Simple Summary
Acute lymphoblastic leukemia (ALL) is characterized by a large number of chromosomal, structural aberrations associated with risk stratification and treatment outcome. However, conventional karyotyping, FISH and PCR have many limitations in detecting chromosomal aberrations. The aim of our study was to assess the potential added value of optical genomic mapping (OGM) for identifying chromosomal aberrations. The chromosomal aberrations of 46 children with B-cell ALL were determined by OGM, and the results of OGM were compared with those of conventional techniques. We found that OGM could detect most clinically significant chromosomal aberrations, and that it has a strong ability to detect complex chromosomal aberrations and refine complex karyotypes. In addition, several novel fusion genes and single-gene mutations, associated with important clinical features, were also identified. Our results show that OGM is highly effective in identifying chromosomal aberrations and has important implications for risk stratification of ALL and the pathogenesis of leukemia.
Abstract
Purpose: To assess the potential added value of Optical Genomic Mapping (OGM) for identifying chromosomal aberrations. Methods: We utilized Optical Genomic Mapping (OGM) to determine chromosomal aberrations in 46 children with B-cell Acute lymphoblastic leukemia ALL (B-ALL) and compared the results of OGM with conventional technologies. Partial detection results were verified by WGS and PCR. Results: OGM showed a good concordance with conventional cytogenetic techniques in identifying the reproducible and pathologically significant genomic SVs. Two new fusion genes (LMNB1::PPP2R2B and TMEM272::KDM4B) were identified by OGM and verified by WGS and RT-PCR for the first time. OGM has a greater ability to detect complex chromosomal aberrations, refine complicated karyotypes, and identify more SVs. Several novel fusion genes and single-gene alterations, associated with definite or potential pathologic significance that had not been detected by traditional methods, were also identified. Conclusion: OGM addresses some of the limitations associated with conventional cytogenomic testing. This all-in-one process allows the detection of most major genomic risk markers in one test, which may have important meanings for the development of leukemia pathogenesis and targeted drugs.
1. Introduction
Acute lymphoblastic leukemia (ALL) is the most common malignancy in children, of which acute B-lymphoblastic leukemia accounts for about 85% of the patients [1]. Studies have shown that there are numerous structural variations (SVs) in leukemic cells. Many SVs correlate to the drug resistance of leukemic cells, or disease occurrence and progression [2,3]. For example, KMT2A (MLL) translocations, t(17;19)/TCF3::HLF, haploidy or low hypodiploidy are high-risk biomarkers, t(9;22)/BCR::ABL1 patients require targeted treatment (imatinib/dasatinib), whereas iAMP21 patients achieve better outcomes when treated intensively. Clarification of cancer cell behavior and further personalization of treatment require precise identification of SVs.
Nowadays, there are some cytogenetic and molecular methods used in SV detection, with different advantages and limitations. Methods used to detect genetic aberrations vary in their resolutions, which may affect the more precise assignment of the gene being tested [4]. For instance, karyotyping analysis allows for the identification of balanced and unbalanced structural anomalies, limited by the proliferation index of the blast, as well as by the poor quality of the metaphases obtained and the low resolution (about 5 Mbp) [5]. Especially in ALL, the low proliferative index of blasts is the cause of common karyotype failure [5]. FISH has a resolution of about 200 Kbp. However, it is targeted and limited in known regions, and cannot achieve a comprehensive genome-wide detection, especially in ALL. The next-generation sequencing (NGS) technology has greatly enhanced resolution and throughput of detection and the promoted discovery of mutations at single or several base pairs. However, the detection of large SVs, such as inversion, is still difficult due to the short reads sequenced. Due to the various forms of gene variation, the determination of genes involved in known, recurrent karyotype abnormalities, mainly comes from the inference of prior knowledge. For example, t (10;11) (p12; q23) mostly forms the KMT2A::MLLT10 fusion gene, but in rare cases may also be cause KMT2A::NEBL fusion. Thus, a single diagnostic assay that easily identifies clinically significant SVs is highly desirable.
Optical genomic mapping (OGM) is a technology for high-resolution genome reconstruction from single enzyme-labelled DNA molecules. It can detect balanced and unbalanced translocations, CNVs in a range of few kb up to whole chromosomes (aneuploidies), as well as genomic insertions and inversions [6,7,8]. OGM is based on the imaging of labeled and linearized ultra-high molecular weight (UHMW) DNA. Accurate and precise patterns of labels allow us to de novo assemble the human genome, which is compared to the reference genome map, and extract aberrant molecules from alignments, followed by the generation of local consensus, in order to detect SVs.
Here, we describe a clinical validation study to investigate the genetic aberrations of 46 pediatric B-ALL samples using OGM, karyotyping, FISH, WGS and PCR. By analyzing the detection results of OGM, we demonstrate the feasibility of OGM for the detection of well-established, as well as new putative SVs, in ALL.
2. Materials and Methods
2.1. Study Design
Forty-six patients with newly diagnosed B-ALL, admitted into Hematology/Oncology Center of Beijing Children’s Hospital, Capital Medical University from June 2019 to June 2020, were enrolled in this study. The children were divided into low-, intermediate- and high-risk groups according to the Chinese Children’s Leukemia Collaborative Group (CCLG)-ALL-2008 regimen [9]. Heparinized bone marrow (BM) samples were subjected to routine genetic diagnostic testing (karyotyping, FISH, and PCR). Clinical data, such as patients’ clinical characteristics, treatment and outcome, were retrieved from medical records. This study has been approved by Institutional Review Board of Beijing Children’s Hospital (IEC-C-008-A08-V.05.1) and all patients signed informed consent.
2.2. UHMW DNA Isolation, Quantification and Labeling for Optical Genome Mapping
UHMW gDNA extraction from frozen bone marrow aspirates (BMA) was performed following the manufacturer’s protocols (Bionano Genomics, San Diego, CA, USA). For each sample, 1 mL frozen BMA was used as initial material, and a minimum of 1.5 million white blood cells was used as input to isolate UHMW DNA. Briefly, WBC were centrifuged and lysed by Proteinase K, RNase A in Lysis and Binding Buffer (LBB). DNA was precipitated with isopropanol and bound with a nanobind magnetic disk. Bound UHMW DNA was resuspended in the elution buffer. We used QubitTM dsDNA BR Assay Kit with a Qubit 3.0 Fluorometer (ThermoFisher Scientific, Waltham, MA, USA) to quantify the gDNA. The gDNA isolation was considered successful when the DNA concentration was equal to or above 36 ng/μL and the coefficient of variation (CV) was <0.3. A total of 750 ng UHMW gDNA was labeled specifically, according to the manufacturer’s guidelines, by using the Bionano Prep Direct Label and Stain (DLS) Protocol. The labeled UHMW gDNA was loaded onto the Saphyr chip for linearization and imaging, and the Saphyr chip was operated at maximum capacity with real-time throughput and quality metrics. According to the manufacturer’s instructions, quality and run parameters included: (1) the total DNA collected ≥150 kb; (2) the map rate (the % of Bionano molecules that align to the reference); (3) the N50 (≥150 kb); (4) the average label density (in labels/100 kb); (5) the positive and negative label variance (indicating the percentage of the labels absent in the reference and the percentage of reference labels absent in the molecules, respectively); (6) the effective coverage of the reference.
2.3. Structural Variant Calling and Variant Filtering
Variant calling was executed, enabling SV and CNV detection, with the rare variant pipeline (RVP) included in Bionano Solve (v.3.7). The results were analyzed through two distinct pipelines: a CNV pipeline that allows for the detection of large, unbalanced aberrations, based on normalized molecule coverage, and an SV pipeline that compares the labeling patterns between the constructed sample genome maps and a reference genome map. Reporting and direct visualization of SVs were performed with Bionano Access software v.1.7. In order to assess rare SVs only, we filtered out calls present in an OGM dataset of 180 human control samples provided by Bionano Genomics. The software represents the results from both pipelines in a circos plot, a tool which allows for an easy overview of the detected variants at a glance. Of note, the software calls ‘duplications’ that are smaller than 30 kb ‘insertions’, and ‘inversions’ involving segments of 5 Mb or larger are called ‘intra-chromosomal translocations’.
2.4. Comparison of Clinically Significant SVs/CNVs Identified by Conventional Testing
To compare OGM data with standard workflows, we used a visual data presentation consisting of circos plots and individual genome browser views. For data filtering, the variant hg38 DLE-1 SV mask, which blocks difficult-to-map regions and common artifacts, was turned on and the following recommended confidence scores were applied: insertion, 0; deletion, 0; inversion, 0.7; duplication, −1; intra- and inter-translocation, 0.05; and copy number, 0.99 (low stringency, filter set to 0). Per sample, prefiltered data were downloaded as SMAP files for SVs and CNVs separately. These SMAP files were used to determine the number and types of aberrations per sample. With the a priori knowledge that OGM reveals structural complexity undiscernible by karyotyping [10,11], we sought to focus exclusively on SVs and CNVs of potential clinical significance. SVs with a variant allele frequency (VAF) of <10% (equivalent to the presence of SVs in 20% of cell fraction) were considered outside the scope of this study. Of note, ‘Whole genome SV and CNV’ views were only enabled in the latest Bionano Access software version, 1.7, to show SVs and CNVs on different chromosomes (Figure 1).

Figure 1.
View of the distribution of SVs and CNVs on chromosomes of sample 48. Chromosome 7 and 21 missed or gained one chromatid, respectively. Each chromosome is divided into color-coded bands. The short horizontal lines in different colors on the left side of the chromosome represent the SV composition, including deletion, insertion, duplication, translocation, and inversion, respectively, while those on the right side of the chromosome represent gain or loss of CNV in different bands.
2.5. Confirmation of Additional SVs with Whole-Genome Sequencing
Whole-genome sequencing (WGS) was used to confirm the existence of extra SVs. These were identified by OGM, have with potentially clinical significance and were found via the MGI Tech (DNBSEQ-T7) platform [12]. A MGIEasy FS DNA prep kit (BGI, Beijing, China) was used for WGS library construction according to the manufacturer’s instructions. Paired-end sequencing was performed on a DNBSEQ-T7 sequencing instrument, yielding ~150 bp-sized sequencing reads. A raw data quality check was conducted using FastQC (version 0.11.9). Base quality information was obtained from the FastQC results. Then, the filtered files were mapped to the reference human genome (hg38), and the output BAM files were sorted using samtools sort. The following criteria were used to determine whether SVs, detected independently by OGM and WGS, refer to the same event: (1) deletions, insertions, and duplications, detected by WGS, must overlap with the SV interval defined by optical mapping by at least 50%, and the difference in size predicted by the two methods must be less than 30%; (2) For translocation and inversion, the breakpoint detected by WGS must be within 500 Kb of the breakpoint detected by optical mapping, and the SV direction determined by the two methods must be consistent.
Then, GeneFuse software was used to detect gene fusions directly from the original FastQC files, eliminating the influence of alignment results. GeneFuse was able to visualize the detected fusion with the supported reads and inferred fusion protein structures [13].
2.6. Verification of LMNB1::PPP2R2B and TMEM272::KDM4B Putative Fusion Genes
IGVTools (version 2.5.1, http://www.broadinstitute.org/igv, accessed on date 4 January 2019) was used to check the MGI Tech (DNBSEQ-T7) BAM files and extract LMNB1-PPP2R2B exon sequences. Based on the predicted sequence of the fusion mRNA, a plasmid, encoding adjacent sequences of the fusion site, was constructed with GV219, and its digested product with BamHI was used as a positive control. The LMNB1-PPP2R2B fusion sequence was amplified by cDNA-based PCR in 396 ALL cDNA samples, and agarose gel (concentration: 2%) electrophoresis was performed. The sequences of PCR primers were as follows: F: 5′-AGCTGCTCCTCAAGCTATGC-3′; R: 5′-AAGCTGTGGAAAGTCAGCGA-3′ (product size: 220bp). We verified the amplified products with Sanger sequencing.
TMEM272::KDM4B fused mRNA was detected by cDNA-based PCR amplification in sample 66. The sequences of PCR primers were as follows: F: 5′-ACAATGCCAGGAGGTCTGGA-3′; R: 5′-AGGATTTGTCAGGTGCCTCC-3′ (product size: 98bp). The expression of KDM4B mRNA was quantified by the SYBR Green method, with primers as follows: F: 5′-AAGGCCAAGTTCATCTCCTCCGTC-3′; R: 5′-TGCTCAGTGACAGCCGAGAGCGGA-3′ [14].
2.7. Statistics
Associations among categorical values were examined using the Chi-square test or a two-sided Fisher’s exact test. The correlation between SVs and clinical features was analyzed by a Spearman correlation test. Analyses and chart production were performed by using R version 3.4.1. p < 0.05 was considered statistically significant.
3. Results
3.1. Clinical Characteristics of Patients and Technical Characteristics of OGM
3.1.1. Patient Characteristics
This study enrolled 20 male (43.5%) and 26 female (56.5%) patients with median age of 4.17 years (1.4–14.7 years). There were 41 patients (90.7%) with the diagnosis of common-B-ALL and 5 patients (9.3%) with pre-B-ALL. There were 13 cases with the ETV6::RUNX1 fusion gene. IKZF1 deletion, BCR::ABL and TCF3::PBX1 fusion genes were detected in 3, 2, and 2 cases, respectively. Three patients carried KMT2A::MLLT3, KMT2A::AF4 fusion genes, and iAMP21, respectively. Based on the CCLG-ALL-2008 protocol, patients were divided into standard- (12 cases), intermediate- (24 cases) and high-risk (10 cases) groups. Two cases in the intermediate-risk group died of acute intracranial hemorrhage and bone marrow suppression with severe infection, respectively.
3.1.2. Raw Data Quality and SV/CNV Callings in OGM
We first evaluated the technical performance of the OGM analysis, which resulted in an average N50 of 260 Kbp (212–351 Kbp), average map rate of 82.6% (60.1–92.0%) and average effective coverage of 420.5X (95.15–667.87X). Thus, all the samples conformed with the requirements of quality control of OGM (Supplementary Table S1). In total, we identified 71,534 SVs and 1592 CNVs in 46 leukemia samples (Table 1 and Table S2).

Table 1.
SVs and CNVs calling in OGM.
Per sample, an average of 1555.1 SVs was detected in total, comprising 671.6 insertions, 632.6 deletions, 67.5 inversions, 179.5 duplications, 1.5 intra-chromosomal translocations and 2.3 inter-chromosomal translocations. Of all identified SVs in the total cohort, 2204 were rare (Table 1 and Table S2). This represented, on average 47.0 per sample, including 11.2 insertions, 28.3 deletions, 0.7 inversion, 3.1 duplications, 2.2 intra-chromosomal translocations and 1.5 inter-chromosomal translocations. Regarding the 1592 CNVs calls (1189 gains and 403 losses), there were 34.5 CNVs per sample including 25.8 (range: 0–131) non-masked gains and 8.7 (range 0–137) non-masked losses (Table 1 and Table S2). It was noteworthy that the RVP analysis automatically masked regions of the genome with unusually high variance in their relative coverage across control datasets (including centromeric and telomeric regions), assuming that high variance regions may be regions of high CNV occurrence in normal healthy individuals [15].
3.2. Concordance between OGM and Conventional Cytogenetic Results
We determined molecular and cytogenetic aberrations in the enrolled patients with conventional technologies, including FISH, PCR, and karyotyping. All 46 cases had FISH/PCR results; 45 cases had G-banded karyotype results. Conventional cytogenetic technologies detected 9 types of fusion genes in 19 samples, IKZF1 gene deletions in 3 cases, and copy number gains of 5 genes in 15 samples, as shown in Table 2, Table 3 and Table 4.

Table 2.
Additional SVs and CNVs, Identified by OGM but not by Conventional Technologies.

Table 3.
Refining Cytogenetic Breakpoints and Resolving Unknown Cytogenetic Elements by OGM.

Table 4.
OGM identified all SVs detected by conventional technologies.
3.2.1. OGM Reaches 100% True Positive Rate for Known Aberrations, except for Specific Gene Regions
We divided the cytogenetic status, revealed by conventional technologies, into three categories (Table 5): negative karyotype (17 cases), aneuploidies only (18 cases) and trans locations and/or aneuploidies (11 cases). OGM identified most of the clinically relevant genomic abnormalities detected by the conventional technologies (Table 2, Table 3 and Table 4), except for P2RY8::CRLF2 fusion gene resulting from a micro-deletion in three patients (case 48, 51, 76; Table 3 and Table 4). The microdeletion-induced P2RY8::CRLF2 fusion gene occurred in the pseudoautosomal region “PAR” in Xp22.33, which was not contiguously covered by OGM.

Table 5.
Result comparison between OGM and conventional technologies.
3.2.2. Refinement of Abnormal Karyotypes and Resolution of Complex Genome by OGM
In line with the advantage of higher sensitivity and resolution of OGM, our data showed that OGM was able to refine the karyotype (Table 2 and Table 3). In sample 48, besides iAMP21 revealed by FISH, OGM further detected a chromothripsis of chromosome 21 (Figure 2). Intra-chromosomal amplification of chromosome 21 (iAMP21) defines a subgroup of pediatric B-ALL, characterized by multiple structural abnormalities of amplification, inversion and deletion, and which has a poor prognosis with standard therapy [16]. The circus plot illustrated the shattering of chromosome 21, resulting in large-scale intra-chromosomal rearrangements. Furthermore, the overall copy number of chr21 was more than 3, and the copy number in the region from 33.2Mb to 39.2Mb, where RUNX1 lies, was 6. Array-based comparative genomic hybridization (aCGH) revealed that the gene amplification/deletion patterns of abnormal chromosome 21 were significantly different among patients [17]. Consistent with previous studies [17,18], at the OGM-CNV interface, we also see a classical stepwise rise in copy number around the 33–45 Mb region, followed by a sharp drop off to deletion or normal diploid levels. These patterns were consistent with the classical breakage—fusion—bridge (BFB) cycle of oncogene amplification [19]. At the same time, we also observed some secondary genetic changes in this sample, including −7 (5%) [18], P2RY8::CRLF2 (17%) and IKZF1 (22%) deletion [20], etc. By comparing copy number profiles from the OGM with those from previous microarray studies, the altered complexity observed by the OGM brings an intuitive visual illustration of the BFB mechanism.
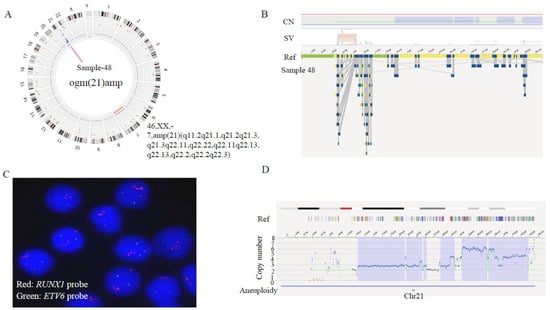
Figure 2.
Identification of chromothripsis structures of chromosome 21 in sample 48 by OGM. (A) The circus plot illustrated the shattering of chromosome 21, leading to large-scale intra-chromosomal rearrangements. In addition, a monosomy of chromosome 7 was identified (indicated by the red line at the innermost layer of the circus). (B) Zoom-in to chromosome 21, showing the aberrant CNV profile(top) and maps of the rare variants (bottom). (C) Metaphase-FISH indicated 5 or more RUNX1 signals (red). (D) OGM-CNV profile: the green line presents the copy number metrics of each fragment, which means different levels of gain at various breakpoints: 33.2–38.1 Mb fragment with CN = 6, 39.2–39.9 Mb fragment with CN = 4, 39.9–40.8 Mb fragment with CN = 3, 40.8–45.5 Mb fragment with CN = 4.
On the other hand, high-resolution OGM uncovered the complexity of structural variations and heterogeneity in breakpoint regions that were difficult to resolve with conventional cytogenetic technologies. In five cases with normal karyotype (case 41, 80, 97, 101, 109), OGM showed large copy number changes, ranging from 18,514 bp to 133,785,261 bp on different chromosomes (Table 2). In case 47, karyotyping showed the additional genetic material of unknown origin at 1q21. OGM revealed that some fragments of chromosome 1 amplified and translocated intra-chromosomally. Thus, OGM refined the karyotype as dup(1)(q21.2q24.3-q24.3q32.3) and der(1)t(1;1)(q41;q43) (Figure 3; Table 2). In summary, OGM provides a more accurate and comprehensive insight into the genomic origin of complex karyotypes.
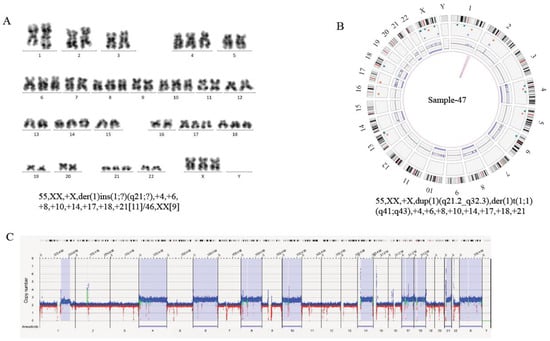
Figure 3.
Production of an Accurate Cytogenomic Karyotype by Refining Cytogenetic Breakpoints and Resolving Unknown Cytogenetic Elements by OGM. (A) Standard G-banded karyotyping in sample 47 show the karyotype as 55, XX, +X, der(1)ins(1;?)(q21;?),+4,+6,+8,+10,+14,+17,+18,+21[11]/46,XX[9]. (B) Circus plot, showing a whole-genome view of a derivative chromosome 1 (pink lines) and copy number profiles (inner circle blue boxes indicate gains and red boxes indicate deletions). Thus, OGM identified additional intra-chromosomal translocation in chromosome 1 and refined the karyotype as 55, XX, +X, dup(1)(q21.2q24.3-q24.3q32.3), der(1) t(1;1)(q41;q43), +4,+6,+8,+10,+14,+17,+18,+21. (C) Whole-genome CNV profiles, generated with Bionano Access, indicates gains of chromosome 4, 6, 8, 10, 14, 17, 18, 21, 23. Some regions of chromosome 1 were amplified, and the y axis shows copy numbers ranging from 0–8 for each chromosome. The x axis shows increased copy numbers in blue and decreased copy numbers in red. The light blue region, highlighted for the above chromosomes, indicates a significant difference from the baseline, thus flagging a gain in those chromosomes. All chromosomes, except the sex chromosomes (Y)×0 and chromosomes (4, 6, 8, 10, 14, 17, 18, 21, 23)×3, are present in 2 copies.
3.2.3. Identification of Novel Chromosomal Alterations or Gene Fusions by OGM
OGM identified several novel chromosomal alterations or gene fusions (Table 2 and Table 3). In case 66, G-banded karyotyping revealed two distinct translocations—t(11;22) (q24.3; q12.2) and t(13;19)(q14.13;q13.3) (Figure 4C). The former leads to a FLI1::EWSR1 fusion, 15. However, the results of the latter have not been reported. OGM revealed a fusion of TEME272 on chromosome 13 and KDM4B on chromosome 19 in an inverted orientation (Figure 4). Furthermore, OGM detected three-way translocations in samples 83, 103 and 114, which were t(4;7;21) (q21.21; p15.3;q11.2), t (12;16;21) (p13.2;q24.3;q22.12) and t (5;12;21) (q11.2;p11.23; q22.12) (Figure 5). In samples 83 and 103, the above-mentioned inter-chromosomal translocations produced OSBPO3::NRIP1 and SPG7::RUNX1 fusion genes, respectively. As far as we know, the above-mentioned putative fusion genes have not been reported. These findings indicate the presence of cryptic or more complex translocations, which may be under-ascertained with current conventional detection methods. Unfortunately, due to running out of previously clinical samples collected, no raw BMA nor DNA is available for verification now. We will focus on these rare three-way translocations in future studies and further verify their clinical significance. Additionally, the above chromosomal SVs, listed in Supplementary Table S3, affect several cellular biological processes (Figure A1, detailed in Appendix A) in pediatric B-ALL.
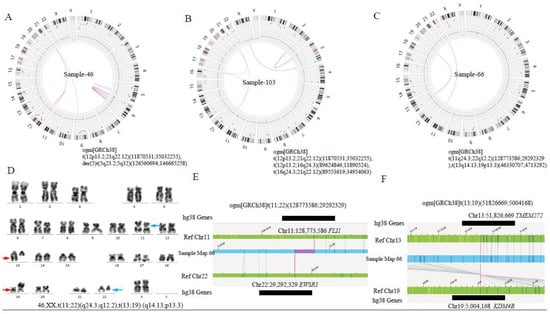
Figure 4.
Identification of additional SV findings by OGM. (A) Circus plot showed a translocation between chromosomes 12 and 21 and the internal rupture of chr5, resulting in the formation of a derivative chromosome in case 46. (B) Circus plot showed three-way translocations between chr12 and 21, chr12 and 16, and chr16 and 21, respectively, in case 103. (C) Circus plot showed two translocations between chr11 and 22, chr13 and 19 in case 66, respectively. (D) Standard G-banded karyotyping showed t (11;22) (q24.3; q12.2) (blue arrow) and t(13;19) (q14.13;q13.3) (red arrow). (E,F) Linear genome browser representation of known FLI1::EWSR1 fusion and a novel TMEM272::KDM4B fusion in case 66. Green represents GRCh38 reference chromosomes with OGM label patterns. Light blue represents assembled sample maps with label patters. Grey lines represent label alignments between two maps. Purple represents translocation breakpoints. Black lines represent the genes in this region.
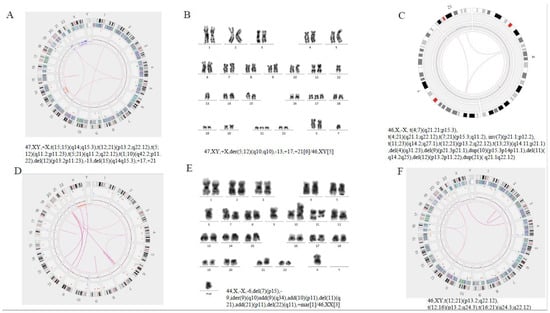
Figure 5.
(A,B) Sample 114: A G-banded karyotype was performed and initially reported as the derivative chromosomes 5 and 12, add (17), add (21), add(X), del (13) in 6 metaphases with five metaphases with a normal karyotype. Optical genome mapping data confirm all known aberrations and show the presence of additional translocations t(15;15)(q14;q15.3), t(5;12)(q11.2;p11.23), t(5;21)(q11.2;q22.12), t(1;10)(q42.2;p11. 22). (C–E) Sample 83: Optical genome mapping data showing a three-way translocation 46,XX,t(4;7;21)(q21.21;p15.3;q11.2); multiple deletions (4,9,11,12,X) and duplications (10,21) on chromosome segments were also shown, as well as translocation between multiple chromosomes. Karyotype did not indicate multiple translocations between chromosomes. (F) Sample 103: Optical genome mapping confirm a three-way translocation on chromosomes 12, 16 and 21, while the G-banded karyotype (not represented in this plot) was normal.
Taken together, OGM serves as a single-platform assay that can identify different types of chromosomal structure variations, which may be difficult to identify with conventional technologies.
3.3. Clinical Values of OGM Detection
Difference in SV Numbers among the Three Risk Groups
Accurate risk stratification is of great significance for patients’ treatment and prognosis evaluation. We divided the 46 patients with B-ALL into 3 groups based on accurate MICM and their clinical manifestations. There were 12, 24, and 10 cases in standard-, intermediate-, and high-risk groups, respectively. We found that the average number of different types of SVs were similar in different risk groups (p > 0.05). Similarly, there were no significant difference in the mean amount of CNV and aneuploidy among the three risk groups (p = 0.484 and 0.263, respectively, Figure 6). Thus, the number of chromosomal aberrations is not fully related to patients’ risk stratification, suggesting the important role of some SVs in leukemogenesis and clinical-biological features.
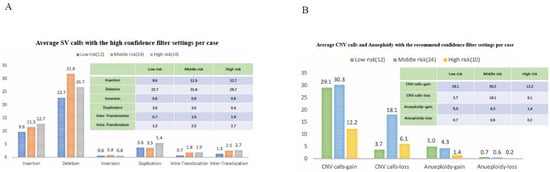
Figure 6.
Chromosomal aberrations in 46 samples, analyzed by rare variant analysis using DLS marker. (A) Average SV with high filter settings per case. Different color bars represent different risk groups. The x axis showed different SV types, including insertion, missing, inversion, repetition, and translocation. The y axis showed the number of cases. (B) Average CNV and aneuploidy, with the recommended filter settings per case. The x axis showed the gain and loss of copy numbers and aneuploidy, while the y axis corresponded to the number of cases.
As there was no significant difference in the number of SVs among different risk groups, we further investigated the correlations between recurrent SVs and common clinical characteristics such as MRD at day 15, 33 and 78, age and white blood cell count at diagnosis of the enrolled patients (Table 6 and Table 7). Regarding gene fusions, we found that AC141586.1::KCTD5, ATP10A::AC016266.1, CALCOCO2::SUMO2P17, and MIR4435.2HG::AC017002.5 were significantly positively correlated with d33 MRD; the former three fusions and PDCD6IPP1::AC138649.1 were related to risk stratification. We also found positive correlations of ETV6-AP000331.1 with d15 MRD, and of AL034430.1::SLX4IP and MKKS::SLX4IP with d78 MRD, respectively. Some candidate fusion genes, such as GRAPL::KYNUP3, ARL8B::EDEM1 and GPN3::FAM216A, were related to patients’ age, white blood cell count and the percentage of leukemic cells in peripheral blood at diagnosis (Table 6).

Table 6.
Correlations of recurrent putative fusion genes with clinical features of B-ALL in children.

Table 7.
Correlation of recurrent single-gene aberrations annotated to cosmic database with clinical features of B-ALL in children.
In the aspect of single-gene aberrations, d15 MRD was positively correlated with aberrations in NF1 and ERG. The gene d33MRD was positively correlated with abnormalities in NF1, SH2B3, IKZF1, ERG and CREBBP; d78 MRD was also positively correlated with abnormalities in KMT2A, CREBBP, BTG1 and PIK3CA. Moreover, abnormalities in CREBBP, ERG, KMT2A and SH2B3 were all correlated with risk stratification. Aberrations in BCR, ABL1 and TCF3 were all positively correlated with the number of leukocytes found at diagnosis (Table 7). In addition, IKZF1 aberrations were positively correlated with age, and the IKZF1 deletion site were, the same as 7p12.2(50324504_50399656), detected by OGM in patient maps (sample 48, 58, 101) (Figure 7). Patients are classified as IKZF1plus positive if they harbor an IKZF1 deletion plus a deletion involving CDKN2A/B, PAX5, or PAR1 (positive for P2RY8::CRLF2), without a concurrent ERG deletion [21]. We detected IKZF1 deletion in 3 cases, among whom patient #48 met the criteria of IKZF1plus, and also harbored iAMP21. The patient was treated with high-risk regimen, with d15MRD of 3.20 × 10−2 and negative MRD at the end of induction. The other two non-IKZF1plus patients (Case #101 and #58) had increased copy number of AML1 and BCR::ABL1 fusion, respectively. Case #101 received high-risk treatment with negative MRD at both day 15 and 33. Case#58 was classified into the intermediate-risk group, and died of acute intracranial hemorrhage during induction. Due to the limited number of samples, we could not obtain clinical characteristics of the IKZF1plus cases. Importantly, this study suggests the potential of OGM to detect the genetic aberrations of IKZF1plus in an all-in-one process. Its clinical value would be further verified in future studies with a large sample size.

Figure 7.
IKZF1 deletion in 7p12.2 detected by OGM. The green bar indicates the reference map, patient maps (sample 48, 58, 101) were displayed in blue, and deletions were marked in red.
In conclusion, we found that some OGM-detected recurrent fusion genes or single-gene aberrations were correlated with clinical risk stratification indicators. Some of these genes have not been reported, and their clinical significance need to be further verified.
3.4. Successful Validation of New SVs with Combination of OGM and NGS
3.4.1. NGS Validation of SVs Detected by OGM
Next, based on the amount of partner genes expressed in bone marrow, their relevance to the pathogenesis of leukemia, and whether the promoter region is preserved, five possible fusion genes were selected for WGS verification: PSPC1::ZMYM2 (deletion), SH2B3::ATXN2(deletion), LMNB1::PPP2R2B (deletion), CWH43::TPTE and TMEM272::KDM4B (inter-chromosomal translocation). The WGS results confirmed the existence of these five gene fusions. However, further analysis suggested that the SH2B3::ATXN2 and PSPC1::ZMYM2 fusion result from the deletion of a 0.02–0.2 Mb region containing promoter sequences of the fusion partners (Figure S1A,B). Thus, these fusions do not lead to the transcription of fused mRNA. With regard to CWH43::TPTE fusion, neither of the promoters of the two genes remained in the fusion (CWH43 lost its promoter and 1-9 exon regions, while TPTE lost its promoter and 1–13 exons), and so the CWH43::TPTE fusion could not produce a fused mRNA and or protein (Figure S1C). Regarding the TMEM272::KDM4B fusion gene, resulting from an inter-chromosomal translocation between chr13 and chr19 in sample 66, WGS that revealed the breakpoints are in intron 2 and intron 1 of TMEM272 and KDM4B, respectively. The RT-PCR result confirmed the existence of TMEM272::KDM4B fused mRNA (Figure S2A,B). Thus, the entire coding sequences of KDM4B were under the control of the TMEM272 promoter in this rearrangement (Figure S2C). The mRNA expression of KDM4B in sample 66 was 1.69 times higher than that in other patients with newly diagnosed ALL.
In sample 46, the deletion of about a 20Mb region (Chr5: 126,720,525-146,759,262), containing 5′ sequences of LMNB1 and PPP2R2B, leads to fusion of LMNB1 and PPP2R2B on chromosome 5. The breakpoints are located at LMNB1 intron 2 and PPP2R2B intron 6, respectively (Figure 8D). The fusion retains 5′ regulatory regions of both genes, exon 1–2 of LMNB1 and exon 1–6 of PPP2R2B (Figure 8D). Further RT-PCR result confirmed the existence of LMNB1::PPP2R2B fused mRNA (Figure S3B,C). However, as the two fused mRNA were predicted to produce truncated LMNB1 (N terminal 172 amino acids encoded by exon 1 and exon 2) and PPP2R2B (N terminal 45 amino acids coded by a range from exon1 to exon 6), respectively, we cannot ascertain the existence of the two truncated proteins due to unavailability of leukemic samples. Therefore, we have validated the existence of the above four gene fusions at the genome DNA level (except for CWH43::TPTE, which lost their respective promoter regions), and further at the mRNA level, for two of them.
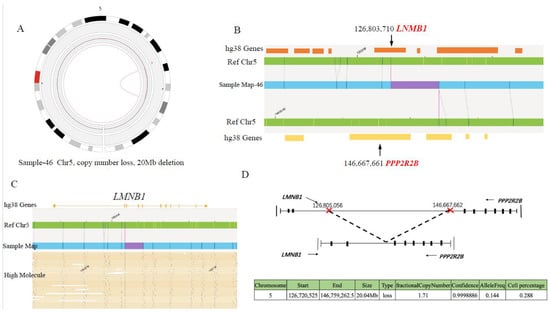
Figure 8.
LMNB1-PPP2R2B fusion gene identified by OGM. (A) Zoom-in circus plot of chromosome 5 only, showing the fusion called by the SV and CNV tool. (B) Chromosome map view, navigated from circus plot, supports the existence of the fusion. The lower track shows the fusion call from the map, specifying the deletion between chr5:126,803,710 and chr5:146,667,661 (size: 20 Mbp). (C) OGM provides direct evidence for LMNB1::PPP2R2B fusion: aberrant molecules supporting the translocation. (D) Diagram of LMNB1::PPP2R2B fusion pattern.
3.4.2. Determination of LMNB1::PPP2R2B Fusion mRNA in Another Cohort of B-ALL Patients
Since LMNB1 plays an important role in nuclear structure and PPP2R2B has phosphatase activity inhibiting oncogenesis, we continued to explore the incidence of LMNB1::PPP2R2B fusion gene in a new cohort of patients. We determined LMNB1::PPP2R2B fusion of mRNA in diagnostic bone marrow samples of 396 children with B-ALL (diagnosed from October 2018 through March 2021). LMNB1::PPP2R2B fused mRNA was finally detected in 1 patient (Figure S3D). The incidence of LMNB1::PPP2R2B fusion is estimated at 0.25%. It is interesting that both the patients with LMNB1::PPP2R2B fusion carried ETV6::RUNX1 fusion, implying that LMNB1::PPP2R2B fusion played a role in the pathogenesis of ETV6::RUNX1-positive leukemia.
Taken together, OGM combined with WGS can play an active role in the identifying new genetic alterations, affecting cellular signaling pathways, and in laying a solid foundation for subsequent research.
4. Discussion
In this study, we compared the role of OGM with that of conventional cytogenetic technologies in the genotyping of pediatric ALL. The results showed a good concordance between conventional cytogenetic techniques and OGM in identification of abnormalities of various types (balanced or complex translocation, duplication, deletion, insertion, inversion and aneuploidies). The exception was P2RY8::CRLF2 fusion, involved in the pseudoautosomal region of X/Y chromosomes. Noteworthily, P2RY8::CRLF2 fusion alterations have been repeatedly reported to have high clinical significance and are included in the stratification of patient in clinical trials since they enable treatment by TKI inhibitors [22,23,24,25]. In fact, among the 3 patients (cases #48, #51 and #76) in this study, two were in the high-risk group and one was in the intermediate-risk group, and all had poor treatment responses. Thus, identification of P2RY8::CRLF2 fusion is critical in clinical practice. Though OGM has been shown to have the potential to be a routine tool in hematology malignancies [26,27], some technical limitations remain to be further addressed by anticipated software improvements, especially in PAR regions. However, it is also possible that P2RY8::CRLF2 fusion can be missed by VAF which is lower than 5%. Importantly, our results highlight several advantages of OGM. Firstly, OGM allows for the high-throughput, accurate detection of different types of anomalies. OGM has greater sensitivity and resolution than karyotypes, theoretically allowing the analysis of whole genomes and the identification of the aberrations (insertions between 5–50 kbp, deletions > 7 kbp, invertions > 70 kbp, duplications > 150 kbp and transpositions where the translocated fragments are >70 kbp) compared with FISH and PCR. At the same time, OGM is a non-time-consuming method, with only 3.5 days required from sample preparation to standard analysis output. Due to these advantages, OGM can refine the karyotype with high-resolution and uncover the complexity and heterogeneity of structural variations in breakpoint regions, as we can see in the details in Table 2 and Table 3. These may disrupt/impact genes within breakpoint regions, leading to subtle genotype-phenotype differences [26] which were difficult to solve by conventional cytogenetic technologies. Our study also highlights the ability of OGM to identify common fusion genes and reveal novel structural variants. Almost every study using OGM for leukemia showed that a large number of SV that could not be identified by conventional methods could be detected in every sample. Dozens of new inversions, duplications, and hundreds of new insertions and deletions were identified in each patient [28,29]. These SVs involve many genes involved in cell growth, differentiation, and tumorigenesis. Some SVs are located in the inter-gene region, which may lead to abnormal gene expression regulation through the cis-acting elements affecting gene expression regulation.
Accurate detection of known fusion genes is of great significance for risk stratification, and the discovery of new aberrations may lead to important biological insights and new therapeutic methods [30]. Our results show that OGM can detect almost all clinically significant SVs (except for some specific chromosome regions, such as pseudoautosomal region on X/Y chromosomes) reported by cytogenetic methods, as well as those that cannot be identified by conventional methods, and provide a more accurate and comprehensive genomic SV analysis of complex karyotypes. For example, OGM detected a large number of CNVs in 4 cases (case 41, 97, 101, 109) with normal karyotype reports. Most of these CNVs were involved in increased numbers of chromosomes 4, 6, 10, 14, 17, 18, 21 and changes of some segments of chromosome 9, 21. In addition, OGM identified a monosomy of chromosome 7 in case 101, and we also noticed the same presence in samples 48, 71 and 95. Therefore, we speculate that chromosome 7 may have instability factors. These changes may lead to the reduction of important tumor suppressor genes, the damage of gene structural stability or gene expression regulation, which may play an important role in tumorigenesis.
In our study, OGM detected a variety of possible gene fusion events and SVs affected single genes, and some of the recurrent SVs were related to clinical characteristics and risk stratification, as we presented in the results section (see Table 6 and Table 7 for details). These variants lead to destruction or loss of the involved genes, or the formation of gene fusion. However, due to the limited samples included in our study, we have not ascertained the effects of these gene fusions and SVs on the response to treatment and patient management. In future studies with larger sample size, we would focus on the clinical significance of these gene alterations. Since the translocations and corresponding fusion genes analyzed by OGM are mainly conjectural on the basis of gene structure, whether fusion transcripts are generated remains to be confirmed after further verification. For example, in case 47, OGM showed an amplification of a segment and the formation of a derivative chromosome 1 [dup (1) (q21.2q24.3-q24.3q32.3) (149910330_213101514), der(1)t(1;1) (q41;q43)] (Figure 3; Table 3). Genome mapping indicated the duplication and inversion of the ESRRG gene in this event. It was reported that ESRRG, a transcriptional activator, regulated the proliferation of breast cancer cells by directly binding to the response element in the promoter of DNA cytosine 5-methyltransferase 1 (DMNT1) [31]. Therefore, the mutation of the ESRRG gene found in our study may be involved in the occurrence and development of leukemia as an important driving factor. Therefore, OGM would play an important role in clinical practice by further optimizing risk stratification and prognosis evaluation in ALL.
In this study, OGM identified two unreported novel fusions (LMNB1::PPP2R2B, TMEM272::KDM4B) in samples 46 and 66, simultaneously, with two known fusions (ETV6::RUNX1, FLI1::ESWR1), respectively. TMEM272::KDM4B results from a translocation between chromosomes 13 and 19, which makes the coding region of KDM4B under the control of the regulatory elements of TMEM272 (Figure S2). RT-PCR confirmed the presence of the fusion mRNA. The mRNA expression of KDM4B was higher than that in other newly diagnosed ALL, suggesting an overexpression of the KDM4B caused by the fusion. A number of studies have shown that KDM4B is often overexpressed in breast, colorectal, ovarian, lung, gastric and prostate cancer cells, resulting in H3K9me3 demethylation, subsequent gene expression changes and genomic instability to induce tumors [32,33,34,35,36]. Whether KDM4B plays the same role in ALL and other hematological malignancies remains to be further explored. In addition, the existence of TMEM272::KDM4B fusion should be confirmed in a large number of samples in a future study.
The LMNB1::PPP2R2B fusion is caused by a large fragment deletion (about 20Mbp) in chromosome 5. Both genes retain their 5′ promoter regions. However, LMNB1 only retains exon 1 and 2, encoding 172 amino acids, and loses its main domains, while the fused PPP2R2B sequence encodes 45 amino acid residues and terminates at a stop codon in the reverse direction. PPP2R2B contains 7 repeated WD-40 motifs (protein–protein and protein–DNA interaction sites). Deletion results in the retention of only two WD-40 motifs (149 amino acid residues) and the loss of the kinase domain (295th–298th amino acid), while the fused LMNB1 sequence encodes 5 amino acid residues and terminates at a stop codon in the reverse direction. Although our study confirmed that the fusion produces two fusion mRNAs, cells may directly recognize and degrade the two truncated proteins, resulting in the haplo-insufficiency of the two genes. PPP2R2B, a serine/threonine protein phosphatase, is implicated in the negative control of cell growth and division. Tan et al. reported that PPP2R2B inactivation could target PDK1/MYC signaling to promote growth and resistance to rapamycin of colorectal cancer cells [37]. Recent studies have reported that PPP2R2B is a robust tumor suppressor and plays an important role in anti-tumor immune responses, and that its dysregulation could contribute to the onset and progression of breast cancer [38]. As a nuclear structural protein, LMNB1 contributes to maintaining nuclear morphology and hematopoietic stem cell function. Down-regulation of LMNB1 expression causes genomic instability due to defective DNA damage repair. Thus, this fusion possibly leads to down-regulation of the expression of the two proteins, which may be related to leukemogenesis. It is worth noting that 2 out of 396 B-ALL patients carried the fusion, and both were ETV6::RUNX1-positive, suggesting that LMNB1::PPP2R2B may be involved in the role of ETV6::RUNX1 in leukemogenesis. The underlying mechanism needs to be further explored.
5. Conclusions
In summary, two new fusion genes (LMNB1::PPP2R2B and TMEM272::KDM4B) were identified by OGM and verified by WGS and RT-PCR for the first time. OGM addresses some of the limitations associated with conventional cytogenomic testing, as this all-in-one process allows the detection of most major genomic risk markers in one test, which has important meanings for the development of leukemia pathogenesis and targeted drugs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15010035/s1, Figure S1: WGS Validation of SVs Detected by OGM. Figure S2: RT-PCR confirmed the existence of SVs. Figure S3: WGS and Clonal sequencing confirmed the presence of LMNB1::PPP2R2B. Table S1: The quality control of samples. Table S2: SVs and CNVs calling in each sample using different confidence filter settings. Table S3: Chromosomal SVs affected genes annotated in COSMIC database.
Author Contributions
Conceptualization, Z.L., T.W. and R.Z.; Writing—original draft preparation, H.G.; Data curation, H.X.; Resources, C.W.; Formal analysis, L.C.; Software, X.H. and W.L.; Methodology, Z.Y., S.T., X.Z., J.L. and Y.W. Validation, T.X. (Tianlin Xue) and T.X. (Tianyu Xing); Writing—review and editing, Z.L. and T.W. Investigation and Supervision, Z.L., T.W. and R.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Science and Technology Major Project of the Ministry of Science and Technology of China (No. 2017ZX09304029003), the National Natural Science Foundation of China (No. 81870114 and 81970135), the Beijing Natural Science Foundation of China (No. 7202044 and 7214242), the Fundamental Research Funds for the Central Universities (2021RC207) and Funding for Reform and Development of Beijing Municipal Health Commission.
Institutional Review Board Statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Institutional Review Board (IRB) of Beijing Children’s Hospital, Capital Medical University (IEC-C-008-A08-V.05.1). All parents signed informed consent forms.
Informed Consent Statement
Not applicable.
Data Availability Statement
Please contact author for data requests.
Acknowledgments
We thank the medical staffs of Hematology Center, Beijing Children’s Hospital, Capital Medical University, and all participated patients and their families.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
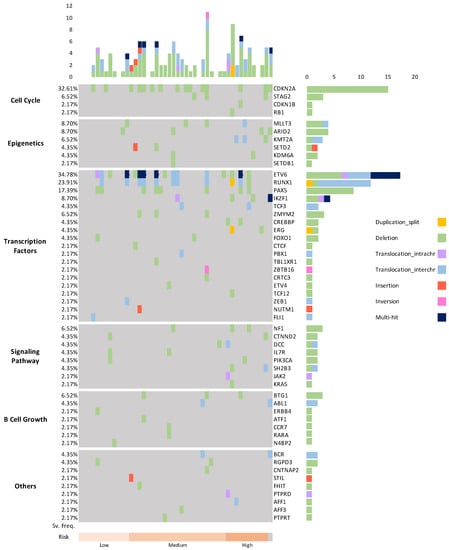
Figure A1.
An overview of the genome-wide alterations identified by OGM in 46 pediatric B-ALL samples. The samples are sorted into 3 risk groups, as indicated at the bottom of the panel. The relative mutation frequencies (SV. Freq.) of the genes in different biological processes are indicated on the left. Gene names and types of mutation are shown on the right. The numbers of mutations are shown at the top in a bar plot. The types of mutations are highlighted in different colors.
Appendix B. Genes and Cellular Biological Processes Affected by Chromosomal SVs
We identified 131 genomic alterations, affecting 97 genes including several known targets of mutation in B-ALL (Supplement Table S3). The most commonly affected genes were ETV6 (n = 16, 34.8%), CDKN2A (n = 15, 32.6%), RUNX1 (n = 11, 23.9%), PAX5 (n = 8, 17.4%), MLLT3, IKZF1 and ARID2 (each in 4 cases, 8.7%), BTG1, NF1, STAG2, ZMYM2 and KMT2A (each in 3, 6.5%). Other affected genes and alteration frequencies were specified in Supplement Table S3. Some of the 97 putative driver genes, for example, STAG2, CTNND2, MDM2, BRCA1, CAMTA1, SSX2, have not been previously described in pediatric B-ALL. These alterations mainly involved in genes encoding tumor suppressors, cell cycle regulators, and hematopoietic transcription regulators. 12.2% (16/134) of alterations were found in genes encoding epigenetic regulators and chromatin modifiers including NUTM1, NCOR1, and SUZ12, in this study. In summary, we identified 88 deletion variants (67.2%), more than other types of alterations.
We next tried to identify the cellular biological processes which were affected by the above SVs and that were detected by OGM (Supplement Figure S1). We could classify the affected genes into six categories, including cell cycle (such as CDKN2A, PTEN, RB1), epigenetic regulation and chromatin modification (such as MLLT3, ARID2, KMT2A), transcription regulation (such as RUNX1, PAX5, IKZF1), signaling pathways (such as PIK3CA, IL7R), B lymphocyte growth (such as BTG1, ERBB4), and others (such as CNTNAP2 and BCR).
References
- Hunger, S.P.; Mullighan, C.G. Acute lymphoblastic leukemia in children. N. Engl. J. Med. 2015, 373, 1541–1552. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, I.; Mullighan, C.G. Genetic basis of acute lymphoblastic leukemia. J. Clin. Oncol. 2017, 35, 975–983. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-F.; Wang, B.-Y.; Zhang, W.-N.; Huang, J.-Y.; Li, B.-S.; Zhang, M.; Jiang, L.; Li, J.-F.; Wang, M.-J.; Dai, Y.-J.; et al. Genomic profiling of adult and pediatric B-cell acute lymphoblastic leukemia. EBioMedicine 2016, 8, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Suttorp, J.; Lühmann, J.L.; Behrens, Y.L.; Göhring, G.; Steinemann, D.; Reinhardt, D.; Neuhoff, N.V.; Schneider, M. Optical Genome Mapping as a Diagnostic Tool in Pediatric Acute Myeloid Leukemia. Cancers 2022, 14, 2058. [Google Scholar] [CrossRef] [PubMed]
- Lafage-Pochitaloff, M.; Baranger, L.; Hunault, M.; Cuccuini, W.; Lefebvre, C.; Bidet, A.; Tigaud, I.; Eclache, V.; Delabesse, E.; Bilhou-Nabéra, C.; et al. Impact of cytoge- netic abnormalities in adults with Ph-negative B-cell precursor acute lymphoblastic leukemia. Blood 2017, 130, 1832–1844. [Google Scholar] [CrossRef]
- Chan, S.; Lam, E.; Saghbini, M.; Bocklandt, S.; Hastie, A.; Cao, H.; Holmlin, E.; Borodkin, M. Structural Variation Detection and Analysis Using Bionano Optical Mapping. Methods Mol. Biol. 2018, 1833, 193–203. [Google Scholar]
- Chan, E.K.; Cameron, D.L.; Petersen, D.C.; Lyons, R.J.; Baldi, B.F.; Papenfuss, A.T.; Thomas, D.M.; Hayes, V.M. Optical mapping reveals a higher level of genomic architecture of chained fusions in cancer. Genome Res. 2018, 28, 26–738. [Google Scholar] [CrossRef]
- Eisfeldt, J.; Pettersson, M.; Vezzi, F.; Wincent, J.; Käller, M.; Gruselius, J.; Nilsson, D.; Lundberg, E.S.; Carvalho, C.M.B.; Lindstrand, A. Comprehensive structural variation genome map of individuals carrying complex chromosomal rearrangements. PLoS Genet. 2019, 15, e1007858. [Google Scholar] [CrossRef]
- Hematology Group, Scientific Society, Chinese Medical Association. Recommendations for Diagnosis and Treatment of pediatric Acute lymphoblastic Leukemia (Fourth Revision). Chin. J. Pediatr. 2014, 52, 641–644. [Google Scholar]
- Dixon, J.R.; Xu, J.; Dileep, V.; Zhan, Y.; Song, F.; Le, V.T.; Yardımcı, G.G.; Chakraborty, A.; Bann, D.V.; Wang, Y.; et al. Integrative detection and analysis of structural variation in cancer genomes. Nat. Genet. 2018, 50, 1388–1398. [Google Scholar] [CrossRef]
- Broach, J.R.; Xu, J.; Schleicher, E.; Pool, C.; Hennessy, M.; Sheldon, K.; Annageldiyev, C.; Sharma, A.; Chang, Y.; Hastie, A.; et al. An integrated framework for genome analysis reveals numerous previously unrecognizable structural variants in leukemia patients’ samples. Cancer Res. 2019, 79, 1708. [Google Scholar] [CrossRef]
- Kumar, K.R.; Cowley, M.J.; Davis, R.L. Next-Generation Sequencing and Emerging Technologies. Semin. Thromb. Hemost. 2019, 45, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Liu, M.; Huang, T.; Liao, W.; Xu, M.; Gu, J. GeneFuse: Detection and visualization of target gene fusions from DNA sequencing data. Int. J. Biol. Sci. 2018, 14, 843–848. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Wu, L.; Wang, Q.; Li, Y.; Wang, X. KDM4B promotes epithelial-mesenchymal transition through up-regulation of ZEB1 in pancreatic cancer. Acta Biochim. Biophys. Sin. 2015, 47, 997–1004. [Google Scholar] [CrossRef]
- Gerding, W.M.; Tembrink, M.; Nilius-Eliliwi, V.; Mika, T.; Dimopoulos, F.; Ladigan-Badura, S.; Eckhardt, M.; Pohl, M.; Wünnenberg, M.; Farshi, P.; et al. Optical genome mapping reveals additional prognostic information compared to conventional cytogenetics in AML/MDS patients. Int. J. Cancer 2022, 150, 1998–2011. [Google Scholar] [CrossRef]
- Moorman, A.V.; Richards, S.M.; Robinson, H.M.; Strefford, J.C.; Gibson, B.E.S.; Kinsey, S.E.; Eden, T.O.B.; Vora, A.J.; Mitchell, C.D.; Harrison, C.J.; et al. Prognosis of children with acute lymphoblastic leukemia (ALL) and intrachromosomal amplification of chromosome 21 (iAMP21). Blood 2006, 109, 2327–2330. [Google Scholar] [CrossRef]
- Strefford, J.C.; van Delft, F.W.; Robinson, H.M.; Worley, H.; Yiannikouris, O.; Selzer, R.; Richmond, T.; Hann, I.; Bellotti, T.; Raghavan, M.; et al. Complex genomic alterations and gene expression in acute lymphoblastic leukemia with intrachromosomal amplification of chromosome 21. Proc. Natl. Acad. Sci. USA 2006, 103, 8167–8172. [Google Scholar] [CrossRef]
- Sinclair, P.B.; Parker, H.; An, Q.; Rand, V.; Ensor, H.; Harrison, C.J.; Strefford, J.C. Analysis of a breakpoint cluster reveals insight into the mechanism of intrachromosomal amplification in a lymphoid malignancy. Hum. Mol. Genet. 2011, 20, 2591–2602. [Google Scholar] [CrossRef]
- McClintock, B. The stability of broken ends of chromosomes in Zea Mays. Genetics 1941, 26, 234–282. [Google Scholar] [CrossRef]
- Rand, V.; Parker, H.; Russell, L.J.; Schwab, C.; Ensor, H.; Irving, J.; Jones, L.; Masic, D.; Minto, L.; Morrison, H.; et al. Genomic characterization implicates iAMP21 as a likely primary genetic event in childhood B-cell pre-cursor acute lymphoblastic leukemia. Blood 2001, 117, 6848–6855. [Google Scholar] [CrossRef]
- Stanulla, M.; Dagdan, E.; Zaliova, M.; Möricke, A.; Palmi, C.; Cazzaniga, G.; Eckert, C.; Te Kronnie, G.; Bourquin, J.P.; Bornhauser, B.; et al. International BFM Study Group. IKZF1plus defines a new minimal residual disease-dependent very-poor prognostic profile in pediatric B-Cell precursor acute lymphoblastic leukemia. J. Clin. Oncol. 2018, 36, 1240–1249. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.G.; Li, Y.; Payne-Turner, D.; Harvey, R.C.; Yang, Y.-L.; Pei, D.; McCastlain, K.; Ding, L.; Lu, C.; Song, G.; et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N. Engl. J. Med. 2014, 371, 1005–1015. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.G.; Gu, Z.; Payne-Turner, D.; McCastlain, K.; Harvey, R.; Chen, I.-M.; Pei, D.; Iacobucci, I.; Valentine, M.; Pounds, S.B.; et al. High frequency and poor outcome of Philadelphia chromosome-like acute lymphoblastic leukemia in adults. J. Clin. Oncol. 2017, 35, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, I.; Li, Y.; Roberts, K.G.; Dobson, S.M.; Kim, J.C.; Payne-Turner, D.; Harvey, R.C.; Valentine, M.; McCastlain, K.; Easton, J.; et al. Truncating erythropoietin receptor rearrangements in acute lymphoblastic leukemia. Cancer Cell 2016, 29, 186–200. [Google Scholar] [CrossRef]
- Schwab, C.; Ryan, S.L.; Chilton, L.; Elliott, A.; Murray, J.; Richardson, S.; Wragg, C.; Moppett, J.; Cummins, M.; Tunstall, O.; et al. EBF1-PDGFRB fusion in pediatric B-cell precursor acute lymphoblastic leukemia (BCP-ALL): Genetic profile and clinical implications. Blood 2016, 127, 2214–2218. [Google Scholar] [CrossRef]
- Yang, H.; Garcia-Manero, G.; Sasaki, K.; Montalban-Bravo, G.; Tang, Z.; Wei, Y.; Kadia, T.; Chien, K.; Rush, D.; Nguyen, H.; et al. High-resolution structural variant profiling of myelodysplastic syndromes by optical genome mapping uncovers cryptic aberrations of prognostic and therapeutic significance. Leukemia 2022, 36, 2306–2316. [Google Scholar] [CrossRef]
- Balducci, E.; Kaltenbach, S.; Villarese, P.; Duroyon, E.; Zalmai, L.; Friedrich, C.; Suarez, F.; Marcais, A.; Bouscary, D.; Decroocq, J.; et al. Optical genome mapping refines cytogenetic diagnostics, prognostic stratification and provides new molecular insights in adult MDS/AML patients. Blood Cancer J. 2022, 12, 126. [Google Scholar] [CrossRef]
- Finlay, D.; Murad, R.; Hong, K.; Lee, J.; Pang, A.; Lai, C.; Burian, C.; Mason, J.R.; Hastie, A.; Yin, J.; et al. Optical mapping uncovers multiple novel genomic structural variants in patient leukemias. Blood 2020, 136 (Suppl. S1), 33–34. [Google Scholar] [CrossRef]
- Levy, B.; Baughn, L.B.; Chartrand, S.; LaBarge, B.; Claxton, D.; Lennon, A.; Akkari, Y.; Cujar, C.; Kolhe, R.; Kroeger, K.; et al. A national multicenter evaluation of the clinical utility of optical genome mapping for assessment of genomic aberrations in acute myeloid leukemia. medRxiv 2020. [Google Scholar] [CrossRef]
- Hastie, A.R.; Lam, E.T.; Chun Pang, A.W.; Zhang, X.; Andrews, W.; Lee, J.; Liang, T.Y.; Wang, J.; Zhou, X.; Zhu, Z.; et al. Rapid Automated Large Structural Variation Detection in a Diploid Genome by NanoChannel Based Nest-Generation Mapping. bioRxiv 2017. [CrossRef]
- Elfassihi, L.; Giroux, S.; Bureau, A.; Laflamme, N.; Cole, D.E.; Rousseau, F. Association with replication between estrogen-related receptor gamma (ESRRgamma) polymorphisms and bone phenotypes in women of European ancestry. J. Bone Miner. Res. 2010, 25, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Jubb, A.M.; Pike, L.; Buffa, F.M.; Turley, H.; Baban, D.; Leek, R.; Gatter, K.C.; Ragoussis, J.; Harris, A.L. The histone demethylase JMJD2B is regulated by estrogen receptor alpha and hypoxia, and is a key mediator of estrogen induced growth. Cancer Res. 2010, 70, 6456–6466. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.; Chen, L.; Yang, J.; Ye, T.; Chen, Y.; Fang, J. HIF-1alpha-induced histone demethylase JMJD2B contributes to the malignant phenotype of colorectal cancer cells via an epigenetic mechanism. Carcinogenesis 2012, 33, 1664–1673. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Qiu, L.; Hong, Y.; Karnik, T.; Tadros, G.; Mau, B.; Ma, T.; Mu, Y.; New, J.; Louie, R.J.; et al. The histone demethylase KDM4B regulates peritoneal seeding of ovarian cancer. Oncogene 2017, 36, 2565–2576. [Google Scholar] [CrossRef]
- Li, W.; Zhao, L.; Zang, W.; Liu, Z.; Chen, L.; Liu, T.; Xu, D.; Jia, J. Histone demethylase JMJD2B is required for tumor cell proliferation and survival and is over-expressed in gastric cancer. Biochem. Biophys. Res. Commun. 2011, 416, 372–378. [Google Scholar] [CrossRef]
- Slee, R.B.; Steiner, C.M.; Herbert, B.S.; Vance, G.H.; Hickey, R.J.; Schwarz, T.; Christan, S.; Radovich, M.; Schneider, B.P.; Schindelhauer, D.; et al. Cancer-associated alteration of pericentromeric heterochromatin may contribute to chromosome instability. Oncogene 2012, 31, 3244–3253. [Google Scholar] [CrossRef]
- Tan, J.; Lee, P.L.; Li, Z.; Jiang, X.; Lim, Y.C.; Hooi, S.C.; Yu, Q. B55β-associated PP2A complex controls PDK1-directed MYC signaling and modulates rapamycin sensitivity in colorectal cancer. Cancer Cell 2010, 18, 459–471. [Google Scholar] [CrossRef]
- Li, Z.; Li, Y.; Wang, X.; Yang, Q. PPP2R2B downregulation is associated with immune evasion and predicts poor clinical outcomes in triple-negative breast cancer. Cancer Cell Int. 2021, 21, 13. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).