Simple Summary
Although multiple actionable genes have been identified in melanoma, 38–42% of patients are still not druggable based on current research. In our study, DNA-NGS and RNA-NGS were utilized to construct molecular profiles of a Chinese cohort of 469 melanoma patients. Up to 11.7% (7/60) of patients in the undruggable group could be recognized as actionable by DNA and RNA sequential sequencing. Additionally, the use of RNA-NGS enhanced the proportion of druggable fusions from 2.56% to 17.27%. In total, the use of RNA-NGS increased the druggable proportion from 75% to 78%. Our study systemically analyzed the genetic landscape of Asian melanoma and demonstrated how DNA and RNA sequential sequencing is essential in bringing clinical benefits to more melanoma patients.
Abstract
Background: In contrast to Caucasian melanoma, which has been extensively studied, there are few studies on melanoma in Asian populations. Sporadic studies reported that only 40% of Asian melanoma patients could be druggable, which was much lower than that in Caucasians. More studies are required to refine this conclusion. Methods: Chinese melanoma patients (n = 469) were sequentially sequenced by DNA-NGS and RNA-NGS. The genomic alterations were determined, and potentially actionable targets were investigated. Results: Patients with potential druggable targets were identified in 75% of Chinese melanoma patients by DNA-NGS based on OncoKB, which was much higher than in a previous Asian study. NRG1 fusions were first identified in melanoma. In addition, up to 11.7% (7/60) of patients in the undruggable group could be recognized as actionable by including RNA-NGS analysis. By comparing the fusion detection rate between DNA-NGS and RNA-NGS, all available samples after DNA-NGS detection were further verified by RNA-NGS. The use of RNA-NGS enhanced the proportion of druggable fusions from 2.56% to 17.27%. In total, the use of RNA-NGS increased the druggable proportion from 75% to 78%. Conclusions: In this study, we systemically analyzed the actionable landscape of melanoma in the largest Asian cohort. In addition, we first demonstrated how DNA and RNA sequential sequencing is essential in bringing clinical benefits to more patients with melanoma.
1. Background
The morbidity rate of melanoma has rapidly increased in recent years, causing more than 57,000 deaths worldwide in 2020 [1]. DNA-based next-generation sequencing (NGS) has been widely applied to decode the genetic landscape of melanoma [2,3,4,5]. With comprehensive molecular profiling, multiple actionable targets have been discovered; for example, BRAF V600E could be targeted with dabrafenib and trametinib, which have been approved by the FDA for pan-cancer treatment [6]. Furthermore, numerous oncogenic driver fusions could be inhibited with inhibitors in melanoma [7,8,9]. For example, tumors with NRTK fusions could be inhibited by TRK inhibitors [9]. Additionally, patients with a high tumor mutation burden (TMB-H) or high microsatellite instability (MSI-H) could be referred to immunotherapy [10,11,12,13].
Although multiple actionable genes have been identified in melanoma, 38–42% of patients are still not druggable based on current research [14,15]. In non-small-cell lung cancer (NSCLC), DNA and RNA sequential sequencing could bring clinical benefits to an additional 14% of patients in the driver-negative cohort [16]. This suggests that additional screening techniques need to be utilized. RNA-NGS is a great candidate to extend the druggable map. However, distinct from Western countries where RNA-NGS utility is now routine diagnosis, RNA sequencing is less common in China. Therefore, few studies have been performed on melanoma.
To identify underlying genomic targets with clinical significance, DNA-NGS and RNA-NGS were utilized to construct molecular profiles of a Chinese cohort of 469 melanoma patients. We systematically delineated the molecular genetic landscape of Chinese melanoma patients and stratified patients into druggable or undruggable groups based on actionable genes, TMB status, and MSI status. Furthermore, potential actionable genes were investigated in the undruggable group, and we demonstrated how RNA sequencing is essential in bringing benefits to more melanoma patients.
2. Methods
2.1. Patients Population
Tumor histological specimens (formalin-fixed paraffin-embedded tissues or frozen tissues) obtained from multi-centers (Harbin Medical University Cancer Hospital, Zhejiang Provincial People’s Hospital and Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital)) between March 2018 and May 2020 were used for NGS detection. This study has been approved by the Ethics Committee of the Zhejiang Cancer Hospital (IRB-2021-3). All patients were diagnosed via pathology, imaging, and clinical findings.
2.2. DNA Isolation and Targeted Sequencing
Genomic DNA was extracted from tumor samples using a QIAamp DNA FFPE Tissue Kit (Qiagen, Valencia, CA, USA) and fragmented to construct a library using KAPA Hyper Prep kits (KAPA, KK8504). DNA fragments of 200~400 bp were purified by an Agencourt AMPure XP Kit (Beckman Coulter, CA, USA). Fragmented genomic DNA libraries were constructed with the KAPA HTP Library Preparation Kit (Kapa Biosystems, Wilmington, MA, USA) according to the instruction manual. After determining the concentration by a Qubit dsDNA HS Assay kit, DNA libraries were analyzed using Onco PanScan™ (GenetronHealth; Beijing, China), which is an 825-gene panel that includes major tumor-related genes.
2.3. RNA Sequencing
Total RNA was isolated from FFPE samples using the AllPrep DNA/RNA Mini Kit (QIAGEN, Hilden, Germany). The RNA quality and quantity were determined by the Qubit RNA HS Assay (Thermo Fisher Scientific, Waltham, MA, USA) and the 4200 TapeStation System (Thermo Fisher Scientific, Waltham, MA, USA), respectively. Qualified RNA was used to synthesize double-stranded cDNA using a SuperScript VILO cDNA Synthesis kit (Thermo Fisher Scientific, Waltham, MA, USA) and an Abclonal Second Strand Synthesis Module (Abclonal, Wuhan, China) according to the recommended protocol. To prepare genomic libraries, the double-stranded cDNA was fragmented and constructed by a KAPA HTP Library Preparation Kit (KAPA Biosystems, Wilmington, MA, USA). The prepared libraries were captured with FusionCapture™ (Genetron Health, Beijing, China), which contains 395 cancer-related genes, and then subjected to Illumina NovaSeq6000 (Illumina, Inc., San Diego, CA, USA) for paired-end sequencing. The sequencing reads were mapped to UCSC hg19 through HISAT2-2.0.5, and FusionMap was used to identify gene fusions.
2.4. Statistical Analysis
All statistical analyses were conducted using GraphPad Prism (version 8.02) software. The results are expressed as the mean ± SEM. The data were analyzed using a t test or one-way ANOVA, followed by the appropriate post-hoc tests (GraphPad Prism 7, GraphPad, CA, USA).
3. Results
3.1. The Landscape of Somatically Actionable Genes Identified by DNA-NGS
Melanoma samples were obtained from 256 men (55%) and 213 women (45%) with ages ranging from 5 to 89 years (average age = 57 years). Collectively, 469 resected melanoma samples were screened by DNA sequencing. Numerous pathways were involved in the landscape. The BRAF mutation, RAS mutation (NRAS, KRAS, HRAS), and NF1 mutation, which are included in the KRAS/RAF pathway, were found in 120 (26%), 115 (25%), and 46 (10%) samples, respectively. Mutations in PTEN and PIK3CA from the PIK3CA pathway were identified in 28 (6%) and 14 (3%) patients, respectively. The genes from the CDKN2A pathway were also analyzed, and TP53 (8%, n = 36), CDK4 (7%, n = 32), MDM2 (6%, n = 27), and CDKN2A (4%, n = 20) were the most frequently mutated genes. Furthermore, two receptor tyrosine kinase fusion genes not related to the above pathways, ALK and NRG1, were found in our cohort, accounting for 1.5% (n = 7) and 0.6% (n = 3), respectively. In addition, other top six mutated genes in the cohort were KIT (11%, n = 50), CCND1 (8%, n = 38), FAT3 (7%, n = 32), LRP1B (6%, n = 26), MYC (5%), and TERT (4%) (Figure 1A). Apart from the mutated genes, the tumor mutation burden (TMB) was also calculated in all patients (mean TMB = 8.7), and 96 (20.5%, 96/469) participants presented high TMB (≥10). Microsatellite instability (MSI) status was identified in 374 patients, including 1 patient (0.3%, 1/374) with MSI-H.
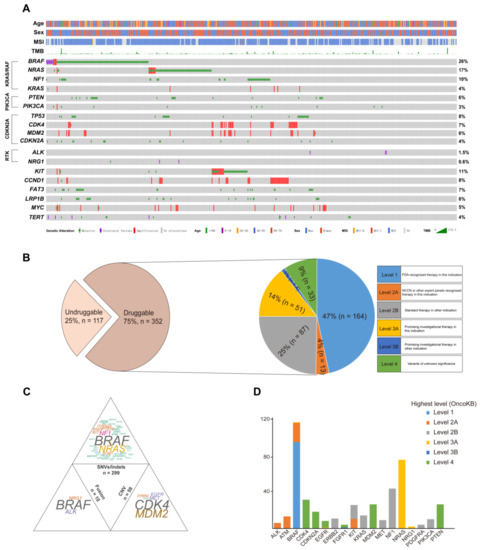
Figure 1.
The characteristics of the actionable genes in melanoma patients identified by DNA sequencing. (A) The spectrum of the variations in the melanoma; (B) the percentage of patients that were actionable in the cohort; (C) the distribution of targetable mutations based on mutation types; and (D) the number of patients was plotted against the most actionable genes in the cohort.
To broadly and systematically evaluate the clinical utility of molecular profiling to guide therapy decisions in melanoma patients, OncoKB, a curated knowledge base of the oncogenic effects and treatment implications of somatic mutations that were clarified by the FDA, was used to group all mutations into levels of clinical actionability (http://oncokb.org/, accessed on 1 January 2022). Seventy-five percent (352/469) of patients had actionable genes after matching those genes with actionable mutations in OncoKB. The druggable patients were then grouped based on the highest level of drug response evidence in OncoKB. Approximately 47% (164/352) of patients could be classified as OncoKB level 1. This was followed by level 2 (29%, 100/352), level 3 (15%, 55/352), and level 4 (9%, 33/352) (Figure 1B).
To explore the specific variations involved in the druggable group, the potential actionable mutations were classified in a mutation-type-specific manner: SNV (single nucleotide variant)/Indel, CNV (copy number variant), and fusion, which were found in 85% (299/352), 17% (59/352), and 3% (10/352) of patients, respectively (Figure 1C). The patients with BRAF, NRAS, and NF1 mutations were the top three in the SNV/Indel group, accounting for more than half of the total. In the CNV group, the proportions of CDK4 and MDM2 amplification were obviously predominant. Only three druggable tyrosine kinase fusion genes (ALK, BRAF, and NRG1) were identified at the DNA level, and the BRAF fusions were dominant (60%, n = 6) in the fusion group. Furthermore, the top genes in the three groups were analyzed, and the number of gene mutations was counted based on the highest actionable level from OncoKB. The top actionable mutation genes in the study were BRAF, NRAS, NF1, and CDK4. Most of the genes only had one highest level due to a single variation. However, BRAF, FGFR1, and KIT presented two highest levels because of various mutations (Figure 1D).
3.2. RNA-NGS Could Identify Actionable Fusions in Undruggable Melanoma Patients
To explore additional opportunities for targeted therapy, the undruggable melanoma group identified by DNA-NGS was further sequenced by RNA-NGS. In total, 117 melanoma patients were identified as undruggable based on the DNA-NGS results. Sixty patients with enough samples were sequenced by RNA-NGS, and positive fusions were found in 15 samples (25%) (Figure 2A). These fusions involved multiple driver genes: BRAF (5%, n = 3), RAF1 (3%, n = 2), ALK (2%, n = 1), FGFR1 (2%, n = 1), ETV6 (2%, n = 1), AKT3 (2%, n = 1), TCF12 (2%, n = 1), ATF (2%, n = 1), ZCCHC7 (2%, n = 1), ARHGEF2 (2%, n = 1), KDM6A (2%, n = 1), and WHSC1L1 (2%, n = 1) (Figure 2B). Based on the RNA-NGS results, 47% (7/15) of the fusions could be druggable in the fusion-positive group, including PUM2-ALK, FGFR1-TACC1, AGAP3-BRAF, AGK-BRAF, ZKSCAN1-BRAF, SOX6-RAF1, and CTDSPL-RAF1. Most of the rearrangements occurred on the same chromosome, except for SOX6-RAF1 (chr11-chr3). All fusions with druggable potential could be targeted by drugs from clinical trials (Figure 2C). In total, RNA-seq identified 11.7% (7/60) of patients in the undruggable group who carried potentially actionable targets.
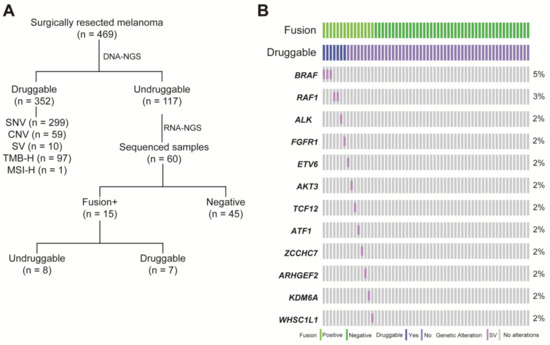
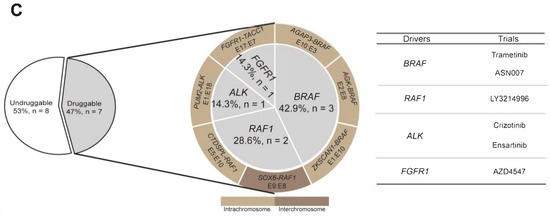
Figure 2.
The fusion landscape of undruggable patients by RNA sequencing. (A) Description of the melanoma cohort by comprehensive DNA-seq and RNA-seq; (B) OncoPrint depicting fusions in the undruggable patients by Fusioncapture panel; and (C) pie chart summarizing the actionable fusions and matched therapy on RNA-level.
3.3. RNA-NGS Could Significantly Enhance the Proportion of Fusions in Melanoma
At the DNA level, 27 fusions involving multiple fusion genes from the whole cohort were identified in 23 patients, which accounted for 4.9% of the cohort (23/469): ALK (n = 4), BRAF (n = 10), NRG1 (n = 2), RAF1 (n = 2), BCR (n = 1), NAB2 (n = 2), RELA (n = 2), and TERT (n = 2) (Figure 3A). We explored how integrated DNA-NGS and RNA-NGS sequencing are essential in enhancing the proportion of druggable patients with melanoma through fusion detection. Patients with enough samples after DNA sequencing were detected by RNA-NGS (n = 110), regardless of whether they were druggable. At the RNA level, positive fusions were validated in 38 patients: ALK (n = 3), BRAF (n = 10), FGFR1 (n = 1), NTRK2 (n = 1), NTRK3 (n = 1), RAF1 (n = 3), AKT3 (n = 1), ATF1 (n = 1), CBFB (n = 1), ETS1 (n = 2), ETV6 (n = 2), JAZF1 (n = 2), KDM6A (n = 1), MAG (n = 1), METFA (n = 1), NF1 (n = 1), PAX5 (n = 1), RB1 (n = 1), SMARCB1 (n = 1), TCF12 (n = 1), WHSC1L1 (n = 1), and ZCCHC7 (n = 1) (Figure 3A). RNA-NGS enhanced the total fusion proportion from 4.9% (23/469) to 34.6% (38/110) (Figure 3A). If only the fusions with clinical potential were considered, the fusion proportion was 2.1% (10/469) at the DNA level; however, RNA-GS increased the percentage to 17.27% (19/110) (Figure 3A). Overall, RNA-NGS increased the percentage of druggable patients from 75% to 78% (Figure 3B).
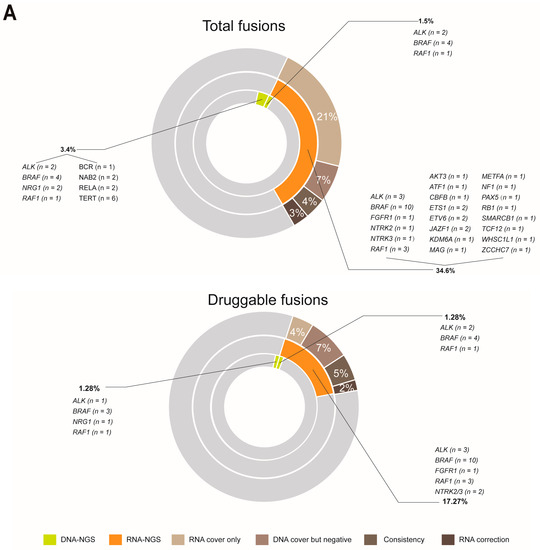
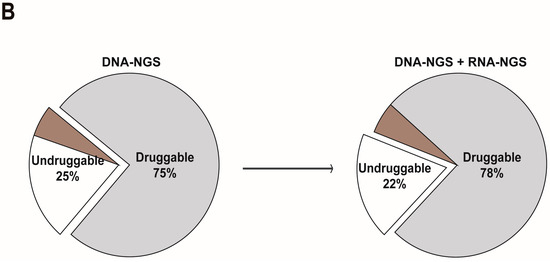
Figure 3.
The comparison of fusion detection rates between DNA and RNA sequencing. (A) RNA sequencing increased the detection proportion of total and druggable fusions; and (B) RNA sequencing expanded the scope (enlarged the proportion) of druggable patients in melanoma.
The significant enhancement of fusion detection by RNA-NGS was attributed to the different designs of the DNA-NGS and RNA-NGS panels, which could be categorized into four groups: breakpoints that were covered by only the RNA-NGS panel (RNA cover only), breakpoints that were covered by DNA-NGS but failed to identify fusions (DNA cover but negative), the fusions that were verified by both DNA-NGS and RNA-NGS (Consistency), and the different partners that were validated by DNA-NGS and RNA-NGS (RNA correction) (Figure 3A). In the total fusion-positive cohort (34.6%), RNA cover accounted for only 21%, DNA cover but negative accounted for 7%, Consistency accounted for 4%, and the last 3% could be attributed to RNA correction (Figure 3A). Comparatively, in the druggable fusions group (17.27%), 4% was due to RNA cover only, 7% was due to DNA cover but negative, and Consistency and RNA correction accounted for 5% and 2%, respectively (Figure 3A).
3.4. Unreliability of the Fusion Breakpoint in Predicting Relevant Transcripts Based on DNA-NGS
Two fusions were detected by both DNA-NGS and RNA-NGS; however, their partners were different (Figure 4), namely, BRAF-WDFY1 to CUX1-BRAF and LMCD1-RAF1 to ERC1-RAF1 (Table S1, P1–P2). Patient 1 harbored a BRAF-WDFY1 fusion detected by DNA NGS with “antisense rearrangements” juxtaposing the 5′ portion of the kinase genes and the 3′ portion of intergenic partners (Figure 4A). This indicated that BRAF lacked the tyrosine kinase domain (TKD), and the patient could not be referred to RAF tyrosine kinase domain (TKIs). However, the RNA-NGS results revealed that CUX1 exon 11 was fused to BRAF exon 9 with an intact TKD. The inconsistency between the DNA and RNA results was perhaps due to the reciprocal rearrangement in which DNA-NGS detected only the opposite breakpoint downstream of BRAF rather than the primary junction constituting CUX1 and BRAF. In Patient 2, DNA-NGS revealed that the partner LMCD1 was fused to the 3′ portion of RAF1 and that the breakpoint was in intron 9 of RAF1. At the RNA level, CUX1-BRAF was found, in which exon 8 of RAF1 was involved (Figure 4B). Alternative splicing may contribute to functional expression.
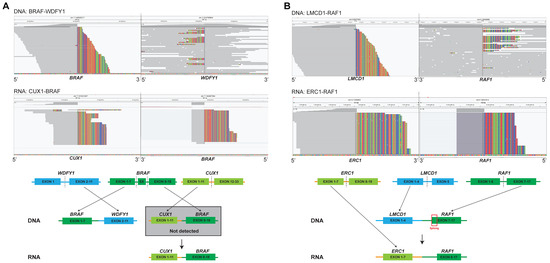
Figure 4.
RNA sequencing can identify and correct the putative fusion transcripts of DNA sequencing. (A) BRAF fusion identified by both DNA-seq and RNA-seq; and (B) RAF1 fusion identified by both DNA-seq and RNA-seq.
3.5. Novel Fusions Detected in Melanoma Patients
Ten novel potential druggable fusions were found in the cohort, which involved ALK, BRAF, RAF1, and NTRK2 (Figure 5A). SLC9A3-BRAF, PTPRM-BRAF, and DYNC2H1-BRAF were identified by both DNA-NGS and RNA-NGS. ERC1-RAF1, PUM2-ALK, ZNF131-BRAF, COBL-BRAF, CTDSPL-RAF1, and MYO5A-NTRK2 were identified by RNA-NGS only, although the breakpoint of ERC1-RAF1 was covered by DNA-NGS. USP20-RAF1 was identified and sequenced only by DNA-NGS due to the limited amount of specimen. The H&E staining images of each fusion are presented in Figure 5A and Figure S1. Samples with enough specimens were further validated by FISH, and break-apart signals were found in both DYNC2H1-BRAF and MYO5A-NTRK2 (Figure 5B).
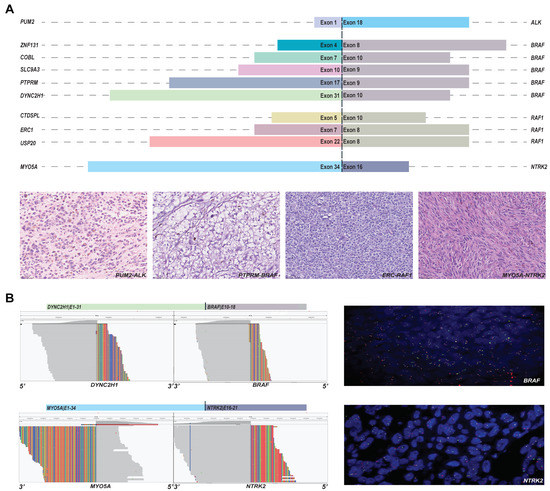
Figure 5.
The novel fusions detected by panels. (A) The partners and drivers of novel fusions and representative H&E images; and (B) the further validation by FISH.
4. Discussion
In this study, the genetic landscape was systematically analyzed in a Chinese melanoma cohort. Seventy-five percent of Chinese melanoma patients were found to be targetable, and the predominant druggable gene was BRAF based on DNA-NGS. Furthermore, RNA-NGS identified actionable fusions in 11.67% of patients in the undruggable group, which further enhanced the number of druggable patients from 75% to 78%. These results demonstrated the importance of integrated DNA and RNA sequencing in improving druggable patients with melanoma, which could accurately guide personalized melanoma therapy.
Melanoma has been studied extensively in the Caucasian population [7,17,18,19,20], but only a few studies with limited cohort sizes could be found in Asia [3,21,22,23,24]. Annotated by the OncoKB database, the results from the MSKCC and AACR studies indicated that approximately 60% of Caucasian melanoma patients could be treatable after DNA-NGS testing [14,15]. In contrast, Park et al. discovered that DNA-seq could help approximately 40% of Asian melanoma patients [25]. It seems that there is a pathological split between Caucasian and Asian populations. However, around 75% of Asian melanoma patients could be treatable with medication based on our findings. TMB-H (20.5%), annotated as L1 in OncoKB, was analyzed in the cohort and may contribute to the high actionable proportion. Nonetheless, around 55% patients were druggable after excluding TMB-H patients. Furthermore, up to 5% of patients in the cohort had fusions with potential clinical significance, which may be another reason. To our knowledge, druggable fusions in Asian melanoma have not yet been characterized. No fusions were reported in the largest known Asian cohort (n = 178) from the Park et al. study [22]. Only one BRAF fusion and one ALK fusion were identified in the largest Chinese cohort (n = 81) from the Huang et al. study [21]. In contrast, 16 fusions with potential clinical significance were detected in our study, which is the largest Chinese melanoma cohort (n = 469). The leading number of fusions with clinical significance in our cohort was that of BRAF (1.7%, 8/469), which was slightly higher than that in the Caucasian population (1.1%) [14]. It is interesting to note that in contrast to glioma, KIAA1549-BRAF is the most common BRAF fusion [26]. The partners of BRAF fusions in our present study were discrete. This result demonstrated that there was no enriched BRAF fusion type in melanoma. Furthermore, the RAF1 fusions (0.4%, 2/469) in the same pathway as BRAF were first reported in the Asian cohort. Among these fusions, four (three BRAF fusions and one RAF1 fusion) were novel. Additionally, our study identified NRG1 fusions in melanoma, which have never been found before. Surprisingly, we found high numbers of ALK rearrangements in melanoma (n = 4) for the first time. However, compared to the results in lung cancer [27], the partners of the ALK fusions in our study were totally different, and none of them were the canonical type (EML4-ALK). Couts et al. demonstrated that melanoma expressing EML4-ALK was sensitive to ALK inhibitors in vitro and in vivo [28]. This result emphasized the widespread beneficial potential for melanoma patients with ALK fusions from our study.
Although more than half of the population could be targetable based on DNA-NGS results from our findings and the abovementioned studies [14,15], approximately 25% to 42% of patients were still nondruggable. Benayed et al. found that extra fusions were identified by RNA-NGS in 14% of NSCLC patients from a driver-negative group, and 91.6% of the fusions were targetable [16]. This suggested that RNA-NGS is an effective way to expand the druggable targets by identifying actionable fusions. However, few studies have explored how RNA-NGS is essential in enhancing the druggable proportion in melanoma. Hutchinson et al. [7] reported only two fusions from a driver-negative melanoma cohort, which was limited to BRAF fusions. In our study, RNA-NGS sequencing was systemically performed on patients without potential clinical utility at the DNA level for the first time, and comprehensive fusion types were investigated. In addition to the BRAF fusions, ALK, RAF1, and FGFR1 fusions were detected in the cohort. Briefly, up to 11.7% (7/60) of patients in the undruggable group could benefit from RNA-NGS based on the results. Overall, the use of RNA-NGS increased the proportion of treatable patients from 75% to 78%.
The increased proportion of fusion detection by RNA-seq could be attributed to several causes. Large introns could not be covered by the panel assay due to their length. For example, intron 13 of NTRK3 is close to 100 kb in length (UCSC Genome Browser), which is close to 10% of the total size of the panel. Tiling such introns is not only technically challenging but also not practical in terms of the overall sequencing throughput. Additionally, introns with low frequency will not be tiled due to the cost factor. For example, ALK fusions have been frequently found in introns 18, 19, and 20 [29]. To our knowledge, the ALK breakpoint in intron 17 was first identified in our study, so it is unrealistic to be covered as a frequent intron in gene rearrangement.
On the other hand, eight fusions only detected by RNA-NGS were expected to be identified by DNA-NGS. Due to the complexity of the introns, the rearrangement that takes place in an intron may not be successfully detected by DNA-NGS. Five of those fusions involved BRAF on introns 7, 8, and 9. Similar to ROS1, the repeat sequence in those introns can be present at many other sites in the genome, and the inclusion of baits for these regions would simply result in unmappable reads [16,30]. Therefore, genomic breaks in these regions are usually missed. To enhance the fusion detection rate, the introns of specific partners could be included in the panel. For example, CD74, the most frequent partner in the ROS1 fusion, is usually covered by the DNA-seq panel. However, the partners in the BRAF fusions were too diverse to be covered.
Limitations
Several limitations should be discussed. The clinicopathological parameters, such as clinical subtypes and disease stages were missed in the study. Those parameters could bring more a comprehensive understanding of Asian melanoma. Due to limitations in the amount of specimens, only samples with enough specimens were further sequenced by RNA-NGS. The resulting increased proportion of potential druggable patients in the undruggable group may be biased. However, more than half of the samples in the undruggable group had been analyzed by RNA-NGS, so the results did not induce significant bias. In addition, this study was a retrospective study, and patients with positive fusion detected by RNA-NGS were not able to receive targeted therapy, especially those with novel fusions with potential clinical significance. Therefore, further prospective trials of applying DNA and RNA sequential sequencing in targeted therapy for melanoma patients are needed in the future.
5. Conclusions
This study systemically analyzed the genetic landscape of Asian melanoma and demonstrated how DNA and RNA sequential sequencing is essential in bringing clinical benefits to more melanoma patients.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15010283/s1, Figure S1. The representative H&E images of novel fusions. Table S1. The comparison of fusions at DNA and RNA levels in the melanoma patients.
Author Contributions
Y.L. and B.W. contributed to the experiment performing and manuscript writing. C.W. and D.Z. participated to the data analysis. Y.N., X.W., Z.L., W.L., J.Z. and H.T. provided the clinical samples and relevant information. T.M. and T.L. designed and optimized the experiment. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Zhejiang Province medical and health research projects grant No. 2016KYB027 and Grant No. 2017KY010.
Institutional Review Board Statement
Ethics Committee of the Zhejiang Cancer Hospital (IRB-2021-301).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
All data used in this study were available in the main text and Supplementary Materials.
Acknowledgments
We would like to thank the patients for donating specimens and all participant have consented to the acknowledgement.
Conflicts of Interest
B.W., C.W., D.Z., Y.N., X.W., W.L., and J.Z. were employed by Jichenjunchuang Clinical Laboratory. All other authors have declared no conflict of interest.
List of Abbreviations
| NGS | Next-generation sequencing |
| TKI | Tyrosine kinase inhibitor |
| TMB | Tumor mutation burden |
| MSI | Microsatellite instability |
| NSCLC | Non-small-cell lung cancer |
| FFPE | Formalin-fixed paraffin-embedded |
References
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; de Vries, E.; Whiteman, D.C.; Bray, F. Global Burden of Cutaneous Melanoma in 2020 and Projections to 2040. JAMA Dermatol. 2022, 158, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Manca, A.; Paliogiannis, P.; Colombino, M.; Casula, M.; Lissia, A.; Botti, G.; Caracò, C.; Ascierto, P.A.; Sini, M.C.; Palomba, G.; et al. Mutational concordance between primary and metastatic melanoma: A next-generation sequencing approach. J. Transl. Med. 2019, 17, 289. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Li, J.; Wen, X.; Zhu, B.; Liu, W.; Wang, J.; Jiang, H.; Ding, Y.; Li, D.; Zhang, X. Next-generation sequencing in advanced Chinese melanoma reveals therapeutic targets and prognostic biomarkers for immunotherapy. Sci. Rep. 2022, 12, 9559. [Google Scholar] [CrossRef] [PubMed]
- Afshar, A.R.; Damato, B.E.; Stewart, J.M.; Zablotska, L.B.; Roy, R.; Olshen, A.B.; Joseph, N.M.; Bastian, B.C. Next-Generation Sequencing of Uveal Melanoma for Detection of Genetic Alterations Predicting Metastasis. Transl. Vis. Sci. Technol. 2019, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Diefenbach, R.J.; Lee, J.H.; Menzies, A.M.; Carlino, M.S.; Long, G.V.; Saw, R.P.M.; Howle, J.R.; Spillane, A.J.; Scolyer, R.A.; Kefford, R.F.; et al. Design and Testing of a Custom Melanoma Next Generation Sequencing Panel for Analysis of Circulating Tumor DNA. Cancers 2020, 12, 2228. [Google Scholar] [CrossRef]
- Wen, P.Y.; Stein, A.; van den Bent, M.; De Greve, J.; Wick, A.; de Vos, F.; von Bubnoff, N.; van Linde, M.E.; Lai, A.; Prager, G.W.; et al. Dabrafenib plus trametinib in patients with BRAF(V600E)-mutant low-grade and high-grade glioma (ROAR): A multicentre, open-label, single-arm, phase 2, basket trial. Lancet Oncol. 2022, 23, 53–64. [Google Scholar] [CrossRef]
- Hutchinson, K.E.; Lipson, D.; Stephens, P.J.; Otto, G.; Lehmann, B.D.; Lyle, P.L.; Vnencak-Jones, C.L.; Ross, J.S.; Pietenpol, J.A.; Sosman, J.A.; et al. BRAF fusions define a distinct molecular subset of melanomas with potential sensitivity to MEK inhibition. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 6696–6702. [Google Scholar] [CrossRef]
- Williams, E.A.; Shah, N.; Montesion, M.; Sharaf, R.; Pavlick, D.C.; Sokol, E.S.; Alexander, B.M.; Venstrom, J.M.; Elvin, J.A.; Ross, J.S.; et al. Melanomas with activating RAF1 fusions: Clinical, histopathologic, and molecular profiles. Mod. Pathol. Off. J. United States Can. Acad. Pathol. Inc. 2020, 33, 1466–1474. [Google Scholar] [CrossRef]
- Forschner, A.; Forchhammer, S.; Bonzheim, I. NTRK gene fusions in melanoma: Detection, prevalence and potential therapeutic implications. JDDG J. Dtsch. Dermatol. Ges. 2020, 18, 1387–1392. [Google Scholar] [CrossRef]
- Marcus, L.; Fashoyin-Aje, L.A.; Donoghue, M.; Yuan, M.; Rodriguez, L.; Gallagher, P.S.; Philip, R.; Ghosh, S.; Theoret, M.R.; Beaver, J.A.; et al. FDA Approval Summary: Pembrolizumab for the Treatment of Tumor Mutational Burden-High Solid Tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 4685–4689. [Google Scholar] [CrossRef]
- André, T.; Shiu, K.K.; Kim, T.W.; Jensen, B.V.; Jensen, L.H.; Punt, C.; Smith, D.; Garcia-Carbonero, R.; Benavides, M.; Gibbs, P.; et al. Pembrolizumab in Microsatellite-Instability-High Advanced Colorectal Cancer. N. Engl. J. Med. 2020, 383, 2207–2218. [Google Scholar] [CrossRef]
- Chan, T.A.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.A.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 44–56. [Google Scholar] [CrossRef]
- Kubeček, O.; Kopecký, J. Microsatellite instability in melanoma: A comprehensive review. Melanoma Res. 2016, 26, 545–550. [Google Scholar] [CrossRef]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov. 2017, 7, 818–831. [CrossRef]
- Benayed, R.; Offin, M.; Mullaney, K.; Sukhadia, P.; Rios, K.; Desmeules, P.; Ptashkin, R.; Won, H.; Chang, J.; Halpenny, D.; et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 4712–4722. [Google Scholar] [CrossRef]
- Ghiasvand, R.; Berge, L.A.M.; Andreassen, B.K.; Stenehjem, J.S.; Heir, T.; Karlstad, Ø.; Juzeniene, A.; Larsen, I.K.; Green, A.C.; Veierød, M.B.; et al. Use of antihypertensive drugs and risk of cutaneous melanoma: A nationwide nested case-control study. Int. J. Epidemiol. 2022. [Google Scholar] [CrossRef]
- Nakamura, K.; Ashida, A.; Kiniwa, Y.; Okuyama, R. Immune status and prognosis of stage II and III primary malignant melanoma: An analysis including 77 cases of acral lentiginous melanoma. J. Dermatol. 2022. [Google Scholar] [CrossRef]
- Bowley, T.Y.; Lagutina, I.V.; Francis, C.; Sivakumar, S.; Selwyn, R.G.; Taylor, E.; Guo, Y.; Fahy, B.N.; Tawfik, B.; Marchetti, D. The RPL/RPS gene signature of melanoma CTCs associates with brain metastasis. Cancer Res. Commun. 2022, 2, 1436–1448. [Google Scholar] [CrossRef]
- Khan, M.; Thompson, J.; Kiiskila, L.; Oboh, O.; Truong, T.; Prentice, A.; Assifi, M.M.; Chung, M.; Wright, G.P. Timing and necessity of staging imaging in clinical stage II cutaneous melanoma: Cost-effectiveness and clinical decision analysis. Am. J. Surg. 2022, 225, 93–98. [Google Scholar] [CrossRef]
- Luo, Y.; Zhang, Z.; Liu, J.; Li, L.; Xu, X.; Yao, X.; Dai, Z.; Wang, X.; Yang, S.; Wu, H.; et al. Characterizations of Gene Alterations in Melanoma Patients from Chinese Population. BioMed Res. Int. 2020, 2020, 6096814. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yang, L.; Lai, Y.; Wang, X.; Han, X.; Liu, S.; Wang, D.; Li, X.; Hu, N.; Kong, Y.; et al. Genetic alteration of Chinese patients with rectal mucosal melanoma. BMC Cancer 2021, 21, 623. [Google Scholar] [CrossRef] [PubMed]
- Zheng, A.W.; Jia, D.D.; Zhou, C.C.; Li, T. Mutational profiling of melanomas in patients from the southeast coast of China. Transl. Cancer Res. 2020, 9, 4781–4789. [Google Scholar] [CrossRef] [PubMed]
- Byeon, S.; Cho, H.J.; Jang, K.T.; Kwon, M.; Lee, J.; Lee, J.; Kim, S.T. Molecular profiling of Asian patients with advanced melanoma receiving check-point inhibitor treatment. ESMO Open 2021, 6, 100002. [Google Scholar] [CrossRef] [PubMed]
- Park, K.H.; Choi, J.Y.; Lim, A.R.; Kim, J.W.; Choi, Y.J.; Lee, S.; Sung, J.S.; Chung, H.J.; Jang, B.; Yoon, D.; et al. Genomic Landscape and Clinical Utility in Korean Advanced Pan-Cancer Patients from Prospective Clinical Sequencing: K-MASTER Program. Cancer Discov. 2022, 12, 938–948. [Google Scholar] [CrossRef]
- Bouchè, V.; Aldegheri, G.; Donofrio, C.A.; Fioravanti, A.; Roberts-Thomson, S.; Fox, S.B.; Schettini, F.; Generali, D. BRAF Signaling Inhibition in Glioblastoma: Which Clinical Perspectives? Front. Oncol. 2021, 11, 772052. [Google Scholar] [CrossRef]
- Li, W.; Guo, L.; Liu, Y.; Dong, L.; Yang, L.; Chen, L.; Liu, K.; Shao, Y.; Ying, J. Potential Unreliability of Uncommon ALK, ROS1, and RET Genomic Breakpoints in Predicting the Efficacy of Targeted Therapy in NSCLC. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2021, 16, 404–418. [Google Scholar] [CrossRef]
- Couts, K.L.; Bemis, J.; Turner, J.A.; Bagby, S.M.; Murphy, D.; Christiansen, J.; Hintzsche, J.D.; Le, A.; Pitts, T.M.; Wells, K.; et al. ALK Inhibitor Response in Melanomas Expressing EML4-ALK Fusions and Alternate ALK Isoforms. Mol. Cancer Ther. 2018, 17, 222–231. [Google Scholar] [CrossRef]
- Ou, S.I.; Zhu, V.W.; Nagasaka, M. Catalog of 5’ Fusion Partners in ALK-positive NSCLC Circa 2020. JTO Clin. Res. Rep. 2020, 1, 100015. [Google Scholar] [CrossRef]
- Treangen, T.J.; Salzberg, S.L. Repetitive DNA and next-generation sequencing: Computational challenges and solutions. Nat. Rev. Genet. 2011, 13, 36–46. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).