Gene Expression Profiling Elucidates Cellular Responses to NCX4040 in Human Ovarian Tumor Cells: Implications in the Mechanisms of Action of NCX4040
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture and Drug Treatment
2.3. Global Gene Expression and Data Acquisition
2.4. Preprocessing of the Data
2.5. Statistical Data Analysis
2.6. Pattern-Driven Analysis of Gene Expression
2.7. Real-Time RT-PCR
3. Results
3.1. Analysis of Overlay of DEGs by Vaan diagrams
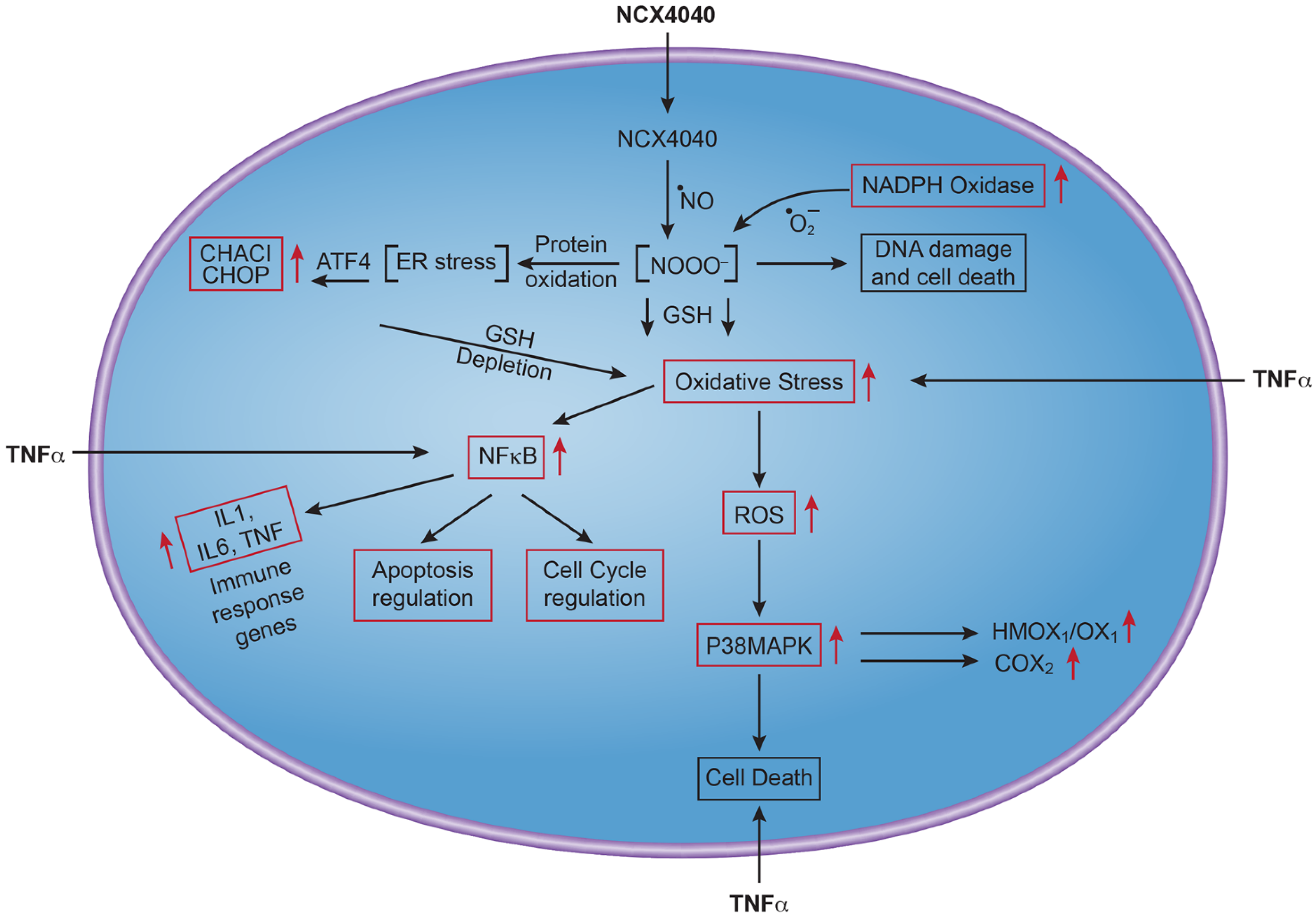
3.2. NCX4040 Induces Oxidative Stress-Related Genes
3.3. NCX4040 Induces Inflammatory Response Genes
3.4. NCX4040 Modulates DNA Damage Response Genes
3.5. Principal Component Analysis
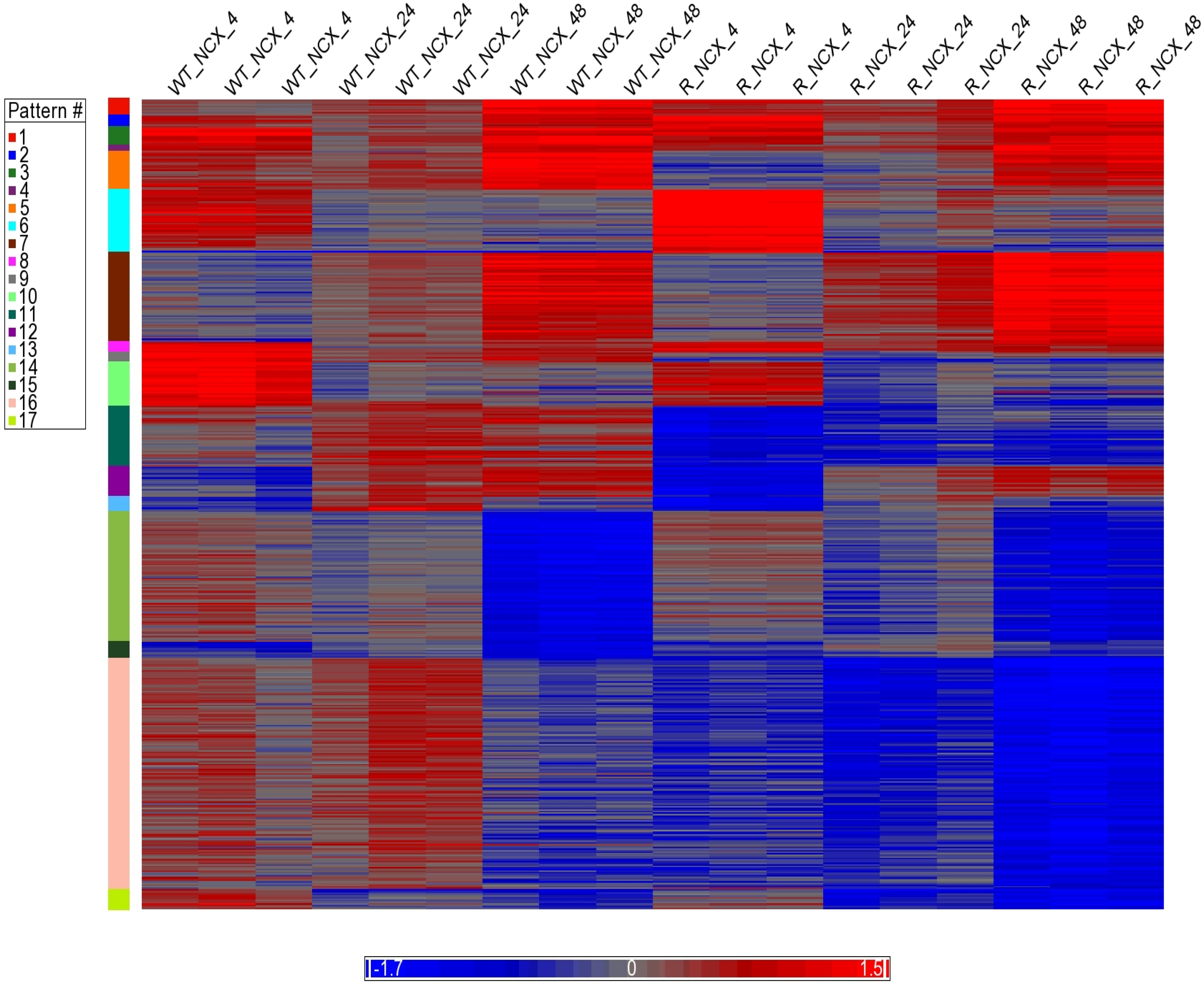
3.6. Analysis of Clustering of DEGs (Heatmap)
3.7. KEGG Pathway Analysis
3.8. Analysis for Co-Expression Pattern
3.9. RT-PCR Analysis
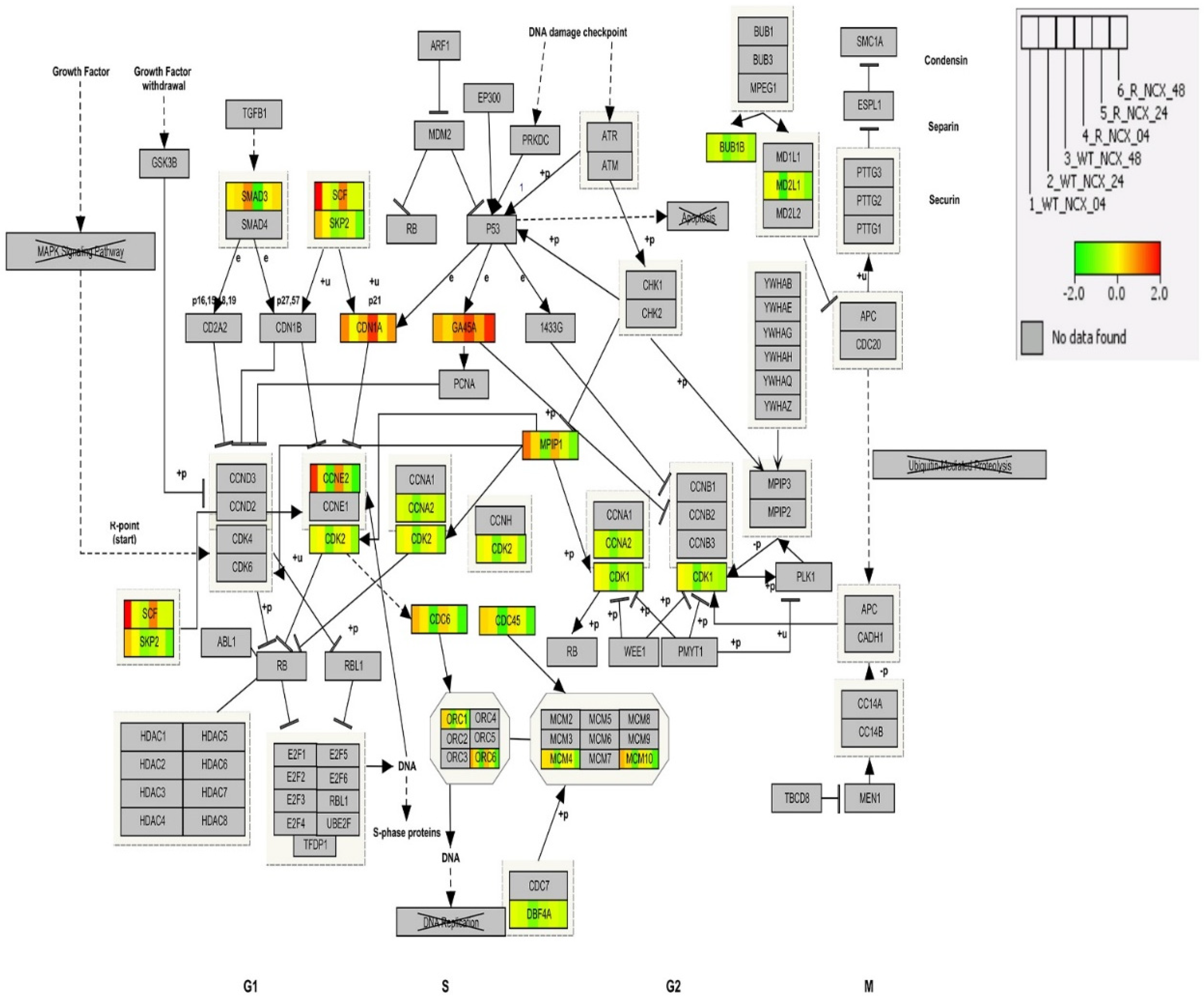
3.10. Cell Cycle Pathway Analysis by IPA
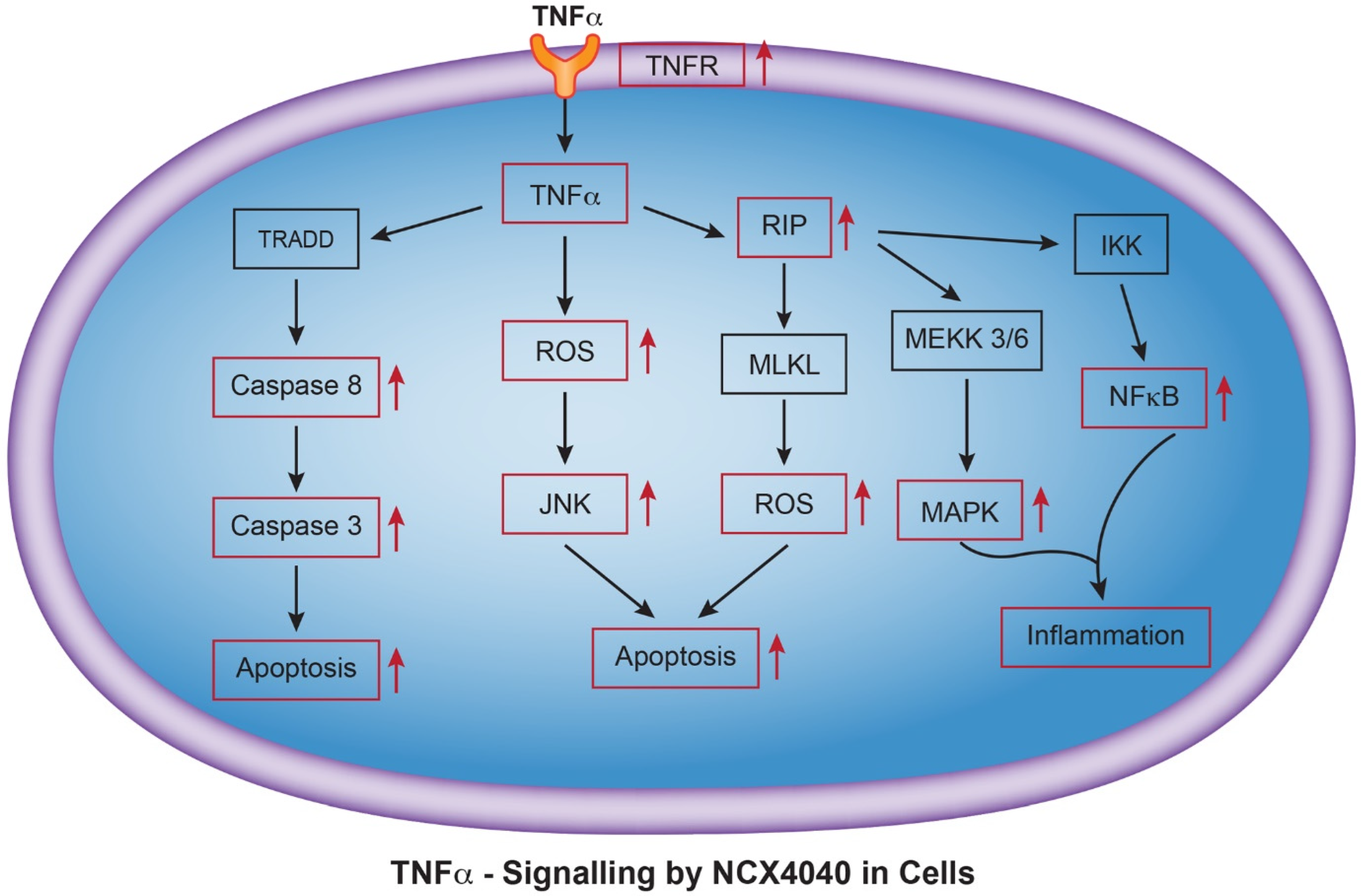
3.11. NCX4040 Induces TNF Signaling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Kossai, M.; Leary, A.; Scoazec, J.Y.; Genestie, C. Ovarian Cancer: A Heterogeneous Disease. Pathobiology 2018, 85, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Parmar, M.K.; Ledermann, J.A.; Colombo, N.; du Bois, A.; Delaloye, J.F.; Kristensen, G.B.; Wheeler, S.; Swart, A.M.; Qian, W.; Torri, V.; et al. Paclitaxel plus platinum-based chemotherapy versus conventional platinum-based chemotherapy in women with relapsed ovarian cancer: The ICON4/AGO-OVAR-2.2 trial. Lancet 2003, 361, 2099–2106. [Google Scholar] [PubMed]
- Sinha, B.K.; Perera, L.; Cannon, R.E. NCX-4040, a Unique Nitric Oxide Donor, Induces Reversal of Drug-Resistance in Both ABCB1- and ABCG2-Expressing Multidrug Human Cancer Cells. Cancers 2021, 13, 1680. [Google Scholar] [CrossRef] [PubMed]
- Sinha, B.K.; Tokar, E.J.; Bortner, C.D. Molecular Mechanisms of Cytotoxicity of NCX4040, the Non-Steroidal Anti-Inflammatory NO-Donor, in Human Ovarian Cancer Cells. Int. J. Mol. Sci. 2022, 23, 8611. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Liu, X.; Rigas, B. Nitric oxide-donating aspirin induces apoptosis in human colon cancer cells through induction of oxidative stress. Proc. Natl. Acad. Sci. USA 2005, 102, 17207–17212. [Google Scholar] [CrossRef] [PubMed]
- Bratasz, A.; Selvendiran, K.; Wasowicz, T.; Bobko, A.; Khramtsov, V.V.; Ignarro, L.J.; Kuppusamy, P. NCX-4040, a nitric oxide-releasing aspirin, sensitizes drug-resistant human ovarian xenograft tumors to cisplatin by depletion of cellular thiols. J. Transl. Med. 2008, 6, 9. [Google Scholar] [CrossRef] [PubMed]
- Sinha, B.K.; Bortner, C.D.; Mason, R.P.; Cannon, R.E. Nitric oxide reverses drug resistance by inhibiting ATPase activity of p-glycoprotein in human multi-drug resistant cancer cells. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2806–2814. [Google Scholar] [CrossRef] [PubMed]
- Sinha, B.K.; Perera, L.; Cannon, R.E. Reversal of drug resistance by JS-K and nitric oxide in ABCB1- and ABCG2-expressing multi-drug resistant human tumor cells. Biomed. Pharmacother. 2019, 120, 109468. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, T.; Chandrasena, R.E.; Wang, Z.; Sinha, V.; Wang, Z.; Thatcher, G.R. Quinone formation as a chemoprevention strategy for hybrid drugs: Balancing cytotoxicity and cytoprotection. Chem. Res. Toxicol. 2007, 20, 1903–1912. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controling the false discovery rate: A practical and powereful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Chou, J.W.; Zhou, T.; Kaufmann, W.K.; Paules, R.S.; Bushel, P.R. Extracting gene expression patterns and identifying co-expressed genes from microarray data reveals biologically responsive processes. BMC Bioinform. 2007, 8, 427. [Google Scholar] [CrossRef] [PubMed]
- Sinha, B.K.; Tokar, E.J.; Bushel, P.R. Elucidation of Mechanisms of Topotecan-Induced Cell Death in Human Breast MCF-7 Cancer Cells by Gene Expression Analysis. Front. Genet. 2020, 11, 775. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Liang, Y.; Zheng, T.; Yang, G.; Zhang, X.; Sun, Z.; Shi, C.; Zhao, S. Inhibition of heme oxygenase-1 enhances anti-cancer effects of arsenic trioxide on glioma cells. J. Neurooncol. 2011, 104, 449–458. [Google Scholar] [CrossRef]
- Yue, Z.; Zhong, L.; Mou, Y.; Wang, X.; Zhang, H.; Wang, Y.; Xia, J.; Li, R.; Wang, Z. Arsenic Trioxide Activate Transcription of Heme Oxygenase-1 by Promoting Nuclear Translocation of NFE2L2. Int. J. Med. Sci. 2015, 12, 674–679. [Google Scholar] [CrossRef]
- Dosunmu-Ogunbi, A.M.; Wood, K.C.; Novelli, E.M.; Straub, A.C. Decoding the role of SOD2 in sickle cell disease. Blood Adv. 2019, 3, 2679–2687. [Google Scholar] [CrossRef]
- Chen, F.; Haigh, S.; Barman, S.; Fulton, D.J. From form to function: The role of Nox4 in the cardiovascular system. Front. Physiol. 2012, 3, 412. [Google Scholar] [CrossRef]
- Crawford, R.R.; Prescott, E.T.; Sylvester, C.F.; Higdon, A.N.; Shan, J.; Kilberg, M.S.; Mungrue, I.N. Human CHAC1 Protein Degrades Glutathione, and mRNA Induction Is Regulated by the Transcription Factors ATF4 and ATF3 and a Bipartite ATF/CRE Regulatory Element. J. Biol. Chem. 2015, 290, 15878–15891. [Google Scholar] [CrossRef]
- Tanaka, T.; Narazaki, M.; Kishimoto, T. IL-6 in inflammation, immunity, and disease. Cold Spring Harb. Perspect. Biol. 2014, 6, a016295. [Google Scholar] [CrossRef]
- Unver, N.; McAllister, F. IL-6 family cytokines: Key inflammatory mediators as biomarkers and potential therapeutic targets. Cytokine Growth Factor Rev. 2018, 41, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhang, X.; Zhu, G.; Liu, H.; Chen, J.; Wang, Y.; He, X. Quercetin inhibits TNF-alpha induced HUVECs apoptosis and inflammation via downregulating NF-kB and AP-1 signaling pathway in vitro. Medicine 2020, 99, e22241. [Google Scholar] [CrossRef] [PubMed]
- Dever, S.M.; White, E.R.; Hartman, M.C.; Valerie, K. BRCA1-directed, enhanced and aberrant homologous recombination: Mechanism and potential treatment strategies. Cell Cycle 2012, 11, 687–694. [Google Scholar] [CrossRef][Green Version]
- Turan, V.; Oktay, K. BRCA-related ATM-mediated DNA double-strand break repair and ovarian aging. Hum. Reprod. Update 2020, 26, 43–57. [Google Scholar] [CrossRef] [PubMed]
- Thacker, J.; Zdzienicka, M.Z. The XRCC genes: Expanding roles in DNA double-strand break repair. DNA Repair 2004, 3, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H.; Pfizenmaier, K.; Scheurich, P. Tumor necrosis factor signaling. Cell Death Differ. 2003, 10, 45–65. [Google Scholar] [CrossRef] [PubMed]
- Cruceriu, D.; Baldasici, O.; Balacescu, O.; Berindan-Neagoe, I. The dual role of tumor necrosis factor-alpha (TNF-alpha) in breast cancer: Molecular insights and therapeutic approaches. Cell Oncol. 2020, 43, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Rhodes, C.J.; Moncol, J.; Izakovic, M.; Mazur, M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006, 160, 1–40. [Google Scholar] [CrossRef]
- Blaser, H.; Dostert, C.; Mak, T.W.; Brenner, D. TNF and ROS Crosstalk in Inflammation. Trends Cell Biol. 2016, 26, 249–261. [Google Scholar] [CrossRef]
- Kamata, H.; Honda, S.; Maeda, S.; Chang, L.; Hirata, H.; Karin, M. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell 2005, 120, 649–661. [Google Scholar] [CrossRef]
- Morgan, M.J.; Liu, Z.G. Reactive oxygen species in TNFalpha-induced signaling and cell death. Mol Cells 2010, 30, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J.B.; Zarrinkar, P.P.; Sapinoso, L.M.; Kern, S.G.; Behling, C.A.; Monk, B.J.; Lockhart, D.J.; Burger, R.A.; Hampton, G.M. Analysis of gene expression profiles in normal and neoplastic ovarian tissue samples identifies candidate molecular markers of epithelial ovarian cancer. Proc. Natl. Acad. Sci. USA 2001, 98, 1176–1181. [Google Scholar] [CrossRef] [PubMed]
- Schaner, M.E.; Ross, D.T.; Ciaravino, G.; Sorlie, T.; Troyanskaya, O.; Diehn, M.; Wang, Y.C.; Duran, G.E.; Sikic, T.L.; Caldeira, S.; et al. Gene expression patterns in ovarian carcinomas. Mol. Biol. Cell 2003, 14, 4376–4386. [Google Scholar] [CrossRef] [PubMed]
- Kannan, K.; Amariglio, N.; Rechavi, G.; Jakob-Hirsch, J.; Kela, I.; Kaminski, N.; Getz, G.; Domany, E.; Givol, D. DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene 2001, 20, 2225–2234. [Google Scholar] [CrossRef]
- Daoud, S.S.; Munson, P.J.; Reinhold, W.; Young, L.; Prabhu, V.V.; Yu, Q.; LaRose, J.; Kohn, K.W.; Weinstein, J.N.; Pommier, Y. Impact of p53 knockout and topotecan treatment on gene expression profiles in human colon carcinoma cells: A pharmacogenomic study. Cancer Res. 2003, 63, 2782–2793. [Google Scholar]
- Januchowski, R.; Sterzynska, K.; Zawierucha, P.; Rucinski, M.; Swierczewska, M.; Partyka, M.; Bednarek-Rajewska, K.; Brazert, M.; Nowicki, M.; Zabel, M.; et al. Microarray-based detection and expression analysis of new genes associated with drug resistance in ovarian cancer cell lines. Oncotarget 2017, 8, 49944–49958. [Google Scholar] [CrossRef]
- Bolon-Canedo, V.; Alonso-Betanzos, A.; Lopez-de-Ullibarri, I.; Cao, R. Challenges and Future Trends for Microarray Analysis. Methods Mol. Biol. 2019, 1986, 283–293. [Google Scholar]
- Batist, G.; Tulpule, A.; Sinha, B.K.; Katki, A.G.; Myers, C.E.; Cowan, K.H. Overexpression of a novel anionic glutathione transferase in multidrug-resistant human breast cancer cells. J. Biol. Chem. 1986, 261, 15544–15549. [Google Scholar] [CrossRef]
- Cowan, K.H.; Batist, G.; Tulpule, A.; Sinha, B.K.; Myers, C.E. Similar biochemical changes associated with multidrug resistance in human breast cancer cells and carcinogen-induced resistance to xenobiotics in rats. Proc. Natl. Acad. Sci. USA 1986, 83, 9328–9332. [Google Scholar] [CrossRef]
- He, S.; Zhang, M.; Ye, Y.; Zhuang, J.; Ma, X.; Song, Y.; Xia, W. ChaC glutathione specific gamma-glutamylcyclotransferase 1 inhibits cell viability and increases the sensitivity of prostate cancer cells to docetaxel by inducing endoplasmic reticulum stress and ferroptosis. Exp. Ther. Med. 2021, 22, 997. [Google Scholar] [CrossRef]
- Chen, P.H.; Shen, W.L.; Shih, C.M.; Ho, K.H.; Cheng, C.H.; Lin, C.W.; Lee, C.C.; Liu, A.J.; Chen, K.C. The CHAC1-inhibited Notch3 pathway is involved in temozolomide-induced glioma cytotoxicity. Neuropharmacology 2017, 116, 300–314. [Google Scholar] [CrossRef] [PubMed]
- Sheida, F.; Razi, S.; Keshavarz-Fathi, M.; Rezaei, N. The role of myeloid-derived suppressor cells in lung cancer and targeted immunotherapies. Expert Rev. Anticancer Ther. 2022, 22, 65–81. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.S.; Wang, S.F.; Hsu, C.Y.; Yin, P.H.; Yeh, T.S.; Lee, H.C.; Tseng, L.M. CHAC1 degradation of glutathione enhances cystine-starvation-induced necroptosis and ferroptosis in human triple negative breast cancer cells via the GCN2-eIF2alpha-ATF4 pathway. Oncotarget 2017, 8, 114588–114602. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Wang, S.; Guo, J.; Liu, S.; Ding, A.; Wang, G.; Li, W.; Zhang, Y.; Bian, X.; Zhao, S.; et al. Ferroptosis-related gene NOX4, CHAC1 and HIF1A are valid biomarkers for stomach adenocarcinoma. J. Cell Mol. Med. 2022, 26, 1183–1193. [Google Scholar] [CrossRef] [PubMed]
- Chaudhari, N.; Talwar, P.; Parimisetty, A.; Lefebvre d’Hellencourt, C.; Ravanan, P. A molecular web: Endoplasmic reticulum stress, inflammation, and oxidative stress. Front. Cell Neurosci. 2014, 8, 213. [Google Scholar] [CrossRef]
- Chong, W.C.; Shastri, M.D.; Eri, R. Endoplasmic Reticulum Stress and Oxidative Stress: A Vicious Nexus Implicated in Bowel Disease Pathophysiology. Int. J. Mol. Sci. 2017, 18, 771. [Google Scholar] [CrossRef]
- Bhattarai, K.R.; Riaz, T.A.; Kim, H.R.; Chae, H.J. The aftermath of the interplay between the endoplasmic reticulum stress response and redox signaling. Exp. Mol. Med. 2021, 53, 151–167. [Google Scholar] [CrossRef]
- Su, L.J.; Zhang, J.H.; Gomez, H.; Murugan, R.; Hong, X.; Xu, D.; Jiang, F.; Peng, Z.Y. Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxid. Med. Cell Longev. 2019, 2019, 5080843. [Google Scholar] [CrossRef]
- Chen, X.; Li, J.; Kang, R.; Klionsky, D.J.; Tang, D. Ferroptosis: Machinery and regulation. Autophagy 2021, 17, 2054–2081. [Google Scholar] [CrossRef]
- Bekric, D.; Ocker, M.; Mayr, C.; Stintzing, S.; Ritter, M.; Kiesslich, T.; Neureiter, D. Ferroptosis in Hepatocellular Carcinoma: Mechanisms, Drug Targets and Approaches to Clinical Translation. Cancers 2022, 14, 1826. [Google Scholar] [CrossRef]




| Time | Gene | OVCAR-8 | NCI/ADR-RES |
|---|---|---|---|
| 4 h | HMOX1/OX1 | +6.7 | +11.1 |
| SOD2 | +2.2 | ND | |
| NOX4 | +2.2 | +2.1 | |
| CHAC1 | ND | +3.5 | |
| 48 h | SOD2 | +2.8 | ND |
| CHAC1 | +3.8 | +6.0 |
| Time | Gene | OVCAR-8 | NCI/ADR-RES |
|---|---|---|---|
| 4 h | IL-6 | +6.1 | +2.2 |
| HSP | +3.5 | +8.2 | |
| MT1M | +3.4 | +2.8 | |
| MT1X | +2.1 | +2.6 | |
| TNFR | +3.3 | +6.0 | |
| TGF | +2.5 | ND | |
| NFKB1 | +2.2 | ND | |
| IL1R | +2.1 | ND | |
| 48 h | IL-6 | +4.5 | +4.2 |
| TNFR | +2.9 | +2.2 | |
| NFKB1 | +3.9 | +2.2 | |
| VEGF | +3.5 | +3.5 | |
| IL-1β | +3.4 | ND |
| Gene | OVCAR-8 | NCI/ADR-RES |
|---|---|---|
| BRCA1 | −2.2 | ND |
| BRCA2 | −2.2 | −2.0 |
| RAD51 | −3.2 | −2.8 |
| XRCC2 | −2.3 | ND |
| XRCC4 | −2.3 | ND |
| XRCC5 | ND | −2.1 |
| XRCC6 | ND | −2.1 |
| GADD45 | ND | +3.2 |
| Cell Line and Treatment | KEGG Pathway | Count | % | p-Value | FDR | |
|---|---|---|---|---|---|---|
| WT 4 h | TNF signaling pathway | 10 | 7 | 1.10 × 10−6 | 7.50 × 10−5 | |
| WT 48 h | Cell cycle | 14 | 3.3 | 5.40 × 10−7 | 3.80 × 10−5 | |
| R 4 h | MAPK signaling pathway | 14 | 5.7 | 4.90 × 10−53 | 7.00 × 10−2 | |
| R 48 h | DNA replication | 7 | 1.4 | 8.10 × 10−5 | 7.50 × 10−3 | |
| 4 h | Gene | OVCAR-8 | NCI/ADR-RES |
|---|---|---|---|
| HMOX1/OX1 | +4.5 *** | +7.3 *** | |
| CHAC1 | 1.0 | 1.0 | |
| NFKB1 | 1.0 | +5.2 *** | |
| VEGFA | 1.0 | +4.9 | |
| IL-6 | +4.6 *** | +16.0 *** | |
| COX2 | 1.0 | +5.8 *** | |
| RAD51 | +2.8 *** | 0.8 | |
| GADD45 | +1.2 | 1.0 | |
| 48 h | HMOX1/OX1 | 0.9 | 0.5 *** |
| CHAC1 | +8.0 *** | +3.6 *** | |
| NFKB1 | 1.0 | +2.3 | |
| VEGFA | +3.0 *** | +10.0 *** | |
| IL-6 | +3.5 *** | +20.0 *** | |
| COX2 | +2.0 * | +10.9 *** | |
| RAD51 | 0.4 ** | 0.23 ** | |
| GADD45 | +3.0 *** | 0.5 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sinha, B.K.; Tokar, E.J.; Li, J.; Bushel, P.R. Gene Expression Profiling Elucidates Cellular Responses to NCX4040 in Human Ovarian Tumor Cells: Implications in the Mechanisms of Action of NCX4040. Cancers 2023, 15, 285. https://doi.org/10.3390/cancers15010285
Sinha BK, Tokar EJ, Li J, Bushel PR. Gene Expression Profiling Elucidates Cellular Responses to NCX4040 in Human Ovarian Tumor Cells: Implications in the Mechanisms of Action of NCX4040. Cancers. 2023; 15(1):285. https://doi.org/10.3390/cancers15010285
Chicago/Turabian StyleSinha, Birandra K., Erik J. Tokar, Jianying Li, and Pierre R. Bushel. 2023. "Gene Expression Profiling Elucidates Cellular Responses to NCX4040 in Human Ovarian Tumor Cells: Implications in the Mechanisms of Action of NCX4040" Cancers 15, no. 1: 285. https://doi.org/10.3390/cancers15010285
APA StyleSinha, B. K., Tokar, E. J., Li, J., & Bushel, P. R. (2023). Gene Expression Profiling Elucidates Cellular Responses to NCX4040 in Human Ovarian Tumor Cells: Implications in the Mechanisms of Action of NCX4040. Cancers, 15(1), 285. https://doi.org/10.3390/cancers15010285






