubMed]
- Yanokura, M.; Banno, K.; Aoki, D. MicroRNA-34b expression enhances chemosensitivity of endometrial cancer cells to paclitaxel. Int. J. Oncol. 2020, 57, 1145–1156. [Google Scholar] [CrossRef]
- Schmidt, L.; Junker, K.; Nakaigawa, N.; Kinjerski, T.; Weirich, G.; Miller, M.; Lubensky, I.; Neumann, H.P.; Brauch, H.; Decker, J.; et al. Novel mutations of the MET proto-oncogene in papillary renal carcinomas. Oncogene 1999, 18, 2343–2350. [Google Scholar] [CrossRef] [PubMed]
| Characteristics | POLE-Mutant | MSI | p53wt/NSMP | p53 Abnormal (CN High) |
|---|---|---|---|---|
| Mutational frequency | >100 mutations/Mb | 100–10 mutations/Mb | <10 mutations/Mb | <10 mutations/Mb |
| Somatic CN alterations | Very low | Low | Low | High |
| Top five recurrent gene mutations (%) | POLE (100%) DMD (100%) CSMD1 (100%) FAT4 (100%) PTEN (94%) | PTEN (88%) PIK3CA (54%) PIK3R1 (42%) RPL22 (37%) ARID1A (37%) | PTEN (77%) PIK3CA (53%) CTNNB1 (52%) ARID1A (42%) PIK3R1 (33%) | TP53 (92%) PIK3CA (47%) FBXW7 (22%) PPP2R1A (22%) PTEN (10%) |
| Associated histological feature | Endometrioid Grade 3 Broad front invasion TILs Giant tumoral cells | Endometrioid Grade 3 LVSI substantial MELF-type invasion TILs | Endometrioid Grade 1–2 Squamous differentiation ER/PR expression | Serous Grade 3 LVSI High cytonuclear atypia Slit-like spaces |
| Associated clinical features | Lower BMI Early Stage (IA/IB) Early onset | Higher BMI Lynch Syndrome | Higher BMI | Lower BMI Advanced stage Late onset |
| Prognosis in early stage (I–II) | Excellent | Intermediate | Excellent/intermediate/poor | Poor |
| Diagnostic test | NGS (exons 9, 13, 14 or 9–14) Tumor mutation burden | MMR-IHC (MLH1, MSH2, MSH6, PMS2) MSI assay Tumor mutation burden | p53-IHC NGS |
| Risk Group | Molecular Classification Unknown | Molecular Classification Known |
|---|---|---|
| Low |
|
|
| Intermediate |
|
|
| High-intermediate |
|
|
| High |
|
|
| Advanced metastatic |
|
|
| miR | Function in EC | Sample | Expression | Target Genes | Clinical Value | Reference |
|---|---|---|---|---|---|---|
| hsa-miR-143-3p | Tumor-suppressive factor by regulating tumorigenesis and progression. | Serum | Up | MAPK1 | Diagnosis | [25] |
| hsa-miR-143-3p | Might inhibit cell proliferation, metastasis, and promote the apoptosis of EC cells. | ET | Down | MAPK1 | Prognosis | [88] |
| hsa-miR-423 | Inhibit cisplatin-induced apoptosis from decreasing the sensitivity of EC. | EC cell lines: HEC-1B and Ishikawa cells | Up | Bcl-2, Caspase 3/7 | Treatment response | [89] |
| hsa-miR-142-3p | Mediation of cell apoptosis by the miR-142-3p-FAM98A signaling pathway (anti-apoptotic and pro-proliferative effects). | Plasma | Up | FAM98A | Diagnosis | [22,23] |
| hsa-miR-146a-5p | Attenuates the effect of NIFK-AS1 on M2 polarization inhibition of macrophages and estrogen-induced EC cell proliferation, migration, and invasion. | Plasma | Up | NIFK-AS1 | Diagnosis | [22,24] |
| hsa-miR-151a-5p * | Induce proliferation, migration, and partial epithelial metastasis. | Plasma | Up | E-cadherin Fibronectin SNAI2 | Diagnosis | [22,90] |
| hsa-miR-195-5p | Suppressing cell migration, proliferation, and promote apoptosis. | Serum | Up | PI3K/AKT and MAPK/ ERK pathways FGFR1, FGF2 | Diagnosis | [25,26,27] |
| hsa-miR-20b-5p | VEGFA transcription. | Serum | Up | HIF1A PTEN STAT3 | Diagnosis | [28,29,30,31,32,33,34,35,36,37] |
| hsa-miR-204 | Mediates the migration and invasion of EC by regulating FOXC1. | - Serum - EC cell lines: HEC1A, HEC1B, AN3CA, KLE, RL95 - ET | Up | FOXC1 | Diagnosis | [38,39] |
| hsa-miR-484 * | Tumor suppressor. | MCF7 and T-47D cells | Down | KLF4 | Treatment response | [91] |
| hsa-miR-499 | Suppressed tumor growth and angiogenesis. | ET | Up/Down | VAV3 | Diagnosis | [20,92] |
| hsa-miR-135b | Promotes proliferation of EC cells. | ET | Up | FOXO1 | Diagnosis | [20,93] |
| hsa-miR-205 | Tumor suppressor through the inhibition of EMT. | ET | Up | PTEN | Prognosis | [94,95] |
| hsa-miR-10b | Inhibits apoptosis and promotes proliferation, migration, and invasion of EC cells. | ET | Down | HOXB3 | Diagnosis | [96] |
| hsa-miR-195 | Inhibits migration, invasion and EMT. | - ET - AN3-CA and Hec1A cells | Down | SOX4 GPER or GPR30 | Diagnosis | [97,98] |
| hsa-miR-30a-5p * | Inhibits cell proliferation and migration. | ET | Down | UBE3C | Diagnosis | [99] |
| hsa-miR-30a-3p | Modulate autophagy. | ET | Down | BECN1 | Diagnosis | [20,100] |
| hsa-miR-let 7a | Inhibits the growth of EC cells. | - ET - HeLa cells | Down | AURKB | Diagnosis | [101] |
| hsa-miR-221 | Act similar to a critical site of the regulatory pathway ERα/HIF1-α/SNAI2. | ET | Up/Down | LMOD1 ERα MDM2 | Diagnosis | [41,102] |
| hsa-miR-193 * | Enhance cell invasion-mediated EMT and improve cell proliferation through the ING5/PI3K/AKT signal pathway. | ET | Up/Down | RAMP1 ING5 | Diagnosis | [41,103] |
| hsa-miR-152 | Inhibits proliferation of EC cells via inducing G2/M phase arrest by suppressing CDC25B expression. | - ET - KLE and HEC-1B cells | Down | ENPP2 SNCAIP CDC25B | Diagnosis | [41,104] |
| hsa-miR-30c | Modulates MTA1, which may promote EC progression through the AKT/mTOR/4E-BP1 pathway. | - ET - EC cell lines: Ishikawa, HEC-1B, and RL-952 | Down | GPRASP2 MTA1 | Prognosis | [41,105] |
| hsa-miR-185 * | Inhibited EMT by targeting Rab25 expression. | ET | Up | KLF2 Rab25 | Diagnosis | [41,106] |
| hsa-miR-106a | Acts as an oncogenic miR in EC by inhibiting tumor suppressor BCL2L11 expression. | ET | Up | TGFB1I1 BCL2L11 | Diagnosis | [41,107] |
| hsa-miR-181a | Acts as an oncogenic miR that negatively regulate tumor suppressor PTEN. | ET | Up | PTEN DPP6 | Diagnosis | [41,108] |
| hsa-miR-210 | Promoted the progression of EC by negative regulation NFIX expression. | ET | Up | ENPP2 C2orf32 NFIX | Diagnosis/Prognosis | [41,109] |
| hsa-miR-103 | - Regulates the progression in EC through ZO-1. - Regulates the growth and invasion of EC cells through the downregulation of TIMP-3 | - ET - Cell lines: HEC-1B and Ishikawa | Up | ZO-1 TIMP-3 | Diagnosis/Prognosis | [110,111] |
| hsa-miR-let 7c | Contributes to paclitaxel resistance via Aurora-B in SEC. | SEC cell lines: USPC1, USPC1-PTXR, USPC1-PTXR2 | Down | Aurora-B | Treatment response | [112] |
| hsa-miR-200c | - It is speculated that the increase in cell proliferation is mediated through repression of KLF9. - Regulated the translocation of β-catenin from the cytoplasm to the nucleus via inhibition of BRD7, resulting in increased expression of its transcriptional target genes, cyclin D1 and c-myc. | - ET - Cell lines: HEC-1A, Ishikawa | Up | PTEN KLF9 BRD7 | Diagnosis | [42,54,113] |
| hsa-miR-183 | Promotes cell proliferation and invasion by targeting MMP-9. | - ET - EC cell lines: KLE, HEC-1-A and HHUA | Up | MMP-9 | Diagnosis | [114] |
| hsa-miR-186 | May reduce the expression of tumor suppressor FOXO1 and thereby deregulates cell cycle control. | - Serum - EC cell lines: HEC-1B and Ishikawa | Up | FOXO1 | Diagnosis | [43,115] |
| hsa-miR-141-3p | Promoter EC cells proliferation, indicating that could act as an oncogenic miR in EC progression. | - ET - EC cell lines: HEC-1 A and KLE cells | Up | DAPK1 | Prognosis | [116,117] |
| hsa-miR-200a | Promotes EMT of EC cells by negatively regulating FOXA2 expression | ET | Up | FOXA2 | Diagnosis | [118] |
| hsa-miR-15a-5p | Inhibits the growth of EC cells via attenuating WNT3A expression in the Wnt/β-catenin signaling pathway. | - ET - Cell lines: HEC-251, AN3CA, RL95-2, HEC-1-A, ISK, Ishikawa, and JEC | Down | WNT3A | Diagnosis | [47] |
| hsa-miR-449a | Might module the EC progression via negative regulation of transcription from RNA polymerase II promoter | ET | Down | LEF1 | Prognosis | [48] |
| hsa-miR-145-5p | Might act as a tumor suppressor and regulate cell cycle associated processes to inhibit the development of EC | ET | Down | SOX11 | Prognosis | [48] |
| hsa-miR-505 | Functions as a tumor suppressor by targeting TGF-α | - ET - EC cell lines: HEC-1B and Ishikawa | Down | TGF-α | Diagnosis | [49] |
| hsa-miR-589-5p | Inhibits cell proliferation, migration, and invasion by targeting TRIP6. | - ET - EC cell lines: HEC-1B and AN3CA | Down | TRIP6 | Diagnosis | [50] |
| hsa-miR-20a-5p | - Inhibits EMT and invasion of EC cells by targeting STAT3 - Inhibits EC progression by targeting Jak1. | - EC cell lines: ECC-1, KLE, HHUA, RL95-2 and Ishikawa - HEK293 cells - Human uterine epithelial cell line HES - ET | Down | STAT3 Jak1 | Diagnosis | [51,52] |
| hsa-miR-23b | It may act as a tumor suppressor miR: suppressed the proliferation of Ishikawa cells. | - ET - Ishikawa EC cells | Down | CCNG1 | Prognosis | [46,119] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oropeza-de Lara, S.A.; Garza-Veloz, I.; Berthaud-González, B.; Martinez-Fierro, M.L. Circulating and Endometrial Tissue microRNA Markers Associated with Endometrial Cancer Diagnosis, Prognosis, and Response to Treatment. Cancers 2023, 15, 2686. https://doi.org/10.3390/cancers15102686
Oropeza-de Lara SA, Garza-Veloz I, Berthaud-González B, Martinez-Fierro ML. Circulating and Endometrial Tissue microRNA Markers Associated with Endometrial Cancer Diagnosis, Prognosis, and Response to Treatment. Cancers. 2023; 15(10):2686. https://doi.org/10.3390/cancers15102686
Chicago/Turabian StyleOropeza-de Lara, Sergio Antonio, Idalia Garza-Veloz, Bertha Berthaud-González, and Margarita L. Martinez-Fierro. 2023. "Circulating and Endometrial Tissue microRNA Markers Associated with Endometrial Cancer Diagnosis, Prognosis, and Response to Treatment" Cancers 15, no. 10: 2686. https://doi.org/10.3390/cancers15102686
APA StyleOropeza-de Lara, S. A., Garza-Veloz, I., Berthaud-González, B., & Martinez-Fierro, M. L. (2023). Circulating and Endometrial Tissue microRNA Markers Associated with Endometrial Cancer Diagnosis, Prognosis, and Response to Treatment. Cancers, 15(10), 2686. https://doi.org/10.3390/cancers15102686







