Potential Impact of Preoperative Circulating Biomarkers on Individual Escalating/de-Escalating Strategies in Early Breast Cancer
Abstract
Simple Summary
Abstract
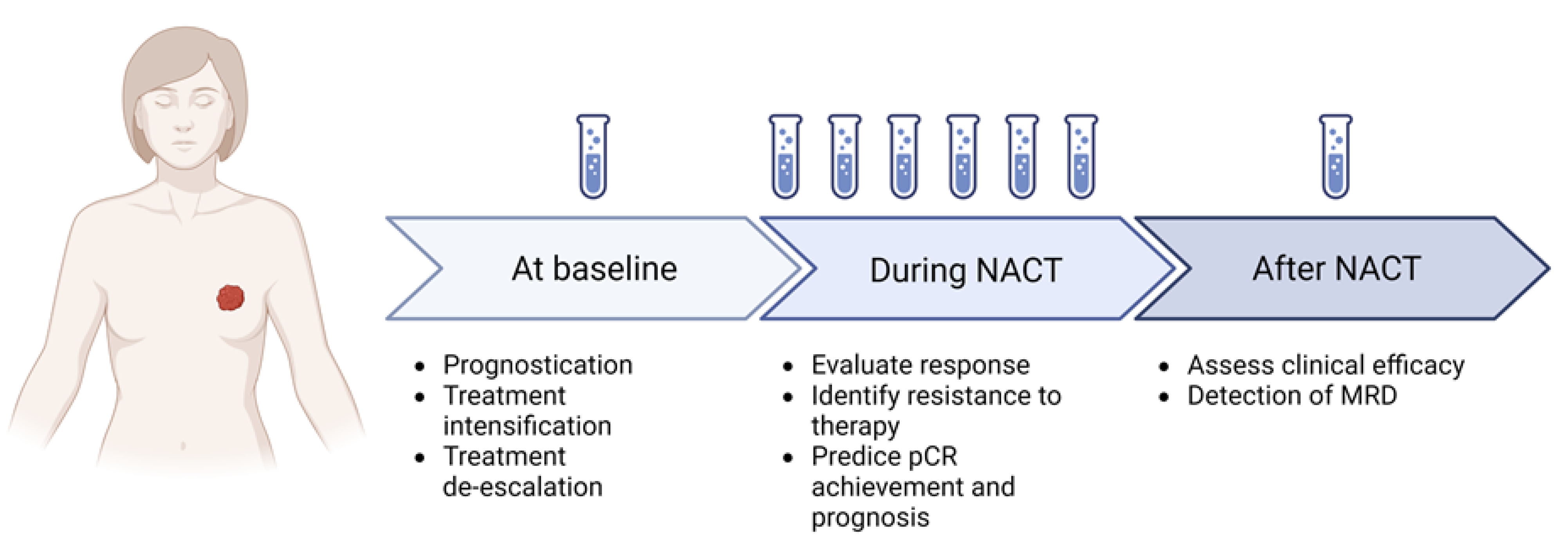
1. Introduction
2. Immune System and Tumor Cells: The Circulating Counterpart
2.1. Circulating Inflammatory Cells
2.2. Circulating Tumor Cells
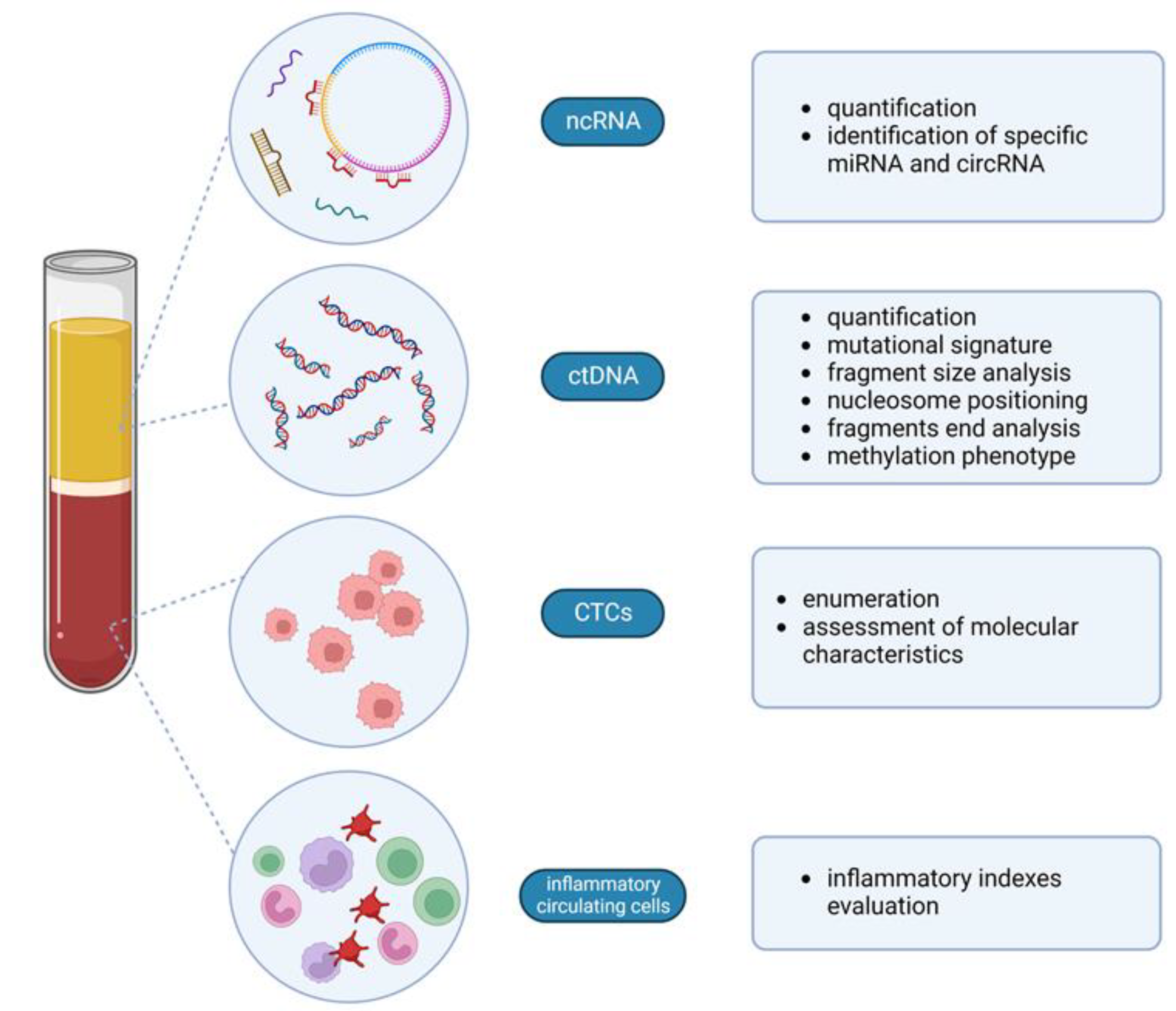
3. Circulating Cell-Free Nucleic Acids
3.1. Circulating Tumor DNA
3.2. Fragmentomics
3.3. DNA Methylation Signature
3.4. Non-Coding RNAs: miRNA and circRNA
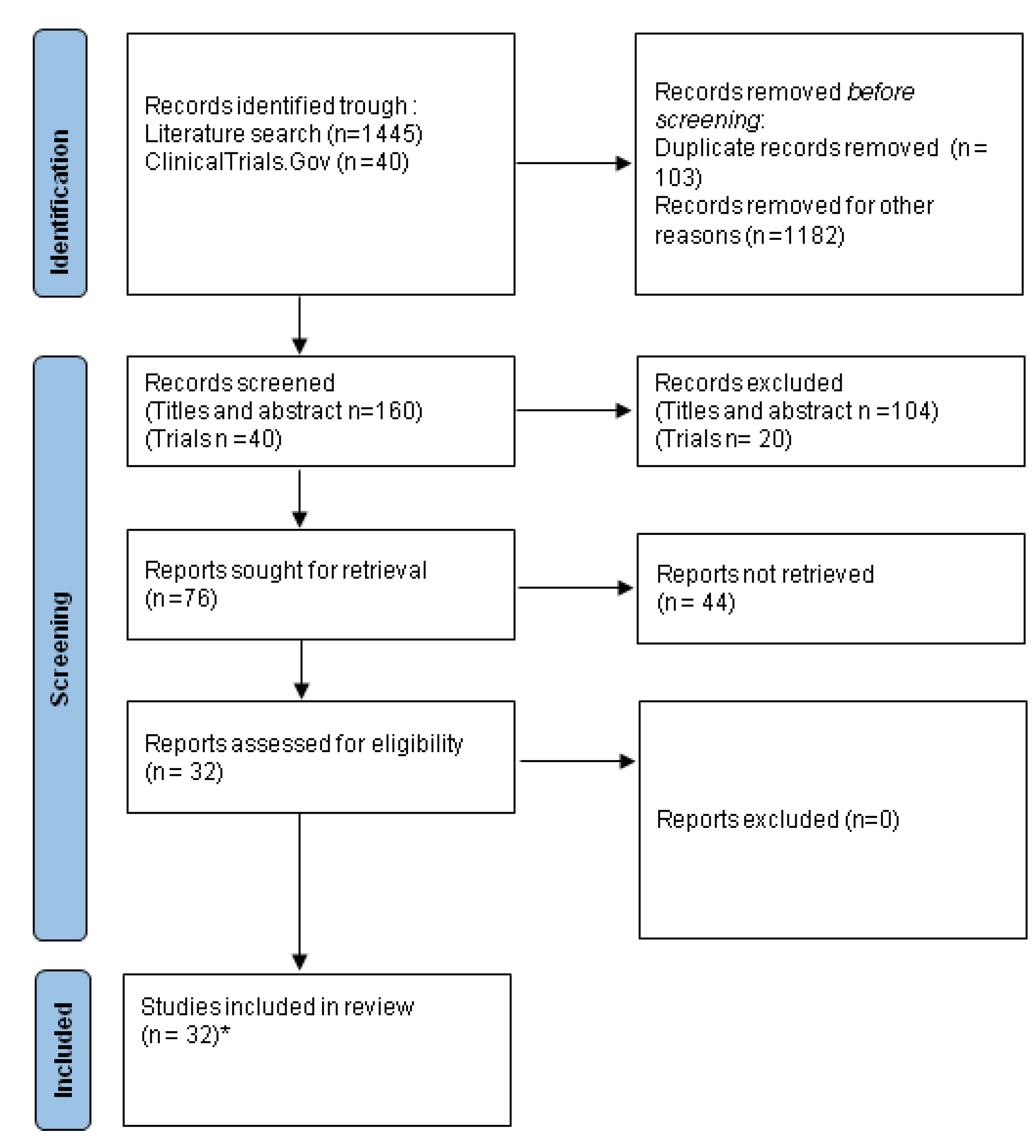
4. Methods
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harbeck, N.; Gnant, M. Breast cancer. Lancet 2017, 389, 1134–1150. [Google Scholar] [CrossRef] [PubMed]
- Andre, F.; Ismaila, N.; Allison, K.H.; Barlow, W.E.; Collyar, D.E.; Damodaran, S.; Henry, N.L.; Jhaveri, K.; Kalinsky, K.; Kuderer, N.M.; et al. Biomarkers for Adjuvant Endocrine and Chemotherapy in Early-Stage Breast Cancer: ASCO Guideline Update. J. Clin. Oncol. 2022, 40, 1816–1837. [Google Scholar] [CrossRef] [PubMed]
- Pietri, E.; Conteduca, V.; Andreis, D.; Massa, I.; Melegari, E.; Sarti, S.; Cecconetto, L.; Schirone, A.; Bravaccini, S.; Serra, P.; et al. Androgen receptor signaling pathways as a target for breast cancer treatment. Endocr. Relat. Cancer 2016, 23, R485–R498. [Google Scholar] [CrossRef]
- van der Poort, E.K.J.; van Ravesteyn, N.T.; Broek, J.J.V.D.; de Koning, H.J. The Early Detection of Breast Cancer Using Liquid Biopsies: Model Estimates of the Benefits, Harms, and Costs. Cancers 2022, 14, 2951. [Google Scholar] [CrossRef]
- Gianni, C.; Palleschi, M.; Merloni, F.; Di Menna, G.; Sirico, M.; Sarti, S.; Virga, A.; Ulivi, P.; Cecconetto, L.; Mariotti, M.; et al. Cell-Free DNA Fragmentomics: A Promising Biomarker for Diagnosis, Prognosis and Prediction of Response in Breast Cancer. Int. J. Mol. Sci. 2022, 23, 14197. [Google Scholar] [CrossRef] [PubMed]
- Schrag, D.; McDonnell, C.; Nadauld, L.; Dilaveri, C.; Klein, E.; Reid, R.; Marinac, C.; Chung, K.; Lopatin, M.; Fung, E.; et al. 903O A prospective study of a multi-cancer early detection blood test. Ann. Oncol. 2022, 33, S961. [Google Scholar] [CrossRef]
- Zigman, M.; Huber, M.; Kepesidis, K.; Voronina, L.; Fleischmann, F.; Fill, E.; Hermann, J.; Koch, I.; Kolben, T.; Schulz, G.B.; et al. 90P Infrared molecular fingerprinting: A new in vitro di-agnostic platform technology for cancer detection in blood-based liquid biopsies. Ann. Oncol. 2022, 33, S580. [Google Scholar] [CrossRef]
- Gao, Q.; Wang, C.; Yang, X.; Fang, S.; Zhang, Y.; Wang, G.; Liu, F.; Wen, X.; Zhao, J.; Zhou, G.; et al. 909P A multi-cancer early detection model based on liquid biopsy of multi-omics biomarkers: A proof of concept study (PROMISE study). Ann. Oncol. 2022, 33, S963–S964. [Google Scholar] [CrossRef]
- Conforti, F.; Pala, L.; Sala, I.; Oriecuia, C.; De Pas, T.; Specchia, C.; Graffeo, R.; Pagan, E.; Queirolo, P.; Pennacchioli, E.; et al. Evaluation of pathological complete response as surrogate endpoint in neoadjuvant randomised clinical trials of early stage breast cancer: Systematic review and meta-analysis. BMJ 2021, 375, e066381. [Google Scholar] [CrossRef]
- Gianni, L.; Pienkowski, T.; Im, Y.-H.; Tseng, L.-M.; Liu, M.-C.; Lluch, A.; Starosławska, E.; De La Haba-Rodríguez, J.R.; Im, S.-A.; Pedrini, J.L.; et al. 5-year analysis of neoadjuvant pertuzumab and trastuzumab in patients with locally advanced, inflammatory, or early-stage HER2-positive breast cancer (NeoSphere): A multicentre, open-label, phase 2 randomised trial. Lancet Oncol. 2016, 17, 791–800. [Google Scholar] [CrossRef]
- Von Minckwitz, G.; Huang, C.-S.; Mano, M.S.; Loibl, S.; Mamounas, E.P.; Untch, M.; Wolmark, N.; Rastogi, P.; Schneeweiss, A.; Redondo, A.; et al. Trastuzumab Emtansine for Residual Invasive HER2-Positive Breast Cancer. N. Engl. J. Med. 2019, 380, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N.; et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Von Minckwitz, G.; Untch, M.; Blohmer, J.-U.; Costa, S.D.; Eidtmann, H.; Fasching, P.A.; Gerber, B.; Eiermann, W.; Hilfrich, J.; Huober, J.; et al. Definition and Impact of Pathologic Complete Response on Prognosis After Neoadjuvant Chemotherapy in Various Intrinsic Breast Cancer Subtypes. J. Clin. Oncol. 2012, 30, 1796–1804. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Guarneri, V.; Pascual, T.; Brasó-Maristany, F.; Sanfeliu, E.; Paré, L.; Schettini, F.; Martínez, D.; Jares, P.; Griguolo, G.; et al. Development and validation of the new HER2DX assay for predicting pathological response and survival outcome in early-stage HER2-positive breast cancer. Ebiomedicine 2022, 75, 103801. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-Y.; Cho, N.; Choi, Y.; Lee, S.H.; Ha, S.M.; Kim, E.S.; Chang, J.M.; Moon, W.K. Factors Affecting Pathologic Complete Response Following Neoadjuvant Chemotherapy in Breast Cancer: Development and Validation of a Predictive Nomogram. Radiology 2021, 299, 290–300. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Y.; Zhao, R.; Ji, Y.; Li, J.; Zhang, Y.; Lu, H. Development and validation of a nomogram based on pretreatment dynamic contrast-enhanced MRI for the prediction of pathologic response after neoadjuvant chemotherapy for triple-negative breast cancer. Eur. Radiol. 2021, 32, 1676–1687. [Google Scholar] [CrossRef]
- Zhang, J.; Xiao, L.; Pu, S.; Liu, Y.; He, J.; Wang, K. Can We Reliably Identify the Pathological Outcomes of Neoadjuvant Chemotherapy in Patients with Breast Cancer? Development and Validation of a Logistic Regression Nomogram Based on Preoperative Factors. Ann. Surg. Oncol. 2020, 28, 2632–2645. [Google Scholar] [CrossRef]
- Gianni, C.; Palleschi, M.; Schepisi, G.; Casadei, C.; Bleve, S.; Merloni, F.; Sirico, M.; Sarti, S.; Cecconetto, L.; Di Menna, G.; et al. Circulating inflammatory cells in patients with metastatic breast cancer: Implications for treatment. Front. Oncol. 2022, 12, 882896. [Google Scholar] [CrossRef]
- De Giorgi, U.; Mego, M.; Scarpi, E.; Giordano, A.; Giuliano, M.; Valero, V.; Alvarez, R.H.; Ueno, N.T.; Cristofanilli, M.; Reuben, J.M. Association between circulating tumor cells and peripheral blood monocytes in metastatic breast cancer. Ther. Adv. Med. Oncol. 2019, 11, 1758835919866065. [Google Scholar] [CrossRef]
- De Giorgi, U.; Mego, M.; Rohren, E.M.; Liu, P.; Handy, B.C.; Reuben, J.M.; Macapinlac, H.A.; Hortobagyi, G.N.; Cristofanilli, M.; Ueno, N.T. 18F-FDG PET/CT Findings and Circulating Tumor Cell Counts in the Monitoring of Systemic Therapies for Bone Metastases from Breast Cancer. J. Nucl. Med. 2010, 51, 1213–1218. [Google Scholar] [CrossRef]
- Linde, N.; Casanova-Acebes, M.; Sosa, M.S.; Mortha, A.; Rahman, A.; Farias, E.; Harper, K.; Tardio, E.; Torres, I.R.; Jones, J.; et al. Macrophages orchestrate breast cancer early dissemination and metastasis. Nat. Commun. 2018, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Lu, X.; Liu, Q.; Zhang, T.; Li, P.; Qiao, W.; Deng, M. Prognostic value of neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio for breast cancer patients: An updated meta-analysis of 17079 individuals. Cancer Med. 2019, 8, 4135–4148. [Google Scholar] [CrossRef] [PubMed]
- Truffi, M.; Piccotti, F.; Albasini, S.; Tibollo, V.; Morasso, C.F.; Sottotetti, F.; Corsi, F. Preoperative Systemic Inflammatory Biomarkers Are Independent Predictors of Disease Recurrence in ER+ HER2- Early Breast Cancer. Front. Oncol. 2021, 11, 773078. [Google Scholar] [CrossRef] [PubMed]
- Moon, G.; Noh, H.; Cho, I.-J.; Lee, J.-I.; Han, A. Prediction of late recurrence in patients with breast cancer: Elevated neutrophil to lymphocyte ratio (NLR) at 5 years after diagnosis and late recurrence. Breast Cancer 2019, 27, 54–61. [Google Scholar] [CrossRef]
- Ferroni, P.; Roselli, M.; Buonomo, O.C.; Spila, A.; Portarena, I.; Laudisi, A.; Valente, M.G.; Pirillo, S.P.; Fortunato, L.; Costarelli, L.; et al. Prognostic Significance of Neutrophil–to–lymphocyte Ratio in the Framework of the 8th TNM Edition for Breast Cancer. Anticancer. Res. 2018, 38, 4705–4712. [Google Scholar] [CrossRef]
- Grassadonia, A.; Graziano, V.; Iezzi, L.; Vici, P.; Barba, M.; Pizzuti, L.; Cicero, G.; Krasniqi, E.; Mazzotta, M.; Marinelli, D.; et al. Prognostic Relevance of Neutrophil to Lymphocyte Ratio (NLR) in Luminal Breast Cancer: A Retrospective Analysis in the Neoadjuvant Setting. Cells 2021, 10, 1685. [Google Scholar] [CrossRef]
- Salat, A.; Gnant, M.; Kwasny, W.; Mlineritsch, B.; Menzel, R.-C.; Schmid, M.; Smola, M.G.; Stierer, M.; Tausch, C.; Galid, A.; et al. Impact of pretreatment thrombocytosis on survival in primary breast cancer. Thromb. Haemost. 2003, 89, 1098–1106. [Google Scholar] [CrossRef]
- Takeuchi, H.; Abe, M.; Takumi, Y.; Hashimoto, T.; Kobayashi, R.; Osoegawa, A.; Miyawaki, M.; Okamoto, T.; Sugio, K. The prognostic impact of the platelet distribution width-to-platelet count ratio in patients with breast cancer. PLoS ONE 2017, 12, e0189166. [Google Scholar] [CrossRef]
- Corbeau, I.; Thezenas, S.; Maran-Gonzalez, A.; Colombo, P.-E.; Jacot, W.; Guiu, S. Inflammatory Blood Markers as Prognostic and Predictive Factors in Early Breast Cancer Patients Receiving Neoadjuvant Chemotherapy. Cancers 2020, 12, 2666. [Google Scholar] [CrossRef]
- Krenn-Pilko, S.; Langsenlehner, T.; Thurner, E.-M.; Stojakovic, T.; Pichler, M.; Gerger, A.; Kapp, K.S. The elevated preoperative platelet-to-lymphocyte ratio predicts poor prognosis in breast cancer patients. Br. J. Cancer 2014, 110, 2524–2530. [Google Scholar] [CrossRef]
- Koh, C.-H.; Bhoopathy, N.; Ng, K.-L.; Jabir, R.S.; Tan, G.-H.; See, M.H.; Jamaris, S.; Taib, N.A. Utility of pre-treatment neutrophil–lymphocyte ratio and platelet–lymphocyte ratio as prognostic factors in breast cancer. Br. J. Cancer 2015, 113, 150–158. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, S.; Ding, N.; Li, N.; Huang, J.; Xiao, Z. Platelet/Lymphocyte Ratio Is Superior to Neutrophil/Lymphocyte Ratio as a Predictor of Chemotherapy Response and Disease-free Survival in Luminal B-like (HER2−) Breast Cancer. Clin. Breast Cancer 2020, 20, e403–e409. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Ye, F.; Huang, X.; Li, S.; Yang, L.; Xiao, X.; Xie, X. Prognostic Significance of Preoperative Circulating Monocyte Count in Patients with Breast Cancer. Medicine 2015, 94, e2266. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, J.; Chen, X. Lymphocyte-to-Monocyte Ratio is Associated with the Poor Prognosis of Breast Cancer Patients Receiving Neoadjuvant Chemotherapy. Cancer Manag. Res. 2021, 13, 1571–1580. [Google Scholar] [CrossRef] [PubMed]
- Talamantes, S.; Xie, E.; Costa, R.L.B.; Chen, M.; Rademaker, A.; Santa-Maria, C.A. Circulating immune cell dynamics in patients with triple negative breast cancer treated with neoadjuvant chemotherapy. Cancer Med. 2020, 9, 6954–6960. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Chen, R.; Qu, F.; Ye, Y.; Fu, Y.; Tang, Z.; Wang, Y.; Zong, B.; Yu, H.; Luo, F.; et al. Low pretreatment lymphocyte/monocyte ratio is associated with the better efficacy of neoadjuvant chemotherapy in breast cancer patients. Cancer Biol. Ther. 2019, 21, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ni, C.; Ma, C.; Zhang, L.; Jing, X.; Li, C.; Liu, Y.; Qu, X. Association of neutrophil/lymphocyte ratio and platelet/lymphocyte ratio with ER and PR in breast cancer patients and their changes after neoadjuvant chemotherapy. Clin. Transl. Oncol. 2017, 19, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Tiainen, S.; Rilla, K.; Hämäläinen, K.; Oikari, S.; Auvinen, P. The prognostic and predictive role of the neutrophil-to-lymphocyte ratio and the monocyte-to-lymphocyte ratio in early breast cancer, especially in the HER2+ subtype. Breast Cancer Res. Treat. 2020, 185, 63–72. [Google Scholar] [CrossRef]
- Zhou, Q.; Dong, J.; Sun, Q.; Lu, N.; Pan, Y.; Han, X. Role of neutrophil-to-lymphocyte ratio as a prognostic biomarker in patients with breast cancer receiving neoadjuvant chemotherapy: A meta-analysis. BMJ Open 2021, 11, e047957. [Google Scholar] [CrossRef]
- Hua, X.; Long, Z.-Q.; Zhang, Y.-L.; Wen, W.; Guo, L.; Xia, W.; Zhang, W.-W.; Lin, H.-X. Prognostic Value of Preoperative Systemic Immune-Inflammation Index in Breast Cancer: A Propensity Score-Matching Study. Front. Oncol. 2020, 10, 580. [Google Scholar] [CrossRef]
- Pang, J.; Wang, S.; Liao, L.; Liu, X. Association between systemic immune-inflammation index and neoadjuvant chemotherapy efficacy as well as prognosis in triple-negative breast cancer. J. Cent. South University. Med. Sci. 2021, 46, 958–965. [Google Scholar] [CrossRef]
- Jiang, C.; Lu, Y.; Zhang, S.; Huang, Y. Systemic Immune-Inflammation Index Is Superior to Neutrophil to Lymphocyte Ratio in Prognostic Assessment of Breast Cancer Patients Undergoing Neoadjuvant Chemotherapy. BioMed. Res. Int. 2020, 2020, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Kong, X.; Wang, Z.; Wang, X.; Fang, Y.; Wang, J. Pre-treatment systemic immune-inflammation index is a useful prognostic indicator in patients with breast cancer undergoing neoadjuvant chemotherapy. J. Cell Mol. Med. 2020, 24, 2993–3021. [Google Scholar] [CrossRef] [PubMed]
- Thery, L.; Meddis, A.; Cabel, L.; Proudhon, C.; Latouche, A.; Pierga, J.-Y.; Bidard, F.-C. Circulating Tumor Cells in Early Breast Cancer. JNCI Cancer Spectr. 2019, 3, pkz026. [Google Scholar] [CrossRef] [PubMed]
- Cristofanilli, M.; Budd, G.T.; Ellis, M.J.; Stopeck, A.; Matera, J.; Miller, M.C.; Reuben, J.M.; Doyle, G.V.; Allard, W.J.; Terstappen, L.W.M.M.; et al. Circulating Tumor Cells, Disease Progression, and Survival in Metastatic Breast Cancer. N. Engl. J. Med. 2004, 351, 781–791. [Google Scholar] [CrossRef]
- Giuliano, M.; Giordano, A.; Jackson, S.; Hess, K.R.; De Giorgi, U.; Mego, M.; Handy, B.C.; Ueno, N.T.; Alvarez, R.H.; De Laurentiis, M.; et al. Circulating tumor cells as prognostic and predictive markers in metastatic breast cancer patients receiving first-line systemic treatment. Breast Cancer Res. 2011, 13, R67. [Google Scholar] [CrossRef]
- Bidard, F.-C.; Peeters, D.J.; Fehm, T.; Nolé, F.; Gisbert-Criado, R.; Mavroudis, D.; Grisanti, S.; Generali, D.; Garcia-Saenz, J.A.; Stebbing, J.; et al. Clinical validity of circulating tumour cells in patients with metastatic breast cancer: A pooled analysis of individual patient data. Lancet Oncol. 2014, 15, 406–414. [Google Scholar] [CrossRef]
- Krell, J.; Stebbing, J. Circulating tumour cells as biomarkers in early breast cancer. Lancet Oncol. 2012, 13, 653–654. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Cristofanilli, M.; Singh, B.; Reuben, J.; Gao, H.; Cohen, E.N.; Andreopoulou, E.; Hall, C.S.; Lodhi, A.; Jackson, S.; et al. Detection of minimal residual disease in blood and bone marrow in early stage breast cancer. Cancer 2010, 116, 3330–3337. [Google Scholar] [CrossRef]
- Boya, M.; Chu, C.-H.; Liu, R.; Ozkaya-Ahmadov, T.; Sarioglu, A.F. Circulating Tumor Cell Enrichment Technologies; Springer: Cham, Switzerland, 2020; Volume 215, pp. 25–55. [Google Scholar]
- Liang, D.H.; Hall, C.; Lucci, A. Circulating Tumor Cells in Breast Cancer. Tumor Liq. Biopsies 2019, 215, 127–145. [Google Scholar] [CrossRef]
- Schneck, H.; Gierke, B.; Uppenkamp, F.; Behrens, B.; Niederacher, D.; Stoecklein, N.H.; Templin, M.F.; Pawlak, M.; Fehm, T.; Neubauer, H.; et al. EpCAM-Independent Enrichment of Circulating Tumor Cells in Metastatic Breast Cancer. PLoS ONE 2015, 10, e0144535. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Wang, C.; Xie, M.; Zhu, C.; Shu, Y.; Tang, J.; Guan, X. Heterogeneity of CTC contributes to the organotropism of breast cancer. Biomed. Pharmacother. 2021, 137, 111314. [Google Scholar] [CrossRef] [PubMed]
- Harouaka, R.A.; Nisic, M.; Zheng, S.-Y. Circulating Tumor Cell Enrichment Based on Physical Properties. SLAS Technol. Transl. Life Sci. Innov. 2013, 18, 455–468. [Google Scholar] [CrossRef]
- Mego, M.; Giordano, A.; De Giorgi, U.; Masuda, H.; Hsu, L.; Giuliano, M.; Fouad, T.M.; Dawood, S.; Ueno, N.T.; Valero, V.; et al. Circulating tumor cells in newly diagnosed inflammatory breast cancer. Breast Cancer Res. 2015, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pierga, J.-Y.; Bidard, F.-C.; Autret, A.; Petit, T.; Andre, F.; Dalenc, F.; Levy, C.; Ferrero, J.-M.; Romieu, G.; Bonneterre, J.; et al. Circulating tumour cells and pathological complete response: Independent prognostic factors in inflammatory breast cancer in a pooled analysis of two multicentre phase II trials (BEVERLY-1 and -2) of neoadjuvant chemotherapy combined with bevacizumab. Ann. Oncol. 2017, 28, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Bidard, F.-C.; Michiels, S.; Riethdorf, S.; Mueller, V.; Esserman, L.J.; Lucci, A.; Naume, B.; Horiguchi, J.; Gisbert-Criado, R.; Sleijfer, S.; et al. Circulating Tumor Cells in Breast Cancer Patients Treated by Neoadjuvant Chemotherapy: A Meta-analysis. Gynecol. Oncol. 2018, 110, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Azim, H.H.A.; Rothé, F., Jr.; Aura, C.M.; Bavington, M.; Maetens, M.; Rouas, G.; Gebhart, G.; Gámez-Cenzano, C.; Eidtmann, H.; Baselga, J.; et al. Circulating tumor cells and response to neoadjuvant paclitaxel and HER2-targeted therapy: A sub-study from the NeoALTTO phase III trial. Breast 2013, 22, 1060–1065. [Google Scholar] [CrossRef]
- Ignatiadis, M.; Litière, S.; Rothe, F.; Riethdorf, S.; Proudhon, C.; Fehm, T.; Aalders, K.; Forstbauer, H.; Fasching, P.; Brain, E.; et al. Trastuzumab versus observation for HER2 nonamplified early breast cancer with circulating tumor cells (EORTC 90091-10093, BIG 1-12, Treat CTC): A randomized phase II trial. Ann. Oncol. 2018, 29, 1777–1783. [Google Scholar] [CrossRef]
- Bidard, F.-C.; Mathiot, C.; Delaloge, S.; Brain, E.; Giachetti, S.; de Cremoux, P.; Marty, M.; Pierga, J.-Y. Single circulating tumor cell detection and overall survival in nonmetastatic breast cancer. Ann. Oncol. 2010, 21, 729–733. [Google Scholar] [CrossRef]
- Riethdorf, S.; Müller, V.; Loibl, S.; Nekljudova, V.; Weber, K.; Huober, J.; Fehm, T.; Schrader, I.; Hilfrich, J.; Holms, F.; et al. Prognostic Impact of Circulating Tumor Cells for Breast Cancer Patients Treated in the Neoadjuvant “Geparquattro” Trial. Clin. Cancer Res. 2017, 23, 5384–5393. [Google Scholar] [CrossRef]
- Riethdorf, S.; Müller, V.; Zhang, L.; Rau, T.; Loibl, S.; Komor, M.; Roller, M.; Huober, J.; Fehm, T.; Schrader, I.; et al. Detection and HER2 Expression of Circulating Tumor Cells: Prospective Monitoring in Breast Cancer Patients Treated in the Neoadjuvant GeparQuattro Trial. Clin. Cancer Res. 2010, 16, 2634–2645. [Google Scholar] [CrossRef] [PubMed]
- Pierga, J.-Y.; Bidard, F.-C.; Mathiot, C.; Brain, E.; Delaloge, S.; Giachetti, S.; de Cremoux, P.; Salmon, R.; Vincent-Salomon, A.; Marty, M. Circulating Tumor Cell Detection Predicts Early Metastatic Relapse After Neoadjuvant Chemotherapy in Large Operable and Locally Advanced Breast Cancer in a Phase II Randomized Trial. Clin. Cancer Res. 2008, 14, 7004–7010. [Google Scholar] [CrossRef] [PubMed]
- Onstenk, W.; Kraan, J.; Mostert, B.; Timmermans, M.M.; Charehbili, A.; Smit, V.T.; Kroep, J.R.; Nortier, J.W.; van de Ven, S.; Heijns, J.B.; et al. Improved Circulating Tumor Cell Detection by a Combined EpCAM and MCAM CellSearch Enrichment Approach in Patients with Breast Cancer Undergoing Neoadjuvant Chemotherapy. Mol. Cancer Ther. 2015, 14, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Hall, C.; Karhade, M.; Laubacher, B.; Anderson, A.; Kuerer, H.; DeSynder, S.; Lucci, A. Circulating Tumor Cells After Neoadjuvant Chemotherapy in Stage I–III Triple-Negative Breast Cancer. Ann. Surg. Oncol. 2015, 22, 552–558. [Google Scholar] [CrossRef] [PubMed]
- Lucci, A.; Hall, C.S.; Lodhi, A.K.; Bhattacharyya, A.; Anderson, A.E.; Xiao, L.; Bedrosian, I.; Kuerer, H.M.; Krishnamurthy, S. Circulating tumour cells in non-metastatic breast cancer: A prospective study. Lancet Oncol. 2012, 13, 688–695. [Google Scholar] [CrossRef]
- Pierga, J.-Y.; Petit, T.; Lévy, C.; Ferrero, J.-M.; Campone, M.; Gligorov, J.; Lerebours, F.; Roché, H.; Bachelot, T.; Charafe-Jauffret, E.; et al. Pathological Response and Circulating Tumor Cell Count Identifies Treated HER2+ Inflammatory Breast Cancer Patients with Excellent Prognosis: BEVERLY-2 Survival Data. Clin. Cancer Res. 2015, 21, 1298–1304. [Google Scholar] [CrossRef]
- Serrano, M.J.; Rovira, P.S.; Martínez-Zubiaurre, I.; Rodriguez, M.D.; Fernández, M.; Lorente, J.A. Dynamics of circulating tumor cells in early breast cancer under neoadjuvant therapy. Exp. Ther. Med. 2012, 4, 43–48. [Google Scholar] [CrossRef]
- Janni, W.J.; Rack, B.; Terstappen, L.W.; Pierga, J.-Y.; Taran, F.-A.; Fehm, T.; Hall, C.; de Groot, M.R.; Bidard, F.-C.; Friedl, T.W.; et al. Pooled Analysis of the Prognostic Relevance of Circulating Tumor Cells in Primary Breast Cancer. Clin. Cancer Res. 2016, 22, 2583–2593. [Google Scholar] [CrossRef]
- Andreopoulou, E.; Yang, L.-Y.; Rangel, K.M.; Reuben, J.M.; Hsu, L.; Krishnamurthy, S.; Valero, V.; Fritsche, H.A.; Cristofanilli, M. Comparison of assay methods for detection of circulating tumor cells in metastatic breast cancer: AdnaGen AdnaTest BreastCancer Select/Detect™ versus Veridex CellSearch™ system. Int. J. Cancer 2011, 130, 1590–1597. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Ahn, S.; Kim, S.; Park, S.; Jung, N.; Park, S.; Han, H.; Sohn, J.; Kim, S.; Lee, H. Detection of circulating tumor cell-specific markers in breast cancer patients using the quantitative RT-PCR assay. Int. J. Clin. Oncol. 2015, 20, 878–890. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Ahn, S.; Kim, S.; Park, S.; Park, S.; Han, H.; Sohn, J.H.; Kim, S.; Lee, H. Detection of circulating tumor cells in patients with breast cancer using the quantitative RT-PCR assay for monitoring of therapy efficacy. Exp. Mol. Pathol. 2014, 97, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Flores, L.M.; Kindelberger, D.W.; Ligon, A.H.; Capelletti, M.; Fiorentino, M.; Loda, M.; Cibas, E.S.; Jänne, P.A.; Krop, I.E. Improving the yield of circulating tumour cells facilitates molecular characterisation and recognition of discordant HER2 amplification in breast cancer. Br. J. Cancer 2010, 102, 1495–1502. [Google Scholar] [CrossRef] [PubMed]
- Bonadio, R.C.; Tarantino, P.; Testa, L.; Punie, K.; Pernas, S.; Barrios, C.; Curigliano, G.; Tolaney, S.M.; Barroso-Sousa, R. Management of patients with early-stage triple-negative breast cancer following pembrolizumab-based neoadjuvant therapy: What are the evidences? Cancer Treat. Rev. 2022, 110, 102459. [Google Scholar] [CrossRef] [PubMed]
- Magbanua, M.J.M.; Gumusay, O.; Kurzrock, R.; Veer, L.J.V.; Rugo, H.S. Immunotherapy in Breast Cancer and the Potential Role of Liquid Biopsy. Front. Oncol. 2022, 12, 802579. [Google Scholar] [CrossRef] [PubMed]
- Rossi, T.; Gallerani, G.; Martinelli, G.; Maltoni, R.; Fabbri, F. Circulating Tumor Cells as a Tool to Untangle the Breast Cancer Heterogeneity Issue. Biomedicines 2021, 9, 1242. [Google Scholar] [CrossRef]
- Rossi, T.; Angeli, D.; Tebaldi, M.; Fici, P.; Rossi, E.; Rocca, A.; Palleschi, M.; Maltoni, R.; Martinelli, G.; Fabbri, F.; et al. Dissecting Molecular Heterogeneity of Circulating Tumor Cells (CTCs) from Metastatic Breast Cancer Patients through Copy Number Aberration (CNA) and Single Nucleotide Variant (SNV) Single Cell Analysis. Cancers 2022, 14, 3925. [Google Scholar] [CrossRef]
- Rossi, T.; Gallerani, G.; Angeli, D.; Cocchi, C.; Bandini, E.; Fici, P.; Gaudio, M.; Martinelli, G.; Rocca, A.; Maltoni, R.; et al. Single-Cell NGS-Based Analysis of Copy Number Alterations Reveals New Insights in Circulating Tumor Cells Persistence in Early-Stage Breast Cancer. Cancers 2020, 12, 2490. [Google Scholar] [CrossRef]
- Heeke, S.; Mograbi, B.; Alix-Panabières, C.; Hofman, P. Never Travel Alone: The Crosstalk of Circulating Tumor Cells and the Blood Microenvironment. Cells 2019, 8, 714. [Google Scholar] [CrossRef]
- Kasimir-Bauer, S.; Karaaslan, E.; Hars, O.; Hoffmann, O.; Kimmig, R. In Early Breast Cancer, the Ratios of Neutrophils, Platelets and Monocytes to Lymphocytes Significantly Correlate with the Presence of Subsets of Circulating Tumor Cells but Not with Disseminated Tumor Cells. Cancers 2022, 14, 3299. [Google Scholar] [CrossRef]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef]
- Miklikova, S.; Minarik, G.; Sedlackova, T.; Plava, J.; Cihova, M.; Jurisova, S.; Kalavska, K.; Karaba, M.; Benca, J.; Smolkova, B.; et al. Inflammation-Based Scores Increase the Prognostic Value of Circulating Tumor Cells in Primary Breast Cancer. Cancers 2020, 12, 1134. [Google Scholar] [CrossRef] [PubMed]
- Schochter, F.; Friedl, T.W.P.; Degregorio, A.; Krause, S.; Huober, J.; Rack, B.; Janni, W. Are Circulating Tumor Cells (CTCs) Ready for Clinical Use in Breast Cancer? An Overview of Completed and Ongoing Trials Using CTCs for Clinical Treatment Decisions. Cells 2019, 8, 1412. [Google Scholar] [CrossRef] [PubMed]
- Addanki, S.; Meas, S.; Sarli, V.N.; Singh, B.; Lucci, A. Applications of Circulating Tumor Cells and Circulating Tumor DNA in Precision Oncology for Breast Cancers. Int. J. Mol. Sci. 2022, 23, 7843. [Google Scholar] [CrossRef] [PubMed]
- Crook, T.; Leonard, R.; Mokbel, K.; Thompson, A.; Michell, M.; Page, R.; Vaid, A.; Mehrotra, R.; Ranade, A.; Limaye, S.; et al. Accurate Screening for Early-Stage Breast Cancer by Detection and Profiling of Circulating Tumor Cells. Cancers 2022, 14, 3341. [Google Scholar] [CrossRef]
- Sant, M.; Bernat-Peguera, A.; Felip, E.; Margelí, M. Role of ctDNA in Breast Cancer. Cancers 2022, 14, 310. [Google Scholar] [CrossRef]
- Strasser, A.; Vaux, D.L. Cell Death in the Origin and Treatment of Cancer. Mol. Cell 2020, 78, 1045–1054. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Liquid biopsy and minimal residual disease—Latest advances and implications for cure. Nat. Rev. Clin. Oncol. 2019, 16, 409–424. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-free nucleic acids as biomarkers in cancer patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef]
- Gerratana, L.; Zhang, Q.; Shah, A.N.; Davis, A.A.; Zhang, Y.; Wehbe, F.; Qiang, W.; Flaum, L.; Finkelman, B.; Gradishar, W.J.; et al. Performance of a novel Next Generation Sequencing circulating tumor DNA (ctDNA) platform for the evaluation of samples from patients with metastatic breast cancer (MBC). Crit. Rev. Oncol. 2019, 145, 102856. [Google Scholar] [CrossRef]
- Palacín-Aliana, I.; García-Romero, N.; Asensi-Puig, A.; Carrión-Navarro, J.; González-Rumayor, V.; Ayuso-Sacido, Á. Clinical Utility of Liquid Biopsy-Based Actionable Mutations Detected via ddPCR. Biomedicines 2021, 9, 906. [Google Scholar] [CrossRef] [PubMed]
- Tay, T.K.Y.; Tan, P.H. Liquid Biopsy in Breast Cancer: A Focused Review. Arch. Pathol. Lab. Med. 2020, 145, 678–686. [Google Scholar] [CrossRef] [PubMed]
- Pascual, J.; Attard, G.; Bidard, F.-C.; Curigliano, G.; De Mattos-Arruda, L.; Diehn, M.; Italiano, A.; Lindberg, J.; Merker, J.; Montagut, C.; et al. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. 2022, 33, 750–768. [Google Scholar] [CrossRef] [PubMed]
- De Mattos-Arruda, L.; Siravegna, G. How to use liquid biopsies to treat patients with cancer. ESMO Open 2021, 6, 100060. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Hruban, R.H.; Boyle, J.O.; Tockman, M.; Sidransky, D. Detection of oncogene mutations in sputum precedes di-agnosis of lung cancer. Cancer Res. 1994, 54, 1634–1637. [Google Scholar]
- Baksh, M.; Mahajan, B.; Dufresne, M.M.; Shoukry, M.M.; Nussbaum, S.; Abbaszadeh-Kasbi, A.; Ashary, M.; Vandenberg, J.; Gabriel, E.M. Circulating tumor DNA for breast cancer: Review of active clinical trials. Cancer Treat. Res. Commun. 2022, 32, 100609. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L.; et al. Circulating Tumor DNA Analysis in Patients with Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef]
- Cailleux, F.; Agostinetto, E.; Lambertini, M.; Rothé, F.; Wu, H.-T.; Balcioglu, M.; Kalashnikova, E.; Vincent, D.; Viglietti, G.; Gombos, A.; et al. Circulating Tumor DNA After Neoadjuvant Chemotherapy in Breast Cancer Is Associated with Disease Relapse. JCO Precis. Oncol. 2022, 6, e2200148. [Google Scholar] [CrossRef]
- Turner, N.C.; Swift, C.; Jenkins, B.; Kilburn, L.; Coakley, M.; Beaney, M.; Fox, L.; Goddard, K.; Garcia-Murillas, I.; Proszek, P.; et al. Results of the c-TRAK TN trial: A clinical trial utilising ctDNA mutation tracking to detect molecular residual disease and trigger intervention in patients with moderate and high-risk early stage triple negative breast cancer. Ann. Oncol. 2022. [Google Scholar] [CrossRef]
- Gögenur, M.; Hadi, N.A.-H.; Qvortrup, C.; Andersen, C.L.; Gögenur, I. ctDNA for Risk of Recurrence Assessment in Patients Treated with Neoadjuvant Treatment: A Systematic Review and Meta-analysis. Ann. Surg. Oncol. 2022, 29, 8666–8674. [Google Scholar] [CrossRef]
- Zhou, Q.; Gampenrieder, S.P.; Frantal, S.; Rinnerthaler, G.; Singer, C.F.; Egle, D.; Pfeiler, G.; Bartsch, R.; Wette, V.; Pichler, A.; et al. Persistence of ctDNA in Patients with Breast Cancer During Neoadjuvant Treatment Is a Significant Predictor of Poor Tumor Response. Clin. Cancer Res. 2021, 28, 697–707. [Google Scholar] [CrossRef]
- Garcia-Murillas, I.; Schiavon, G.; Weigelt, B.; Ng, C.; Hrebien, S.; Cutts, R.J.; Cheang, M.; Osin, P.; Nerurkar, A.; Kozarewa, I.; et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci. Transl. Med. 2015, 7, 302ra133. [Google Scholar] [CrossRef] [PubMed]
- Masuda, N.; Lee, S.-J.; Ohtani, S.; Im, Y.-H.; Lee, E.-S.; Yokota, I.; Kuroi, K.; Im, S.-A.; Park, B.-W.; Kim, S.-B.; et al. Adjuvant Capecitabine for Breast Cancer after Preoperative Chemotherapy. N. Engl. J. Med. 2017, 376, 2147–2159. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Murillas, I.; Chopra, N.; Comino-Mendez, I.; Beaney, M.; Tovey, H.; Cutts, R.J.; Swift, C.; Kriplani, D.; Afentakis, M.; Hrebien, S.; et al. Assessment of Molecular Relapse Detection in Early-Stage Breast Cancer. JAMA Oncol. 2019, 5, 1473–1478. [Google Scholar] [CrossRef]
- Cavallone, L.; Aguilar-Mahecha, A.; Lafleur, J.; Brousse, S.; Aldamry, M.; Roseshter, T.; Lan, C.; Alirezaie, N.; Bareke, E.; Majewski, J.; et al. Prognostic and predictive value of circulating tumor DNA during neoadjuvant chemotherapy for triple negative breast cancer. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Papakonstantinou, A.; Gonzalez, N.S.; Pimentel, I.; Suñol, A.; Zamora, E.; Ortiz, C.; Espinosa-Bravo, M.; Peg, V.; Vivancos, A.; Saura, C.; et al. Prognostic value of ctDNA detection in patients with early breast cancer undergoing neoadjuvant therapy: A systematic review and meta-analysis. Cancer Treat. Rev. 2022, 104, 102362. [Google Scholar] [CrossRef]
- Ortolan, E.; Appierto, V.; Silvestri, M.; Miceli, R.; Veneroni, S.; Folli, S.; Pruneri, G.; Vingiani, A.; Belfiore, A.; Cappelletti, V.; et al. Blood-based genomics of triple-negative breast cancer progression in patients treated with neoadjuvant chemotherapy. ESMO Open 2021, 6, 100086. [Google Scholar] [CrossRef]
- Riva, F.; Bidard, F.-C.; Houy, A.; Saliou, A.; Madic, J.; Rampanou, A.; Hego, C.; Milder, M.; Cottu, P.; Sablin, M.-P.; et al. Patient-Specific Circulating Tumor DNA Detection during Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer. Clin. Chem. 2017, 63, 691–699. [Google Scholar] [CrossRef]
- Chen, Y.-H.; Hancock, B.A.; Solzak, J.P.; Brinza, D.; Scafe, C.; Miller, K.D.; Radovich, M. Next-generation sequencing of circulating tumor DNA to predict recurrence in triple-negative breast cancer patients with residual disease after neoadjuvant chemotherapy. npj Breast Cancer 2017, 3, 1–6. [Google Scholar] [CrossRef]
- Radovich, M.; Jiang, G.; Hancock, B.A.; Chitambar, C.; Nanda, R.; Falkson, C.; Lynce, F.C.; Gallagher, C.; Isaacs, C.; Blaya, M.; et al. Association of Circulating Tumor DNA and Circulating Tumor Cells after Neoadjuvant Chemotherapy with Disease Recurrence in Patients with Triple-Negative Breast Cancer. JAMA Oncol. 2020, 6, 1410–1415. [Google Scholar] [CrossRef]
- McDonald, B.R.; Contente-Cuomo, T.; Sammut, S.-J.; Odenheimer-Bergman, A.; Ernst, B.; Perdigones, N.; Chin, S.-F.; Farooq, M.; Mejia, R.; Cronin, P.A.; et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 2019, 11, eaax7392. [Google Scholar] [CrossRef] [PubMed]
- Rothé, F.; Silva, M.J.; Venet, D.; Campbell, C.; Bradburry, I.; Rouas, G.; de Azambuja, E.; Maetens, M.; Fumagalli, D.; Rodrik-Outmezguine, V.; et al. Circulating Tumor DNA in HER2-Amplified Breast Cancer: A Translational Research Substudy of the NeoALTTO Phase III Trial. Clin. Cancer Res. 2019, 25, 3581–3588. [Google Scholar] [CrossRef] [PubMed]
- Cullinane, C.; Fleming, C.; O’Leary, D.P.; Hassan, F.; Kelly, L.; O’Sullivan, M.J.; Corrigan, M.A.; Redmond, H.P. Association of Circulating Tumor DNA with Disease-Free Survival in Breast Cancer. JAMA Netw. Open 2020, 3, e2026921. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.C.; Lo, Y.D. Cell-Free DNA Fragmentomics in Liquid Biopsy. Diagnostics 2022, 12, 978. [Google Scholar] [CrossRef]
- van der Pol, Y.; Mouliere, F. Toward the Early Detection of Cancer by Decoding the Epigenetic and Environmental Fingerprints of Cell-Free DNA. Cancer Cell 2019, 36, 350–368. [Google Scholar] [CrossRef] [PubMed]
- Keller, L.; Belloum, Y.; Wikman, H.; Pantel, K. Clinical relevance of blood-based ctDNA analysis: Mutation detection and beyond. Br. J. Cancer 2020, 124, 345–358. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Giacona, M.B.; Ruben, G.C.; Iczkowski, K.A.; Roos, T.B.; Porter, D.M.; Sorenson, G.D. Cell-Free DNA in Human Blood Plasma. Pancreas 1998, 17, 89–97. [Google Scholar] [CrossRef]
- Sobhani, N.; Generali, D.; Zanconati, F.; Bortul, M.; Scaggiante, B. Cell-free DNA integrity for the monitoring of breast cancer: Future perspectives? World J. Clin. Oncol. 2018, 9, 26–32. [Google Scholar] [CrossRef]
- Iqbal, S.; Vishnubhatla, S.; Raina, V.; Sharma, S.; Gogia, A.; Deo, S.S.V.; Mathur, S.R.; Shukla, N.K. Circulating cell-free DNA and its integrity as a prognostic marker for breast cancer. SpringerPlus 2015, 4, 1–6. [Google Scholar] [CrossRef]
- Kamel, A.M.; Teama, S.; Fawzy, A.; El Deftar, M. Plasma DNA integrity index as a potential molecular diagnostic marker for breast cancer. Tumor Biol. 2015, 37, 7565–7572. [Google Scholar] [CrossRef]
- Lamminaho, M.; Kujala, J.; Peltonen, H.; Tengström, M.; Kosma, V.-M.; Mannermaa, A. High Cell-Free DNA Integrity Is Associated with Poor Breast Cancer Survival. Cancers 2021, 13, 4679. [Google Scholar] [CrossRef]
- Cheng, J.; Cuk, K.; Heil, J.; Golatta, M.; Schott, S.; Sohn, C.; Schneeweiss, A.; Burwinkel, B.; Surowy, H. Cell-free circulating DNA integrity is an independent predictor of impending breast cancer recurrence. Oncotarget 2017, 8, 54537–54547. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, W.; Su, L.; Sang, J.; Wang, S.; Yao, Y. Plasma cell-free DNA integrity: A potential biomarker to monitor the response of breast cancer to neoadjuvant chemotherapy. Transl. Cancer Res. 2019, 8, 1531–1539. [Google Scholar] [CrossRef] [PubMed]
- Adusei, E.; Ahenkorah, J.; Adu-Aryee, N.; Adutwum-Ofosu, K.; Tagoe, E.; Koney, N.; Nkansah, E.; Aryee, N.; Blay, R.; Hottor, B.; et al. Reduced Serum Circulation of Cell-Free DNA Following Chemotherapy in Breast Cancer Patients. Med. Sci. 2021, 9, 37. [Google Scholar] [CrossRef]
- Lehner, J.; Stötzer, O.J.; Fersching, D.; Nagel, D.; Holdenrieder, S. Circulating plasma DNA and DNA integrity in breast cancer patients undergoing neoadjuvant chemotherapy. Clin. Chim. Acta 2013, 425, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Ulz, P.; Thallinger, G.G.; Auer, M.; Graf, R.; Kashofer, K.; Jahn, S.W.; Abete, L.; Pristauz, G.; Petru, E.; Geigl, J.B.; et al. Inferring expressed genes by whole-genome sequencing of plasma DNA. Nat. Genet. 2016, 48, 1273–1278. [Google Scholar] [CrossRef]
- Murtaza, M.; Caldas, C. Nucleosome mapping in plasma DNA predicts cancer gene expression. Nat. Genet. 2016, 48, 1105–1106. [Google Scholar] [CrossRef]
- Snyder, M.W.; Kircher, M.; Hill, A.J.; Daza, R.M.; Shendure, J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell 2016, 164, 57–68. [Google Scholar] [CrossRef]
- Vanderstichele, A.; Busschaert, P.; Landolfo, C.; Olbrecht, S.; Coosemans, A.; Froyman, W.; Loverix, L.; Concin, N.; Braicu, E.I.; Wimberger, P.; et al. Nucleosome footprinting in plasma cell-free DNA for the pre-surgical diagnosis of ovarian cancer. npj Genom. Med. 2022, 7, 30. [Google Scholar] [CrossRef]
- McAnena, P.; Brown, J.A.L.; Kerin, M.J. Circulating Nucleosomes and Nucleosome Modifications as Biomarkers in Cancer. Cancers 2017, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Elsheikh, S.E.; Green, A.R.; Rakha, E.A.; Powe, D.G.; Ahmed, R.A.; Collins, H.M.; Soria, D.; Garibaldi, J.M.; Paish, C.E.; Ammar, A.A.; et al. Global Histone Modifications in Breast Cancer Correlate with Tumor Phenotypes, Prognostic Factors, and Patient Outcome. Cancer Res 2009, 69, 3802–3809. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, J.; Chen, Y.-Y.; Scott, G.K.; DeVries, S.; Chin, K.; Benz, C.C.; Waldman, F.M.; Hwang, E.S. Protein Acetylation and Histone Deacetylase Expression Associated with Malignant Breast Cancer Progression. Clin. Cancer Res. 2009, 15, 3163–3171. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Cai, G.-X.; Han, B.-W.; Guo, Z.-W.; Wu, Y.-S.; Lyu, X.; Huang, L.-M.; Zhang, Y.-B.; Li, X.; Ye, G.-L.; et al. Association between the nucleosome footprint of plasma DNA and neoadjuvant chemotherapy response for breast cancer. npj Breast Cancer 2021, 7, 1–12. [Google Scholar] [CrossRef]
- Stoetzer, O.J.; Fersching, D.M.; Salat, C.; Steinkohl, O.; Gabka, C.J.; Hamann, U.; Braun, M.; Feller, A.-M.; Heinemann, V.; Siegele, B.; et al. Prediction of response to neoadjuvant chemotherapy in breast cancer patients by circulating apoptotic biomarkers nucleosomes, DNAse, cytokeratin-18 fragments and survivin. Cancer Lett. 2013, 336, 140–148. [Google Scholar] [CrossRef]
- Budhraja, K.K.; McDonald, B.R.; Stephens, M.D.; Contente-Cuomo, T.; Markus, H.; Farooq, M.; Favaro, P.F.; Connor, S.; Byron, S.A.; Egan, J.B. Analysis of fragment ends in plasma DNA from patients with cancer. medRxiv 2021. [Google Scholar] [CrossRef]
- Jiang, P.; Sun, K.; Peng, W.; Cheng, S.H.; Ni, M.; Yeung, P.C.; Heung, M.M.; Xie, T.; Shang, H.; Zhou, Z.; et al. Plasma DNA End-Motif Profiling as a Fragmentomic Marker in Cancer, Pregnancy, and Transplantation. Cancer Discov. 2020, 10, 664–673. [Google Scholar] [CrossRef]
- Zhitnyuk, Y.V.; Koval, A.P.; Alferov, A.A.; Shtykova, Y.A.; Mamedov, I.Z.; Kushlinskii, N.E.; Chudakov, D.M.; Shcherbo, D.S. Deep cfDNA fragment end profiling enables cancer detection. Mol. Cancer 2022, 21, 1–5. [Google Scholar] [CrossRef]
- Szyf, M.; Pakneshan, P.; Rabbani, S.A. DNA methylation and breast cancer. Biochem. Pharmacol. 2004, 68, 1187–1197. [Google Scholar] [CrossRef]
- Hon, G.C.; Hawkins, R.D.; Caballero, O.L.; Lo, C.; Lister, R.; Pelizzola, M.; Valsesia, A.; Ye, Z.; Kuan, S.; Edsall, L.E.; et al. Global DNA hypomethylation coupled to repressive chromatin domain formation and gene silencing in breast cancer. Genome Res. 2011, 22, 246–258. [Google Scholar] [CrossRef]
- Koch, A.; Joosten, S.C.; Feng, Z.; De Ruijter, T.C.; Draht, M.X.; Melotte, V.; Smits, K.M.; Veeck, J.; Herman, J.G.; Van Neste, L.; et al. Analysis of DNA methylation in cancer: Location revisited. Nat. Rev. Clin. Oncol. 2018, 15, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Angeles, A.K.; Janke, F.; Bauer, S.; Christopoulos, P.; Riediger, A.L.; Sültmann, H. Liquid Biopsies beyond Mutation Calling: Genomic and Epigenomic Features of Cell-Free DNA in Cancer. Cancers 2021, 13, 5615. [Google Scholar] [CrossRef]
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; CCGA Consortium. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef]
- Widschwendter, M.; Evans, I.; Jones, A.; Ghazali, S.; Reisel, D.; Ryan, A.; Gentry-Maharaj, A.; Zikan, M.; Cibula, D.; Eichner, J.; et al. Methylation patterns in serum DNA for early identification of disseminated breast cancer. Genome Med. 2017, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fiegl, H.; Millinger, S.; Mueller-Holzner, E.; Marth, C.; Ensinger, C.; Berger, A.; Klocker, H.; Goebel, G.; Widschwendter, M. Circulating Tumor-Specific DNA: A Marker for Monitoring Efficacy of Adjuvant Therapy in Cancer Patients. Cancer Res. 2005, 65, 1141–1145. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.-K.; Su, C.-M.; Lin, S.-Y.; Thu, L.T.A.; Liew, P.-L.; Chen, J.-Y.; Tzeng, H.-E.; Liu, Y.-R.; Chang, T.-H.; Lee, C.-Y.; et al. Hypermethylation of TMEM240 predicts poor hormone therapy response and disease progression in breast cancer. Mol. Med. 2022, 28, 1–20. [Google Scholar] [CrossRef]
- Moss, J.; Zick, A.; Grinshpun, A.; Carmon, E.; Maoz, M.; Ochana, B.; Abraham, O.; Arieli, O.; Germansky, L.; Meir, K.; et al. Circulating breast-derived DNA allows universal detection and monitoring of localized breast cancer. Ann. Oncol. 2020, 31, 395–403. [Google Scholar] [CrossRef]
- Pedersen, C.A.; Cao, M.D.; Fleischer, T.; Rye, M.B.; Knappskog, S.; Eikesdal, H.P.; Lønning, P.E.; Tost, J.; Kristensen, V.N.; Tessem, M.-B.; et al. DNA methylation changes in response to neoadjuvant chemotherapy are associated with breast cancer survival. Breast Cancer Res. 2022, 24, 1–14. [Google Scholar] [CrossRef]
- Chrisanthar, R.; Knappskog, S.; Løkkevik, E.; Anker, G.; Østenstad, B.; Lundgren, S.; Berge, E.O.; Risberg, T.; Mjaaland, I.; Mæhle, L.; et al. CHEK2 Mutations Affecting Kinase Activity Together With Mutations in TP53 Indicate a Functional Pathway Associated with Resistance to Epirubicin in Primary Breast Cancer. PLoS ONE 2008, 3, e3062. [Google Scholar] [CrossRef]
- Klajic, J.; Busato, F.; Edvardsen, H.; Touleimat, N.; Fleischer, T.; Bukholm, I.; Børresen-Dale, A.-L.; Lønning, P.E.; Tost, J.; Kristensen, V.N. DNA Methylation Status of Key Cell-Cycle Regulators Such as CDKNA2/p16 and CCNA1 Correlates with Treatment Response to Doxorubicin and 5-Fluorouracil in Locally Advanced Breast Tumors. Clin. Cancer Res. 2014, 20, 6357–6366. [Google Scholar] [CrossRef]
- Luo, Y.; Huang, J.; Tang, Y.; Luo, X.; Ge, L.; Sheng, X.; Sun, X.; Chen, Y.; Zhu, D. Regional methylome profiling reveals dynamic epigenetic heterogeneity and convergent hypomethylation of stem cell quiescence-associated genes in breast cancer following neoadjuvant chemotherapy. Cell Biosci. 2019, 9, 16. [Google Scholar] [CrossRef] [PubMed]
- Pineda, B.; Diaz-Lagares, A.; Pérez-Fidalgo, J.A.; Burgués, O.; González-Barrallo, I.; Crujeiras, A.B.; Sandoval, J.; Esteller, M.; Lluch, A.; Eroles, P. A two-gene epigenetic signature for the prediction of response to neoadjuvant chemotherapy in triple-negative breast cancer patients. Clin. Epigenetics 2019, 11, 33. [Google Scholar] [CrossRef] [PubMed]
- Constâncio, V.; Nunes, S.P.; Henrique, R.; Jerónimo, C. DNA Methylation-Based Testing in Liquid Biopsies as Detection and Prognostic Biomarkers for the Four Major Cancer Types. Cells 2020, 9, 624. [Google Scholar] [CrossRef] [PubMed]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef]
- Akram, M.; Iqbal, M.; Daniyal, M.; Khan, A.U. Awareness and current knowledge of breast cancer. Biol. Res. 2017, 50, 33. [Google Scholar] [CrossRef]
- Xiong, Q.; Zhang, Y.; Li, J.; Zhu, Q. Small Non-Coding RNAs in Human Cancer. Genes 2022, 13, 2072. [Google Scholar] [CrossRef]
- Zhang, M.; Bai, X.; Zeng, X.; Liu, J.; Liu, F.; Zhang, Z. circRNA-miRNA-mRNA in breast cancer. Clin. Chim. Acta 2021, 523, 120–130. [Google Scholar] [CrossRef]
- Crick, F. Central Dogma of Molecular Biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef]
- Afzal, S.; Hassan, M.; Ullah, S.; Abbas, H.; Tawakkal, F.; Khan, M.A. Breast Cancer; Discovery of Novel Diagnostic Biomarkers, Drug Resistance, and Therapeutic Implications. Front. Mol. Biosci. 2022, 9, 783450. [Google Scholar] [CrossRef]
- Hannafon, B.N.; Ding, W.-Q. Intercellular Communication by Exosome-Derived microRNAs in Cancer. Int. J. Mol. Sci. 2013, 14, 14240–14269. [Google Scholar] [CrossRef] [PubMed]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Motawi, T.M.K.; Sadik, N.A.H.; Shaker, O.G.; El Masry, M.R.; Mohareb, F. Study of microRNAs-21/221 as potential breast cancer biomarkers in Egyptian women. Gene 2016, 590, 210–219. [Google Scholar] [CrossRef] [PubMed]
- Thakur, S.; Grover, R.K.; Gupta, S.; Yadav, A.K.; Das, B.C. Identification of Specific miRNA Signature in Paired Sera and Tissue Samples of Indian Women with Triple Negative Breast Cancer. PLoS ONE 2016, 11, e0158946. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Ferracin, M.; Liu, C.-G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res 2005, 65, 7065–7070. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, Q.; Xu, J.; Guo, L.; Li, X. Clinical significance of serum miR-21 in breast cancer compared with CA153 and CEA. Chin. J. Cancer Res. 2013, 25, 743–748. [Google Scholar] [CrossRef]
- Sueta, A.; Fujiki, Y.; Goto-Yamaguchi, L.; Tomiguchi, M.; Yamamoto-Ibusuki, M.; Iwase, H.; Yamamoto, Y. Exosomal miRNA profiles of triple-negative breast cancer in neoadjuvant treatment. Oncol. Lett. 2021, 22, 819. [Google Scholar] [CrossRef]
- Isca, C.; Piacentini, F.; Mastrolia, I.; Masciale, V.; Caggia, F.; Toss, A.; Piombino, C.; Moscetti, L.; Barbolini, M.; Maur, M.; et al. Circulating and Intracellular miRNAs as Prognostic and Predictive Factors in HER2-Positive Early Breast Cancer Treated with Neoadjuvant Chemotherapy: A Review of the Literature. Cancers 2021, 13, 4894. [Google Scholar] [CrossRef]
- Lü, L.; Sun, J.; Shi, P.; Kong, W.; Xu, K.; He, B.; Zhang, S.; Wang, J. Identification of circular RNAs as a promising new class of diagnostic biomarkers for human breast cancer. Oncotarget 2017, 8, 44096–44107. [Google Scholar] [CrossRef] [PubMed]
- Yin, W.-B.; Yan, M.-G.; Fang, X.; Guo, J.-J.; Xiong, W.; Zhang, R.-P. Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin. Chim. Acta 2018, 487, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, D.; Diermeier, S. Circular RNAs: Potential Applications as Therapeutic Targets and Biomarkers in Breast Cancer. Non-Coding RNA 2021, 7, 2. [Google Scholar] [CrossRef]
- Sobhani, N.; Chahwan, R.; Roudi, R.; Morris, R.; Volinia, S.; Chai, D.; D’Angelo, A.; Generali, D. Predictive and Prognostic Value of Non-Coding RNA in Breast Cancer. Cancers 2022, 14, 2952. [Google Scholar] [CrossRef]
- Sadovska, L.; Zayakin, P.; Eglītis, K.; Endzeliņš, E.; Radoviča-Spalviņa, I.; Avotiņa, E.; Auders, J.; Keiša, L.; Liepniece-Karele, I.; Leja, M.; et al. Comprehensive characterization of RNA cargo of extracellular vesicles in breast cancer patients undergoing neoadjuvant chemotherapy. Front. Oncol. 2022, 12, 1005812. [Google Scholar] [CrossRef] [PubMed]
- Diener, Y.; Walenda, T.; Jost, E.; Brümmendorf, T.H.; Bosio, A.; Wagner, W.; Bissels, U. MicroRNA expression profiles of serum from patients before and after chemotherapy. Genom. Data 2015, 6, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, H.; Yu, J.; Xu, L.; Pang, X.; Xiang, Q.; Liu, Q.; Cui, Y. miRNAs as therapeutic predictors and prognostic biomarkers of neoadjuvant chemotherapy in breast cancer: A systematic review and meta-analysis. Breast Cancer Res. Treat. 2022, 194, 483–505. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, H.; Li, C.; Xiang, Q.; Xu, L.; Liu, Q.; Pang, X.; Zhang, W.; Zhang, H.; Zhang, S.; et al. Circulating microRNAs as indicators in the prediction of neoadjuvant chemotherapy response in luminal B breast cancer. Thorac. Cancer 2021, 12, 3396–3406. [Google Scholar] [CrossRef]
- Di Cosimo, S.; Appierto, V.; Pizzamiglio, S.; Tiberio, P.; Iorio, M.V.; Hilbers, F.; de Azambuja, E.; de la Peña, L.; Izquierdo, M.; Huober, J.; et al. Plasma miRNA Levels for Predicting Therapeutic Response to Neoadjuvant Treatment in HER2-positive Breast Cancer: Results from the NeoALTTO Trial. Clin. Cancer Res. 2019, 25, 3887–3895. [Google Scholar] [CrossRef]
- Stevic, I.; Müller, V.; Weber, K.; Fasching, P.A.; Karn, T.; Marmé, F.; Schem, C.; Stickeler, E.; Denkert, C.; Van Mackelenbergh, M.; et al. Specific microRNA signatures in exosomes of triple-negative and HER2-positive breast cancer patients undergoing neoadjuvant therapy within the GeparSixto trial. BMC Med. 2018, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- Zangouei, A.S.; Alimardani, M.; Moghbeli, M. MicroRNAs as the critical regulators of Doxorubicin resistance in breast tumor cells. Cancer Cell Int. 2021, 21, 213. [Google Scholar] [CrossRef]
- Bahrami, A.; Aledavood, A.; Anvari, K.; Hassanian, S.M.; Maftouh, M.; Yaghobzade, A.; Salarzaee, O.; ShahidSales, S.; Avan, A. The prognostic and therapeutic application of microRNAs in breast cancer: Tissue and circulating microRNAs. J. Cell Physiol. 2017, 233, 774–786. [Google Scholar] [CrossRef]
- Zhu, Z.; Li, X.; Dong, H.; Ke, S.; Zheng, W.-H. Let-7f and miRNA-126 correlate with reduced cardiotoxicity risk in triple-negative breast cancer patients who underwent neoadjuvant chemotherapy. Int. J. Clin. Exp. Pathol. 2018, 11, 4987–4995. [Google Scholar]
- Guarneri, V.; de Azambuja, E. Anthracyclines in the treatment of patients with early breast cancer. ESMO Open 2022, 7, 100461. [Google Scholar] [CrossRef]
- Lisencu, L.A.; Roman, A.; Visan, S.; Bonci, E.-A.; Pașca, A.; Grigorescu, E.; Mustea, E.; Cismaru, A.; Irimie, A.; Lisencu, C.; et al. The Role of miR-375-3p, miR-210-3p and Let-7e-5p in the Pathological Response of Breast Cancer Patients to Neoadjuvant Therapy. Medicina 2022, 58, 1494. [Google Scholar] [CrossRef]
- To, N.H.; Nguyen, H.Q.; Thiolat, A.; Liu, B.; Cohen, J.; Radosevic-Robin, N.; Belkacemi, Y. Radiation therapy for triple-negative breast cancer: Emerging role of microRNAs as biomarkers and radiosensitivity modifiers. A systematic review. Breast Cancer Res. Treat. 2022, 193, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Ryspayeva, D.; Halytskiy, V.; Kobyliak, N.; Dosenko, I.; Fedosov, A.; Inomistova, M.; Drevytska, T.; Gurianov, V.; Sulaieva, O. Response to neoadjuvant chemotherapy in breast cancer: Do microRNAs matter? Discov. Oncol. 2022, 13, 43. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xiao, H.; Li, J.; Yang, Z.; Jiang, J.; Ji, J.; Peng, C.; He, Y. Graphene Oxide-Based Highly Sensitive Assay of Circulating MicroRNAs for Early Prediction of the Response to Neoadjuvant Chemotherapy in Breast Cancer. Anal. Chem. 2022, 94, 16254–16264. [Google Scholar] [CrossRef] [PubMed]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic re-views. BMJ. 2021, 372, 71. [Google Scholar] [CrossRef]
| CTCs Detection Rate before NACT | CTCs Detection Rate after NACT | Results | |
|---|---|---|---|
| IMENEO [57,60] | 25% | 17% | OS (p < 0.001; HR 3.93, 2.00–5.45), DDFS (p < 0.001; HR 3.73, 2.82–4.90), LRFI (p < 0.001, HR 3.02; 1.88–4.75)) |
| GeparQuattro [61,62] | 22% | 10% | DMFS (p < 0.001; HR, 3.72, 1.89–7.32), OS 0 (p < 0.01; HR 4.54, 1.97–10.49) |
| REMAGUS 02 [63] | 23% | 17% | RFS p = 0.013 (HR N/A) |
| NEOALTTO [58] | 11% | N/A | No prognostic results |
| NEOZOTAC [64] | 18% | N/A | No prognostic results |
| MD Anderson [65] | N/A | 27% | RFS (p = 0.03; HR 5.25, 1.34–20.56), OS (p = 0.03; HR 7.04, 1.26–39.35) |
| MD Anderson [55] | 54% | N/A | PFS (p = 0.02; HR 0.60, 0.37–0.98), OS (p = 0.03; HR 0.59, 0.35–1.00) * |
| MD Anderson [66] | 24% | N/A | PFS (p = 0.005; HR 4.62, 1.79–11.9), OS (p = 0.01; HR 4.04, 1.28–12.8) |
| BEVERLY-2 [67] | 35% | 7% | DFS (p = 0.01; HR 3.69, 1.34–10.21) |
| BEVERLY-1/-2 [56] | 39% | 9% | DFS (p < 0.01; HR 2.80, 1.65–4.76) OS (p < 0.01; HR N/A) |
| Serrano et al. [68] | 70.08% (10 mL) | 54.1% (10 mL) | OS (p = 0023; HR N/A) |
| Janni et al. [69] | 20.2% | N/A | OS (p < 0.001; HR 1.44, 1.81–3.29), DFS (p < 0.001; HR 2.08, 1.69–2.56), DDFS(p < 0.001; HR 2.20, 1.74–2.78), BCSS (p < 0.001; HR 2.54, 1.910–3.38) |
| Trial | Patients (n) | Objective | Status |
|---|---|---|---|
| NCT04239105 | N/A | To develop a eBC screening test. To evaluate the efficacy of NACT and prognosis. | Not yet recruiting |
| NCT03511859 | 210 | To develop a eBC screening test. | Unknown, not yet posted |
| NCT01322750 | 3125 | To develop simple, reliable, cost-effective, and clinically relevant breast cancer screening test. | Recruiting |
| NCT03842176 | 90 | To monitor response during neo/adjuvant treatment. | Unknown |
| NCT03709134 | 100 | To investigate the role of CTCs predicting response to NACT. | Recruiting |
| NCT05326295 | 1000 | To predict treatment response of NACT, surgery and adjuvant chemotherapy. To evaluate the prognostic role of CTCs. | Recruiting |
| NCT04993014 | 80 | To predict treatment response of NACT, surgery and adjuvant chemotherapy. To evaluate the prognostic role of CTCs. | Recruiting |
| NCT04059003 | 200 | To evaluate changes of CTCs and the efficacy of NACT for TNBC. | Recruiting |
| Trial | Participant | Patients Characteristics | Biological Rationale | Endpoints | Status |
|---|---|---|---|---|---|
| NCT03881384 | 200 | eBC | ctDNA clearance level during NACT and detection of MRD after surgery | ctDNA detection and clearance during NACT | Enrolling |
| NCT04276337 | 50 | HER-2+ stage III eBC | ctDNA monitoring during during NACT (TCHP Regimen) | pCR rate | Active, Not Recruiting |
| NCT05050890 | 38 | eBC | ctDNA monitoring during during NACT | ctDNA detection and clearance during NACT | Active, Not Recruiting |
| NCT02546232 | 196 | I-IV stage BC | ctDNA analysis and molecular characterization for for the Optimal Selection of Treatment Regimens (NACT or treatments for aBC) | Correlate molecular changes to pathological response | Unknown |
| NCT03709134 | 100 | eBC | role of CTCs and ctDNA in predicting response to NAC, | pCR rate | Recruiting |
| NCT04223492 | 100 | eBC | Combination of standard screening techniques to liquid biopsy (CTCs, ctDNA) | pCR rate | Unknown |
| NCT03973034 | 300 | healthy subjects, benign breast tumors, eBC | ctDNA test model for early screening of breast cancer | Diagnosis rate | Unknown |
| NCT03085888 | 99,481 | healthy subjects involved in mammogram screening | Detection of breast and other invasive cancers analyzing cfDNA | Performance of the detection test | Active, Not Recruiting |
| NCT04241796 | 6662 | healthy subjects | cfDNA and machine learning to detect a common cancer signal across >50 cancer types | Performance of Multi-Cancer Early Detection Test | Completed |
| NCT04972201 | 2305 | healthy subjects | cfDNA mutation, miRNA, DNA methylation assays to detect cancer | Sensitivity for cancer detection and tissue of origin of the assays | Recruiting |
| NCT05227261 | 1643 | healthy subjects | Anticipation of cancer diagnosis | Positive predictive value, Negative predictive value of the blood ctDNA test in early detecting cancers | Recruiting |
| NCT05235009 | 500 | healthy subjects | Multi-cancer early detection | Sensitivity and specificity of the test | Recruiting |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gianni, C.; Palleschi, M.; Merloni, F.; Bleve, S.; Casadei, C.; Sirico, M.; Di Menna, G.; Sarti, S.; Cecconetto, L.; Mariotti, M.; et al. Potential Impact of Preoperative Circulating Biomarkers on Individual Escalating/de-Escalating Strategies in Early Breast Cancer. Cancers 2023, 15, 96. https://doi.org/10.3390/cancers15010096
Gianni C, Palleschi M, Merloni F, Bleve S, Casadei C, Sirico M, Di Menna G, Sarti S, Cecconetto L, Mariotti M, et al. Potential Impact of Preoperative Circulating Biomarkers on Individual Escalating/de-Escalating Strategies in Early Breast Cancer. Cancers. 2023; 15(1):96. https://doi.org/10.3390/cancers15010096
Chicago/Turabian StyleGianni, Caterina, Michela Palleschi, Filippo Merloni, Sara Bleve, Chiara Casadei, Marianna Sirico, Giandomenico Di Menna, Samanta Sarti, Lorenzo Cecconetto, Marita Mariotti, and et al. 2023. "Potential Impact of Preoperative Circulating Biomarkers on Individual Escalating/de-Escalating Strategies in Early Breast Cancer" Cancers 15, no. 1: 96. https://doi.org/10.3390/cancers15010096
APA StyleGianni, C., Palleschi, M., Merloni, F., Bleve, S., Casadei, C., Sirico, M., Di Menna, G., Sarti, S., Cecconetto, L., Mariotti, M., & De Giorgi, U. (2023). Potential Impact of Preoperative Circulating Biomarkers on Individual Escalating/de-Escalating Strategies in Early Breast Cancer. Cancers, 15(1), 96. https://doi.org/10.3390/cancers15010096







