Galectin-3 and Epithelial MUC1 Mucin—Interactions Supporting Cancer Development
Abstract
Simple Summary
Abstract
1. Galectins
Galectin-3
2. Mucins
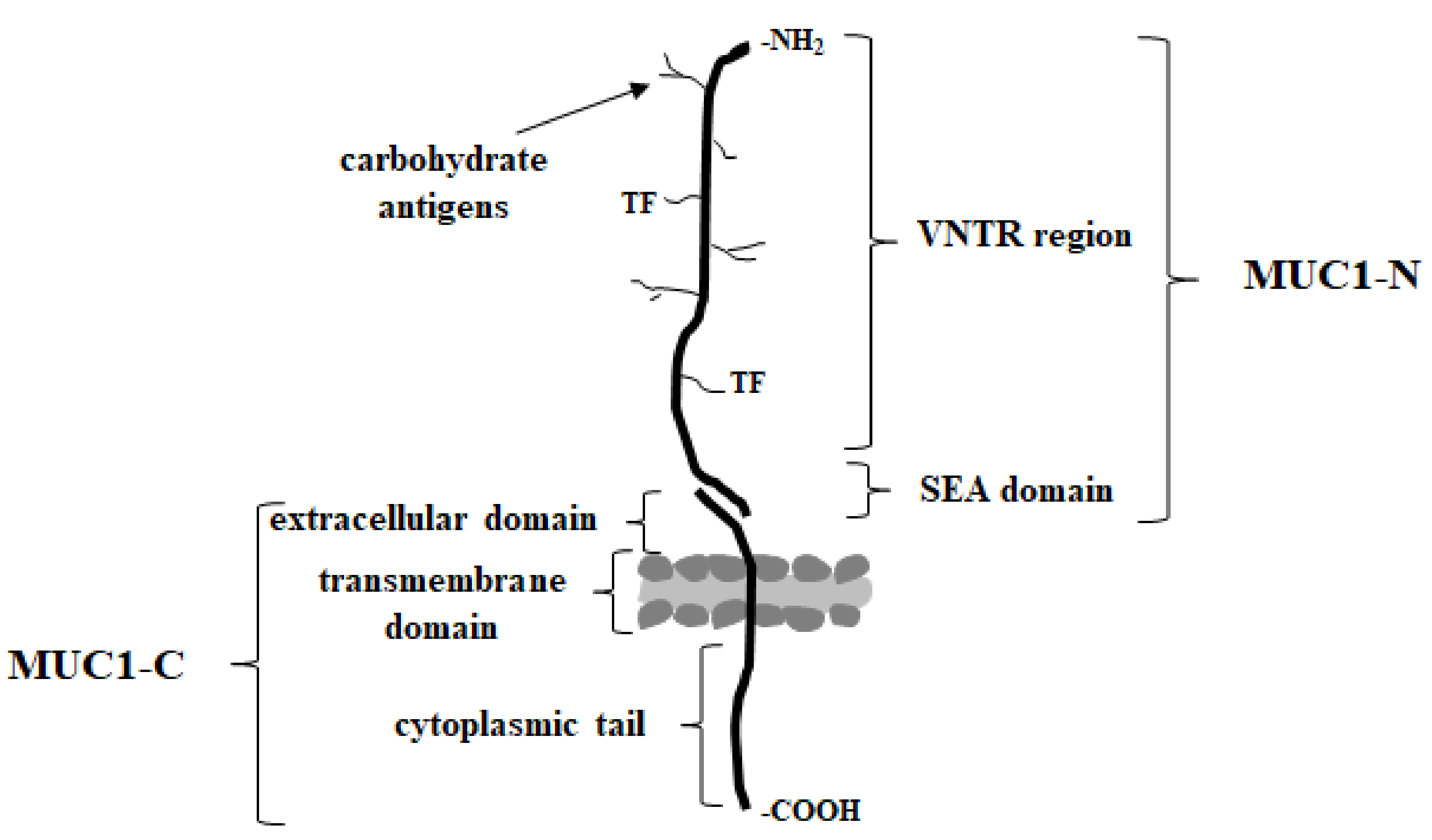
MUC1
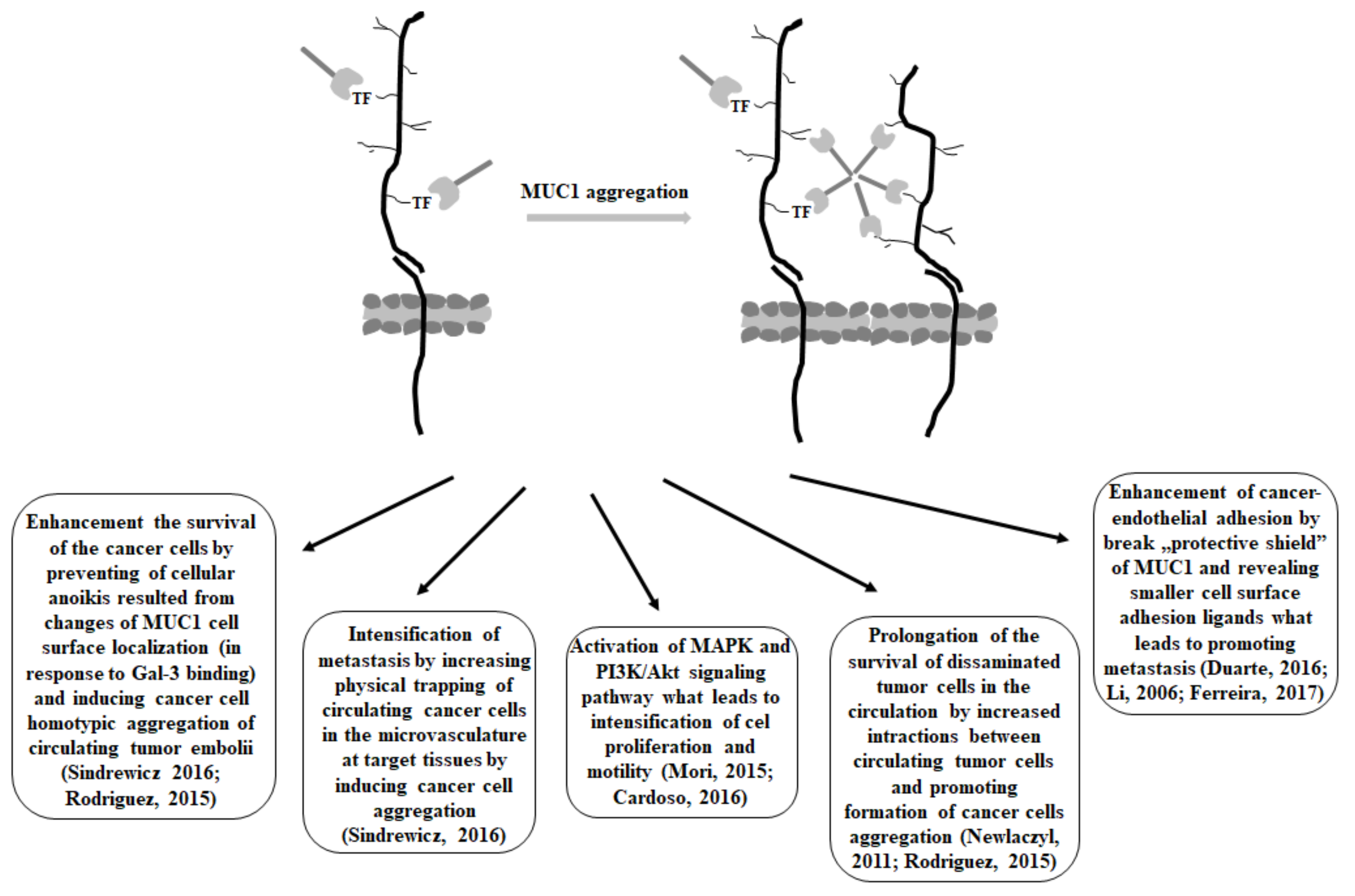
3. Gal-3 and MUC1 Relationships
4. MUC1- Gal-3 Inhibitors as a Therapeutic Strategy
5. Conclusions
Funding
Conflicts of Interest
References
- Brittoli, A.; Fallarini, S.; Zhang, H.; Pieters, R.J.; Lombardi, G. “In vitro” studies on galectin-3 in human natural killer cells. Immunol. Lett. 2018, 194, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Argüeso, P.; Panjwani, N. Focus on molecules: Galectin-3. Exp. Eye Res. 2011, 92, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Caballero, G.G.; Kaltner, H.; Kutzner, T.J.; Ludwig, A.K.; Manning, J.C.; Schmidt, S.; Sinowatz, F.; Gabius, H.J. How galectins have become multifunctional proteins. Histol. Histopathol. 2020, 35, 509–539. [Google Scholar]
- Dumic, J.; Dabelic, S.; Flogel, M. Galectin-3: An open-ended story. Biochim. Biophys. Acta 2006, 1760, 616–635. [Google Scholar] [CrossRef] [PubMed]
- FitzGerald, F.G.; Benavente, M.C.R.; Garcia, C.; Rivero, Y.; Singh, Y.N.; Wang, H.; Fields, G.B.; Cudic, M. TF-containing MUC1 glycopeptides fail to entice Galectin-1 recognition of tumor-associated Thomsen-Freidenreich (TF) antigen (CD176) in solution. Glycoconj. J. 2020, 37, 657–666. [Google Scholar] [CrossRef]
- Sciacchitano, S.; Lavra, L.; Morgante, A.; Ulivieri, A.; Magi, F.; Francesco, G.P.; Bellotti, C.; Salehi, L.B.; Ricci, A. Galectin-3: One molecule for an alphabet of diseases, from A to Z. Int. J. Mol. Sci. 2018, 19, 379. [Google Scholar] [CrossRef]
- Dong, R.; Zhang, M.; Zheng, S.; Soh, A.; Zheng, Y.; Yuan, H. Galectin-3 as a novel biomarker for disease diagnosis and a target for therapy. Int. J. Mol. Med. 2018, 41, 599–614. [Google Scholar] [CrossRef]
- Hong, M.H.; Weng, I.C.; Li, F.Y.; Lin, W.H.; Liu, F.T. Intracellular galectins sense cytosolically exposed glycans as danger and mediate cellular responses. J. Biomed. Sci. 2021, 28, 16. [Google Scholar] [CrossRef]
- Chou, F.C.; Chen, H.Y.; Kuo, C.C.; Sytwu, H.K. Role of galectins in tumors and in clinical immunotherapy. Int. J. Mol. Sci. 2018, 19, 430. [Google Scholar] [CrossRef]
- Johannes, L.; Jacob, R.; Leffler, H. Galectins at a glance. J. Cell Sci. 2018, 131, jsc208884. [Google Scholar] [CrossRef]
- Hughes, R.C. Secretion of the galectin family of mammalian carbohydrate-binding proteins. Biochim. Biophys. Acta 1999, 1473, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Huergo, S.P.; Blidner, A.G.; Rabinovich, G.A. Galectins: Emerging regulatory checkpoints linking tumor immunity and angiogenesis. Curr. Opin. Immunol. 2017, 45, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Dings, R.P.M.; Miller, M.C.; Griffin, R.J.; Mayo, K.H. Galectins as molecular targets for therapeutic intervention. Int. J. Mol. Sci. 2018, 19, 905. [Google Scholar] [CrossRef] [PubMed]
- Thijssen, V.L.; Heusschen, R.; Caers, J.; Griffioen, A.W. Galectin expression in cancer diagnosis and prognosis: A systematic review. Biochim. Biophys. Acta 2015, 1855, 235–247. [Google Scholar] [CrossRef] [PubMed]
- Vladoiu, M.C.; Labrie, M.; St-Pierre, Y. Intracellular galectins in cancer cells: Potential new targets for therapy. Int. J. Oncol. 2014, 44, 1001–1014. [Google Scholar] [CrossRef]
- Li, Y.S.; Li, X.T.; Yu, L.G.; Wang, L.; Shi, Z.Y.; Guo, X.L. Roles of galectin-3 in metabolic disorders and tumor cell metabolism. Int. J. Biol. Macromol. 2020, 142, 463–473. [Google Scholar] [CrossRef]
- Xue, H.; Liu, L.; Zhao, Z.; Zhang, Z.; Guan, Y.; Cheng, H.; Zhou, Y.; Tai, G. The N-terminal tail coordinates with carbohydrate recognition domain to mediate galectin-3 induced apoptosis in T cells. Oncotarget 2017, 8, 49824–49838. [Google Scholar] [CrossRef]
- Ruvolo, P.P. Galectin 3 as a guardian of the tumor microenvironment. Biochim. Biophys. Acta 2016, 1863, 427–437. [Google Scholar] [CrossRef]
- Sindrewicz, P.; Lian, L.Y.; Yu, L.G. Interaction of the oncofetal Thomsen-Friedenreich antigen with galectins in cancer progression and metastasis. Front. Oncol. 2016, 6, 79. [Google Scholar] [CrossRef]
- Ahmad, N.; Gabius, H.J.; Sabesan, S.; Oscarson, S.; Brewer, C.F. Thermodynamic binding studies of bivalent oligosaccharides to galectin-1, galectin-3, and the carbohydrate recognition domain of galectin-3. Glycobiology 2004, 14, 817–825. [Google Scholar] [CrossRef]
- Mori, Y.; Yashiro, M.; Sawada, T.; Hirakawa, K.; Murata, T.; Nakada, H. Binding of galectin-3, a β-galactoside-binding lectin, to MUC1 protein enhances phosphorylation of extracellular signal-regulated Kinase ½ (ERK1/2) and Akt, promoting tumor cell malignancy. J. Biol. Chem. 2015, 290, 26125–26140. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.T.; Rabinovich, G.A. Galectins as modulators of tumour progression. Nat. Rev. Cancer 2005, 5, 29–41. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.Y.; Rabinovich, G.A.; Liu, F.T. Galectins: Structure, function and therapeutic potential. Expert Rev. Mol. Med. 2008, 10, e17. [Google Scholar] [CrossRef] [PubMed]
- Brewer, C.F.; Miceli, M.C.; Baum, L.G. Clusters, bundles, arrays and lattices: Novel mechanisms for lectin-saccharide-mediated cellular interactions. Curr. Opin. Struct. Biol. 2002, 12, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Ochieng, J.; Furtak, V.; Lukyanov, P. Extracellular functions of galectin-3. Glycoconj. J. 2002, 19, 527–535. [Google Scholar] [CrossRef] [PubMed]
- Fermino, M.L.; Polli, C.D.; Toledo, K.A.; Liu, F.T.; Hsu, D.K.; Roque-Barreira, M.C.; Pereira-da-Silva, G.; Bernardes, E.S.; Halbwachs-Mecarelli, L. LPS-induced galectin-3 oligomerization results in enhancement of neutrophil activation. PLoS ONE 2011, 6, e26004. [Google Scholar] [CrossRef] [PubMed]
- Newlaczyl, A.U.; Yu, L.G. Galectin-3—A jack-of-all-trades in cancer. Cancer Lett. 2011, 313, 123–128. [Google Scholar] [CrossRef]
- Michel, A.K.; Nangia-Makker, P.; Raz, A.; Cloninger, M.J. Lactose-functionalized dendrimers arbitrate the interaction galectin-3/MUC1 mediated cancer cellular aggregation. ChemBioChem 2014, 15, 2106–2112. [Google Scholar] [CrossRef] [PubMed]
- Chabre, Y.M.; Roy, R. Design and creativity in synthesis of multivalent neoglycoconjugates. Adv. Carbohydr. Chem. Biochem. 2010, 63, 165–393. [Google Scholar] [PubMed]
- Cardoso, A.C.F.; Andrade, L.N.S.; Bustos, S.O.; Chammas, R. Galectin-3 determines tumor cell adaptive strategies in stressed tumor microenvironments. Front. Oncol. 2016, 6, 127. [Google Scholar] [CrossRef]
- Elad-Sfadia, G.; Haklai, R.; Balan, E.; Kloog, Y. Galectin-3 augments K-Ras activation and triggers a Ras signal that attenuates ERK but not phosphoinositide 3-kinase activity. J. Biol. Chem. 2004, 279, 34922–34930. [Google Scholar] [CrossRef] [PubMed]
- Shimura, T.; Takenaka, Y.; Fukumori, T.; Tsutsumi, S.; Okada, K.; Hogan, V.; Kikuchi, A.; Kuwano, H.; Raz, A. Implication of galectin-3 in Wnt signaling. Cancer Res. 2005, 65, 3535–3537. [Google Scholar] [CrossRef] [PubMed]
- Balan, V.; Nangia-Makker, P.; Kho, D.H.; Wang, Y.; Raz, A. Tyrosine-phosphorylated galectin-3 protein is resistant to prostate-specific antigen (PSA) cleavage. J. Biol. Chem. 2012, 287, 5192–5198. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, Q.; Wang, J.; Liu, X.; Yang, Y.; Zhao, H.; Wang, Y.; Jin, Y.; Zeng, J.; Li, J.; et al. c-Abl and Arg tyrosine kinases regulate lysosomal degradation of the oncoprotein galectin-3. Cell Death Differ. 2010, 17, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Haudek, K.C.; Spronk, K.J.; Voss, P.G.; Patterson, R.J.; Wang, J.L.; Arnoys, E.J. Dynamics of galectin-3 in the nucleus and cytoplasm. Biochim. Biophys. Acta 2010, 1800, 181–189. [Google Scholar] [CrossRef]
- Takenaka, Y.; Fukumori, T.; Raz, A. Galectin-3 and metastasis. Glycoconj. J. 2002, 19, 543–549. [Google Scholar] [CrossRef]
- Yu, L.G.; Andrews, N.; Zhao, Q.; McKean, D.; Williams, J.F.; Connor, L.J.; Gerasimenko, O.V.; Hilkens, J.; Hirabayashi, J.; Kasai, K.; et al. Galectin-3 interaction with Thomsen-Friedenreich disaccharide on cancer-associated MUC1 causes increased cancer cell endothelial adhesion. J. Biol. Chem. 2007, 282, 773–781. [Google Scholar] [CrossRef]
- Ashery, U.; Yizhar, O.; Rotblat, B.; Elad-Sfadia, G.; Barkan, B.; Haklai, R.; Kloog, Y. Spatiotemporal organization of Ras signaling: Rasosomes and the galectin switch. Cell. Mol. Neurobiol. 2006, 26, 471–495. [Google Scholar] [CrossRef]
- Ramasamy, S.; Duraisamy, S.; Barbashov, S.; Kawano, T.; Kharbanda, S.; Kufe, D. The MUC1 and galectin-3 oncoproteins function in a microRNA-dependent regulatory loop. Mol. Cell 2007, 27, 992–1004. [Google Scholar] [CrossRef]
- Zucker, S.; Cao, J.; Chen, W.T. Critical appraisal of the use of matrix metalloproteinase inhibitors in cancer treatment. Oncogene 2000, 19, 6642–6650. [Google Scholar] [CrossRef]
- Harazono, Y.; Kho, D.H.; Balan, V.; Nakajima, K.; Zhang, T.; Hogan, V.; Raz, A. Galectin-3 leads to attenuation of apoptosis through Bax heterodimerization in human thyroid carcinoma cells. Oncotarget 2014, 5, 9992–10001. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Finley, R.L., Jr.; Raz, A.; Kim, H.R. Galectin-3 translocates to the perinuclear membranes and inhibits cytochrome c release from the mitochondria. A role for synexin in galectin-3 translocation. J. Biol. Chem. 2002, 277, 15819–15827. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.M.; Pestell, R.G.; Raz, A.; Kim, H.R. Galectin-3 enhances cyclin D1 promoter activity through SP1 and a cAMP-responsive element in human breast epithelial cells. Oncogene 2002, 21, 8001–8010. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, B.; Subramanian, S.; Mallia, M.B.; Repaka, K.; Kaur, S.; Chandan, R.; Bhardwaj, P.; Dash, A.; Banerjee, R. Multifunctional core-shell glyconanoparticles for galectin-3-targeted, trigger-responsive combination chemotherapy. Biomacromolecules 2020, 21, 2645–2660. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Tang, J.W.; Owusu, L.; Sun, M.Z.; Wu, J.; Zhang, J. Galectin-3 in cancer. Clin. Chim. Acta 2014, 431, 185–191. [Google Scholar] [CrossRef]
- Ogawa, T.; Hirohashi, Y.; Murai, A.; Nishidate, T.; Okita, K.; Wang, L.; Ikehara, Y.; Satoyoshi, T.; Usui, A.; Kubo, T.; et al. ST6GALNAC1 plays important roles in engancing cancer stem phenotypes of colorectal cancer via the Akt pathway. Oncotarget 2017, 8, 112550–112564. [Google Scholar] [CrossRef]
- de Oliveira, J.T.; de Matos, A.J.; Santos, A.L.; Pinto, R.; Gomes, J.; Hespanhol, V.; Chammas, R.; Manninen, A.; Bernardes, E.S.; Reis, C.A.; et al. Sialylation regulates galectin-3/ligand interplay during mammary tumour progression—A case of targeted uncloaking. Int. J. Dev. Biol. 2011, 55, 823–834. [Google Scholar] [CrossRef]
- Huang, Z.; Ai, Z.; Li, N.; Xi, H.; Gao, X.; Wang, F.; Tan, X.; Liu, H. Over expression of galectin-3 associates with short-term poor prognosis in stage II colon cancer. Cancer Biomark. 2016, 17, 445–455. [Google Scholar] [CrossRef]
- Chen, C.; Duckworth, C.A.; Zhao, Q.; Pritchard, D.M.; Rhodes, J.M.; Yu, L.G. Increased circulation of galectin-3 in cancer induces secretion of metastasis-promoting cytokines from blood vascular endothelium. Clin. Cancer Res. 2013, 19, 1693–1704. [Google Scholar] [CrossRef]
- Nakayama, R.; Kuroda, J.; Taniyama, N.; Yamamoto-Sugitani, M.; Wada, S.; Kiyota, M.; Mizutani, S.; Chinen, Y.; Matsumoto, Y.; Nagoshi, H. Suppression of SERPINA1-albumin complex formation by galectin-3 overexpression leads to paracrine growth promotion of chronic myelogenous leukemia cells. Leuk. Res. 2014, 38, 103–108. [Google Scholar] [CrossRef]
- DeRoo, E.P.; Wroblewski, S.K.; Shea, E.M.; Al-Khalil, R.K.; Hawley, A.E.; Henke, P.K.; Myers, D.D., Jr.; Wakefield, T.W.; Diaz, J.A. The role of galectin-3 and galectin-3 binding protein in venous thrombosis. Blood 2015, 125, 1813–1821. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Ni, W.K.; Chen, X.D.; Xiao, M.B.; Chen, B.Y.; He, S.; Lu, C.H.; Li, X.Y.; Jiang, F.; Ni, R.Z. The expressions and clinical significances of tissue and serum galectin-3 in pancreatic carcinoma. J. Cancer Res. Clin. Oncol. 2012, 138, 1035–1043. [Google Scholar] [CrossRef] [PubMed]
- Iacovazzi, P.A.; Notarnicola, M.; Caruso, M.G.; Guerra, V.; Frisullo, S.; Altomare, D.F.; Correale, M. Serum levels of galectin-3 and its ligand 90k/mac-2bp in colorectal cancer patients. Immunopharmacol. Immunotoxicol. 2010, 32, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Tanida, S.; Mori, Y.; Ishida, A.; Akita, K.; Nakada, H. Galectin-3 binds to MUC1-N-terminal domain and triggers recruitment of β-catenin in MUC1-expressing mouse 3T3 cells. Biochim. Biophys. Acta 2014, 1840, 1790–1797. [Google Scholar] [CrossRef]
- Iurisci, I.; Tinari, N.; Natoli, C.; Angelucci, D.; Cianchetti, E.; Iacobelli, S. Concentrations of galectin-3 in the sera of normal controls and cancer patients. Clin. Cancer Res. 2000, 6, 1389–1393. [Google Scholar]
- Beckwith, D.M.; Cudic, M. Tumor-associated O-glycans of MUC1: Carriers of the glycol-code and targets for cancer vaccine design. Semin. Immunol. 2020, 47, 101389. [Google Scholar] [CrossRef]
- Fukumori, T.; Takenaka, Y.; Yoshii, T.; Kim, H.R.C.; Hogan, V.; Inohara, H.; Kagawa, S.; Raz, A. CD29 and CD7 mediate galectin-3-induced type II T-cell apoptosis. Cancer Res. 2003, 63, 8302–8311. [Google Scholar]
- Agrawal, B.; Gupta, N.; Konowalchuk, J.D. MUC1 mucin: A putative regulatory (checkpoint) molecule of T cells. Front. Immunol. 2018, 9, 2391. [Google Scholar] [CrossRef]
- Chen, S.S.; Sun, L.W.; Brickner, H.; Sun, P.Q. Downregulating galectin-3 inhibits proinflammatory cytokine production by human monocyte-derived dendritic cells via RNA interference. Cell. Immunol. 2015, 294, 44–53. [Google Scholar] [CrossRef]
- Cohen, M. Notable aspects of glycan-protein interactions. Biomolecules 2015, 5, 2056–2072. [Google Scholar] [CrossRef]
- Bhatia, R.; Gautam, S.K.; Cannon, A.; Thompson, C.; Hall, B.R.; Aithal, A.; Banerjee, K.; Jain, M.; Solheim, J.C.; Kumar, S.; et al. Cancer-associated mucins: Role in immune modulation and metastasis. Cancer Metastasis Rev. 2019, 38, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Kaur, S.; Kumar, S.; Momi, N.; Sasson, A.R.; Batra, S.K. Mucins in pancreatic cancer and its microenvironment. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Chugh, S.; Gnanapragassam, V.S.; Jain, M.; Rachagani, S.; Ponnusamy, M.P.; Batra, S.K. Pathobiological implications of mucin glycans in cancer: Sweet poison and novel targets. Biochim. Biophys. Acta 2015, 1856, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Brockhausen, I.; Melamed, J. Mucins as anti-cancer targets: Perspectives of the glycobiologist. Glycoconj. J. 2021, 38, 459–474. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, L.; Lei, C.; Li, W.; Han, J.; Zhang, J.; Zhang, Y. A sweet warning: Mucin-type O-glycans in cancer. Cells 2022, 11, 3666. [Google Scholar] [CrossRef]
- Schjoldager, K.T.; Narimatsu, Y.; Joshi, H.J.; Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 729–749. [Google Scholar] [CrossRef]
- Bennett, E.P.; Mandel, U.; Clausen, H.; Gerken, T.A.; Fritz, T.A.; Tabak, L.A. Control of mucin-type O-glycosylation: A classification of the polypeptide GalNAc-transferase gene family. Glycobiology 2012, 22, 736–756. [Google Scholar] [CrossRef]
- Hauselman, I.; Borsig, L. Altered tumor-cell glycosylation promotes metastasis. Front. Oncol. 2014, 4, 28. [Google Scholar] [CrossRef]
- Nath, S.; Mukherjee, P. MUC1: A multifaceted oncoprotein with a key role in cancer progression. Trends Mol. Med. 2014, 20, 332–342. [Google Scholar] [CrossRef]
- Parry, S.; Silverman, H.S.; McDermott, K.; Willis, A.; Hollingsworth, M.A.; Harris, A. Identification of MUC1 proteolytic cleavage sites in vivo. Biochem. Biophys. Res. Commun. 2001, 283, 715–720. [Google Scholar] [CrossRef]
- Chen, W.; Zhang, Z.; Zhang, S.; Zhu, P.; Ko, J.K.S.; Yung, K.K.L. MUC1: Structure, function, and clinic application in epithelial cancers. Int. J. Mol. Sci. 2021, 22, 6567. [Google Scholar] [CrossRef] [PubMed]
- Qing, L.; Li, Q.; Dong, Z. MUC1: An emerging target in cancer treatment and diagnosis. Bull. Cancer 2022, 109, 1202–1216. [Google Scholar] [CrossRef] [PubMed]
- Taylor-Papadimitriou, J.; Burchell, J.M.; Graham, R.; Beatson, R. Latest developments in MUC1 immunotherapy. Biochem. Soc. Trans. 2018, 46, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Kufe, D.W. MUC1-C in chronic inflammation and carcinogenesis; emergence as a target for cancer treatment. Carcinogenesis 2020, 41, 1173–1183. [Google Scholar] [CrossRef]
- Van Putten, J.P.M.; Strijbis, K. Transmembrane mucins: Signaling receptors at the intersection of inflammation and cancer. J. Innate Immun. 2017, 9, 281–299. [Google Scholar] [CrossRef]
- Syrkina, M.S.; Maslakova, A.A.; Potashnikova, D.M.; Veiko, V.P.; Vassetzky, Y.S.; Rubtsov, M.A. Dual role of the extracellular domain of human mucin MUC1 in metastasis. J. Cell. Biochem. 2017, 118, 4002–4011. [Google Scholar] [CrossRef]
- Cascio, S.; Finn, O.J. Intra- and extra-cellular events related to altered glycosylation of MUC1 promote chronic inflammation, tumor progression, invasion, and metastasis. Biomolecules 2016, 6, 39. [Google Scholar] [CrossRef]
- Wang, S.; You, L.; Dai, M.; Zhao, Y. Mucins in pancreatic cancer: A well-established but promising family for diagnosis, prognosis and therapy. J. Cell. Mol. Med. 2020, 24, 10279–10289. [Google Scholar] [CrossRef]
- Carson, D.D. The cytoplasmic tail of MUC1: A very busy place. Sci. Signal. 2008, 1, pe35. [Google Scholar] [CrossRef]
- Ballester, B.; Milara, J.; Cortijo, J. The role of mucin 1 in respiratory diseases. Eur. Respir. Rev. 2021, 30, 200149. [Google Scholar] [CrossRef]
- Kufe, D.W. MUC1-C oncoprotein as a target in breast cancer: Activation of signaling pathways and therapeutic approaches. Oncogene 2013, 32, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Rajabi, H.; Hata, T.; Li, W.; Long, M.D.; Hu, Q.; Liu, S.; Raina, D.; Kui, L.; Yasumizu, Y.; Hong, D.; et al. MUC1-C represses the RASSF1A tumor suppressor in human carcinoma cells. Oncogene 2019, 38, 7266–7277. [Google Scholar] [CrossRef] [PubMed]
- Cascio, S.; Zhang, L.; Finn, O.J. Muc1 protein expression in tumor cells regulates transcription of proinflammatory cytokines by forming a complex with nuclear factor-κb p65 and binding to cytokine promoters importance of extracellular domain. J. Biol. Chem. 2011, 286, 42248–42256. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Raina, D.; Joshi, M.D.; Kawano, T.; Ren, J.; Kharbanda, S.; Kufe, D. MUC1-C oncoprotein functions as a direct activator of the nuclear factor-κB p65 transcriptor factor. Cancer Res. 2009, 69, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Raina, D.; Trivedi, V.; Ren, J.; Rajabi, H.; Kharbanda, S.; Kufe, D. MUC1 oncoprotein activates the IκB kinase β complex and constitutive NF-κB signaling. Nat. Cell Biol. 2007, 9, 1419–1427. [Google Scholar] [CrossRef] [PubMed]
- Apostolopoulos, V.; McKenzie, I.F.C. Cellular mucins: Targets for immunotherapy. Crit. Rev. Immunol. 2017, 37, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Pillai, K.; Pourgholami, M.H.; Chua, T.C.; Morris, D.L. MUC1 as a potential target in anticancer therapies. Am. J. Clin. Oncol. 2015, 38, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Xu, H.; Kufe, D. Human MUC1 oncoprotein regulates p53-responsive gene transcription in the genotoxic stress response. Cancer Cell 2005, 7, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Chia, J.; Goh, G.; Bard, F. Short O-GalNAc glycans: Regulation and role in tumor development and clinical perspectives. Biochim. Biophys. Acta 2016, 1860, 1623–1639. [Google Scholar] [CrossRef] [PubMed]
- Burchell, J.M.; Beatson, R.; Graham, R.; Taylor-Papadimitriou, J.; Tajadura-Ortega, V. O-linked mucin-type glycosylation in breast cancer. Biochem. Soc. Trans. 2018, 46, 779–788. [Google Scholar] [CrossRef] [PubMed]
- Ju, T.; Wang, Y.; Aryal, R.P.; Lehoux, S.D.; Ding, X.; Kudelka, M.R.; Cutler, C.; Zeng, J.; Wang, J.; Sun, X.; et al. Tn and sialyl-Tn antigens, aberrant O-glycomics as human disease markers. Proteom. Clin. Appl. 2013, 7, 618–631. [Google Scholar] [CrossRef] [PubMed]
- Wandall, H.H.; Blixt, O.; Tarp, M.A.; Pedersen, J.W.; Bennett, E.P.; Mandel, U.; Ragupathi, G.; Livingston, P.O.; Hollingsworth, M.A.; Taylor-Papadimitriou, J.; et al. Cancer biomarkers defined by autoantibody signatures to aberrant O-glycopeptide epitopes. Cancer Res. 2010, 70, 1306–1313. [Google Scholar] [CrossRef] [PubMed]
- Bai, R.; Luan, X.; Zhang, Y.; Robbe-Masselot, C.; Brockhausen, I.; Gao, Y. The expression and functional analysis of the sialyl-T-antigen in prostate cancer. Glycoconj. J. 2020, 37, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Kudelka, M.R.; Ju, T.; Heimburg-Molinaro, J.; Cummings, R.D. Simple sugars to complex disease—Mucin-type O-glycans in cancer. Adv. Cancer Res. 2015, 126, 53–135. [Google Scholar]
- Cazet, A.; Julien, S.; Bobowski, M.; Burchell, J.; Delannoy, P. Tumour-associated carbohydrate antigens in breast cancer. Breast Cancer Res. 2010, 12, 204. [Google Scholar] [CrossRef]
- Brockhausen, I.; Gao, Y. Structural Glycobiology: Applications in Cancer Research; Yurevics, E., Ed.; Chapter 8; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group: Abingdon, UK, 2012; pp. 177–213. [Google Scholar]
- Anandkumar, A.; Devaraj, H. Tumour immunomodulation: Mucins in resistance to initiation and maturation of immune response against tumours. Scand. J. Immunol. 2013, 78, 1–7. [Google Scholar] [CrossRef]
- Karsten, U.; Goletz, S. What controls the expression of the core-1 (Thomsen-Friedenreich) glycotope on tumor cells? Biochemistry 2015, 80, 801–807. [Google Scholar] [CrossRef]
- Beatson, R.; Maurstad, G.; Picco, G.; Arulappu, A.; Coleman, J.; Wandell, H.H.; Clausen, H.; Mandel, U.; Taylor-Papadimitriou, J.; Sletmoen, M.; et al. The breast cancer-associated glycoforms of MUC1, MUC1-Tn and sialyl-Tn, are expressed in COSMC wild-type cells and bind the C-type lectin MGL. PLoS ONE 2015, 10, e0125994. [Google Scholar] [CrossRef]
- Duarte, H.O.; Freitas, D.; Gomes, C.; Gomes, J.; Magalhaes, A.; Reis, C.A. Mucin-type O-glycosylation in gastric carcinogenesis. Biomolecules 2016, 6, 33. [Google Scholar] [CrossRef]
- Hollingsworth, M.A.; Swanson, B.J. Mucins in cancer: Protection and control of the cell surface. Nat. Rev. Cancer 2004, 4, 45–60. [Google Scholar] [CrossRef]
- Bafna, S.; Kaur, S.; Batra, S.K. Membrane-bound mucins: The mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene 2010, 29, 2893–2904. [Google Scholar] [CrossRef] [PubMed]
- Raina, D.; Kharbanda, S.; Kufe, D. The MUC1 oncoprotein activates the anti-apoptotic phosphoinositide 3-kinase/Akt and Bcl-xL pathways in rat 3Y1 fibroblasts. J. Biol. Chem. 2004, 279, 20607–20612. [Google Scholar] [CrossRef] [PubMed]
- Agata, N.; Ahmad, R.; Kawano, T.; Raina, D.; Kharbanda, S.; Kufe, D. MUC1 oncoprotein blocks death receptor-mediated apoptosis by inhibiting recruitment of caspase-8. Cancer Res. 2008, 68, 6136–6144. [Google Scholar] [CrossRef] [PubMed]
- Merikhian, P.; Darvishi, B.; Esmailinejad, M.R.; Khatibi, A.S.; Kalbolandi, S.M.; Selehi, M.; Mosayebzadeh, M.; Rahbarizadeh, F.; Farahmand, L. Recombinant nanobody against MUC1 tandem repeats inhibits growth, invasion, metastasis, and vascularization of spontaneous mouse mammary tumors. Mol. Oncol. 2022, 16, 485–507. [Google Scholar] [CrossRef]
- Zhao, Q.; Guo, X.; Nash, G.B.; Stone, P.C.; Hilkens, J.; Rhodes, J.M.; Yu, L.G. Circulating galectin-3 promotes metastasis by modifying MUC1 localization on cancer cell surface. Cancer Res. 2009, 69, 6799–6806. [Google Scholar] [CrossRef]
- Darvishi, B.; Farahmand, L.; Eslami-S, Z.; Majidzadeh-A, K. NF-κB as the main node of resistance to receptor tyrosine kinase inhibitors in triple-negative breast cancer. Tumour Biol. 2017, 39, 1010428317706919. [Google Scholar] [CrossRef]
- Zhao, Q.; Barclay, M.; Hilkens, J.; Guo, X.; Barrow, H.; Rhodes, J.M.; Yu, L.G. Interaction between circulating galectin-3 and cancer-associated MUC1 enhances tumor cell homotypic aggregation and prevents anoikis. Mol. Cancer 2010, 9, 154. [Google Scholar] [CrossRef]
- Rodriguez, M.C.; Yegorova, S.; Pitteloud, J.P.; Chavaroche, A.E.; Andre, S.; Arda, A.; Minond, D.; Jimenez-Barbero, J.; Gabius, H.J.; Cudic, M. Thermodynamic switch in binding of adhesion/growth regulatory human galectin-3 to tumor-associated TF antigen (CD176) and MUC1 glycopeptides. Biochemistry 2015, 54, 4462–4474. [Google Scholar] [CrossRef]
- Hayashi, T.; Takahashi, T.; Motoya, S.; Ishida, T.; Itoh, F.; Adachi, M.; Hinoda, Y.; Imai, K. MUC1 mucin core protein binds to the domain 1 of ICAM-1. Digestion 2001, 63, 87–92. [Google Scholar] [CrossRef]
- Reynolds, I.S.; Fichtner, M.; McNamara, D.A.; Kay, E.W.; Prehn, J.H.; Burke, J.P. Mucin glycoproteins block apoptosis; promote invasion, proliferation, and migration; and cause chemoresistance through diverse pathways in epithelial cancers. Cancer Metastasis Rev. 2019, 38, 237–257. [Google Scholar] [CrossRef]
- Supruniuk, K.; Radziejewska, I. MUC1 is an oncoprotein with a significant role in apoptosis (Review). Int. J. Oncol. 2021, 59, 68. [Google Scholar] [CrossRef] [PubMed]
- Cheever, M.A.; Allison, J.P.; Ferris, A.S.; Finn, O.J.; Hastings, B.M.; Hecht, T.T.; Mellman, I.; Prindiville, S.A.; Viner, J.L.; Weiner, L.M.; et al. The prioritization of cancer antigens: A national cancer institute pilot project for the acceleration of translational research. Clin. Cancer Res. 2009, 15, 5323–5337. [Google Scholar] [CrossRef] [PubMed]
- Dam, T.K.; Gabius, H.J.; André, S.; Kaltner, H.; Lensch, M.; Brewer, C.F. Galectins bind to the multivalent glycoprotein asialofetuin with enhanced affinities and a gradient of decreasing binding constants. Biochemistry 2005, 44, 12564–12571. [Google Scholar] [CrossRef] [PubMed]
- Bian, C.F.; Zhang, Y.; Sun, H.; Li, D.F.; Wang, D.C. Structural basis for distinct binding properties of the human galectins to Thomsen-Friedenreich antigen. PLoS ONE 2011, 6, e25007. [Google Scholar] [CrossRef] [PubMed]
- Arai, K.; Tsutsumi, H.; Mihara, H. A monosaccharide-modified peptide phage library for screening of ligands to carbohydrate-binding proteins. Bioorg. Med. Chem. Lett. 2013, 23, 4940–4943. [Google Scholar] [CrossRef]
- Yongye, A.B.; Calle, L.; Arda, A.; Jimenez-Barbero, J.; Andre, S.; Gabius, H.J.; Martinez-Mayorga, K.; Cudic, M. Molecular recognition of the Thomsen-Friedenreich antigen-threonine conjugate by adhesion/growth regulatory galectin-3: Nuclear magnetic resonance studies and molecular dynamics simulations. Biochemistry 2012, 51, 7278–7289. [Google Scholar] [CrossRef]
- Yu, L.G. The oncofetal Thomsen-Friedenreich carbohydrate antigen in cancer progression. Glycoconj. J. 2007, 24, 411–420. [Google Scholar] [CrossRef]
- Lee, P.C.; Chen, S.T.; Kuo, T.C.; Lin, T.C.; Lin, M.C.; Huang, J.; Hung, J.S.; Hsu, C.L.; Juan, H.F.; Lee, P.H.; et al. C1GALT1 is associated with poor survival and promotes soluble Ephrin A1-mediated cell migration through activation of EPHA2 in gastric cancer. Oncogene 2020, 39, 2724–2740. [Google Scholar] [CrossRef]
- Kumamoto, K.; Goto, Y.; Sekikawa, K.; Takenoshita, S.; Ishida, N.; Kawakita, M.; Kannagi, R. Increased expression of UDP-galactose transporter messenger RNA in human colon cancer tissues and its implication in synthesis of Thomsen-Friedenreich antigen and sialyl Lewis A/X determinants. Cancer Res. 2001, 61, 4620–4627. [Google Scholar]
- Wang, Y.; Ju, T.; Ding, X.; Xia, B.; Wang, W.; Xia, L.; He, M.; Cummings, R.D. COSMC is an essential chaperone for correct protein O-glycosylation. Proc. Natl. Acad. Sci. USA 2010, 107, 9228–9233. [Google Scholar] [CrossRef]
- Liu, F.; Fu, J.; Bergstrom, K.; Shan, X.; McDaniel, M.; McGee, S.; Bai, X.; Chen, W.; Xia, L. Core 1—Derived mucin-type O-glycosylation protects against spontaneous gastritis and gastric cancer. J. Exp. Med. 2020, 217, e20182325. [Google Scholar] [CrossRef] [PubMed]
- Aryal, R.P.; Ju, T.; Cummings, R.D. The endoplasmic reticulum chaperone Cosmc directly promotes in vitro folding of T-synthase. J. Biol. Chem. 2010, 285, 2456–2462. [Google Scholar] [CrossRef] [PubMed]
- Aryal, R.P.; Ju, T.; Cummings, R.D. Tight complex formation between Cosmc chaperone and its specific client non-native T-synthase leads to enzyme activity and client-driven dissociation. J. Biol. Chem. 2012, 287, 15317–15329. [Google Scholar] [CrossRef] [PubMed]
- Baldus, S.E.; Zirbes, T.K.; Hanisch, F.G.; Kunze, D.; Shafizadeh, S.T.; Nolden, S.; Monig, S.P.; Schneider, P.M.; Karsten, U.; Thiele, J.; et al. Thomsen-Friedenreich antigen presents as a prognostic factor in colorectal carcinoma: A clinicopathologic study of 264 patients. Cancer 2000, 88, 1536–1543. [Google Scholar] [CrossRef]
- Pantel, K.; Brakenhoff, R.H. Dissecting the metastatic cascade. Nat. Rev. Cancer 2004, 4, 448–456. [Google Scholar] [CrossRef]
- Li, Y.S.; Kaneko, M.; Sakamoto, D.G.; Takeshima, Y.; Inai, K. The reversed apical pattern of MUC1 expression is characteristics of invasive micropapillary carcinoma of the breast. Breast Cancer 2006, 13, 58–63. [Google Scholar] [CrossRef]
- Senapati, S.; Chaturvedi, P.; Chaney, W.G.; Chakraborty, S.; Gnanapragassam, V.S.; Sasson, A.R.; Batra, S.K. Novel interaction of MUC4 and galectin: Potential pathobiological implications for metastasis in lethal pancreatic cancer. Clin. Cancer Res. 2011, 17, 267–274. [Google Scholar] [CrossRef]
- Kurtenkov, O.; Klaamas, K.; Mensdorff-Pouilly, S.; Miljukhina, L.; Shljapnikova, L.; Chuzmarov, V. Humoral immune response to MUC1 and to the Thomsen-Friedenreich (TF) glycotope in patients with gastric cancer: Relation to survival. Acta Oncol. 2007, 46, 316–323. [Google Scholar] [CrossRef]
- Shimura, T.; Takenaka, Y.; Tsutsumi, S.; Hogan, V.; Kikuchi, A.; Raz, A. Galectin-3, a novel binding partner of beta-catenin. Cancer Res. 2004, 64, 6363–6367. [Google Scholar] [CrossRef]
- Ferreira, J.A.; Magalhaes, A.; Gomes, J.; Peixoto, A.; Gaiteiro, C.; Fernandes, E.; Santos, L.L.; Reis, C.A. Protein glycosylation in gastric and colorectal cancers: Toward cancer detection and targeted therapeutics. Cancer Lett. 2017, 387, 32–45. [Google Scholar] [CrossRef]
- Li, Y.; Ren, J.; Yu, W.H.; Li, Q.; Kuwahara, H.; Yin, L.; Carraway, K.L.; Kufe, D. The EGF receptor regulates interaction of the human DF3/MUC1 carcinoma antigen with c-Src and β-catenin. J. Biol. Chem. 2001, 276, 35239–35242. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Chen, D.; Liu, D.; Yin, L.; Kharbanda, S.; Kufe, D. MUC1 oncoprotein blocks GSK3β-mediated phosphorylation and degradation of β-catenin. Cancer Res. 2005, 65, 10413–10422. [Google Scholar] [CrossRef] [PubMed]
- Heimburg-Molinaro, J.; Almogren, A.; Morey, S.; Glinskii, O.V.; Roy, R.; Wilding, G.E.; Cheng, R.P.; Glinsky, V.V.; Rittenhouse-Olson, K. Development, characterization, and immunotherapeutic use of peptide mimics of the Thomsen-Friedenreich carbohydrate antigen. Neoplasia 2009, 11, 780–792. [Google Scholar] [CrossRef]
- John, C.M.; Leffler, H.; Kahl-Knutsson, B.; Svensson, I.; Jarvis, G.A. Truncated galectin-3 inhibits tumor growth and metastasis in orthotopic nude mouse model of human breast cancer. Clin. Cancer Res. 2003, 9, 2374–2383. [Google Scholar]
- Oberg, C.T.; Leffler, H.; Nilsson, U.J. Arginine binding motifs: Design and synthesis of galactose-derived arginine tweezers as galectin-3 inhibitors. J. Med. Chem. 2008, 51, 2297–2301. [Google Scholar] [CrossRef] [PubMed]
- Duckworth, C.A.; Guimond, S.E.; Sindrewicz, P.; Hughes, A.J.; French, N.S.; Lian, L.Y.; Yates, E.A.; Pritchard, D.M.; Rhodes, J.M.; Turnbull, J.E.; et al. Chemically modified, non-anticoagulant heparin derivatives are potent galectin-3 binding inhibitors and inhibit circulating galectin-3-promoted metastasis. Oncotarget 2015, 6, 23671–23687. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Radziejewska, I. Galectin-3 and Epithelial MUC1 Mucin—Interactions Supporting Cancer Development. Cancers 2023, 15, 2680. https://doi.org/10.3390/cancers15102680
Radziejewska I. Galectin-3 and Epithelial MUC1 Mucin—Interactions Supporting Cancer Development. Cancers. 2023; 15(10):2680. https://doi.org/10.3390/cancers15102680
Chicago/Turabian StyleRadziejewska, Iwona. 2023. "Galectin-3 and Epithelial MUC1 Mucin—Interactions Supporting Cancer Development" Cancers 15, no. 10: 2680. https://doi.org/10.3390/cancers15102680
APA StyleRadziejewska, I. (2023). Galectin-3 and Epithelial MUC1 Mucin—Interactions Supporting Cancer Development. Cancers, 15(10), 2680. https://doi.org/10.3390/cancers15102680