Simple Summary
Prostate cancer is a serious threat to male health worldwide; however, chemotherapy remains an urgent problem because of resistance and insensitivity to androgen-deprivation therapy. Therefore, discovering novel molecules and chemotherapy can provide a new strategy. Pyruvate kinase M2 (PKM2) is not only a vital enzyme in regulating cancer glycolysis, but is also a transcription that regulates gene expression through nuclear translocation, so the targeting PKM2 can be a promising therapy for prostate cancer. In this experiment, we found a novel specific PKM2 inhibitor—compound 3h—and conducted molecular and cellular experiments to evaluate its anticancer effect. Our results indicated that proliferation of LNCaP cells can be inhibited by compound 3h through inducing apoptosis and autophagy, and inhibiting glycolysis and mitochondria respiration. Therefore, our results provide a new way of developing novel chemotherapy for prostate cancer.
Abstract
Pyruvate kinase M2 (PKM2) is a key enzyme involved in the regulation of glycolysis. Although PKM2 is overexpressed in various tumor tissues, its functional role in cancer chemotherapy remains unexplored. In this study, we investigated the anticancer activity of a new PKM2 inhibitor, compound 3h, through the cell metabolism and associated signaling pathways in prostate cancer cells. To evaluate the molecular basis of specific PKM2 inhibitors, the interactions of compounds 3h and 3K with the PKM2 protein were assessed via molecular docking. We found that, compared to compound 3K, compound 3h exhibited a higher binding affinity for PKM2. Moreover, compound 3h significantly inhibited the pyruvate kinase activity and PKM2 expression. Cytotoxicity and colony formation assays revealed the potent anticancer activity of compound 3h against LNCaP cells. Compound 3h significantly increased the apoptotic and autophagic cell death in LNCaP cells. In addition, compound 3h induced AMPK activation along with the inhibition of the mTOR/p70S6K pathway. Furthermore, compound 3h significantly inhibited glycolysis and mitochondrial respiration, as determined by analyzing the extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) production. Our results revealed that compound 3h caused apoptotic and autophagic cell death in LNCaP cells by inhibiting cancer cell metabolism. Therefore, blocking glycolytic pathways using specific PKM2 inhibitors can target cancer cell metabolism in PKM2-overexpressed prostate cancer cells.
1. Introduction
Prostate cancer has the character of malignant growth metastasis, and prostate cancer with bone marrow metastasis, in particular, is a global medical problem with no efficient therapy. Prostate cancer is considered the second greatest threat to the health of American males, with 268,490 new cases and 34,500 deaths predicted to occur in 2022, according to American Cancer Society [1]. Sixty percent of patients with prostate cancer are diagnosed at the age of 65 or older; it is rarely diagnosed in individuals younger than 40 years [2]
Cancer cells require sufficient energy and biosynthetic precursors for rapid proliferation. Unlike in normal cells, active glycolysis occurs in cancer cells even though oxygen is sufficient, which requires abundant glucose [3,4]. Pyruvate kinase (PK), a key rate-limiting enzyme in the Warburg effect, consists of four subtypes (PKM1, PKM2, PKL, and PKR) encoded by two groups of genes. PKM2 in cancer cells has become a key target for cancer treatment. Unlike other pyruvate kinases, PKM2 has tetrameric and dimeric forms (Figure 1). Tetramers, mainly PKM1, function as pyruvate kinase and regulate glycolysis, while dimers act as switches in energy metabolism and biosynthesis, converting the glucose metabolism of producing pyruvate [5]. Dimeric PKM2, as a transcription factor, translocates to the nucleus and activates and regulates the transcription of certain genes, including β-catenin. In addition, c-Myc, glucose transporter 1 (GLUT1), and lactate exporter monocarboxylate transporter 4 (MCT4), as the downstream protein of β-catenin, can be regulated [6,7]; therefore, PKM2-overexpression leads to increased glycolysis by increasing glucose input and lactate acid output, which speeds cancer progression [8,9]. In addition, the ratio of PKM2 determines whether the intracellular glucose metabolism is involved in the biosynthesis for proliferation or oxidative phosphorylation (OXPHOS) in the mitochondria [10].
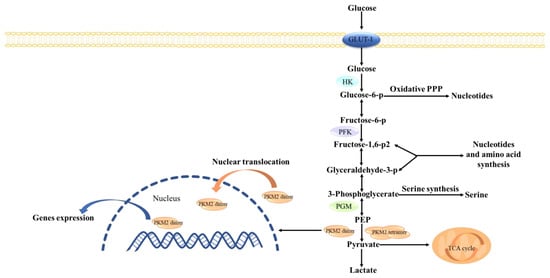
Figure 1.
The tetramer and dimer of PKM2 regulate glycolysis and gene transcription in cancer cells. The active tetramer PKM2 is a glycolytic enzyme that transfers PEP into pyruvate, which enters mitochondria and joins in the TCA cycle. The low activity PKM2 dimer leads to the accumulation of glycolytic intermediates to support the biosynthetic precursors of proliferating cells. In addition, dimeric PKM2, as a transcription factor, translocates to the nucleus and activates and regulates transcription of certain genes. GLUT1: Glucose transporter 1, PPP: Pentose Phosphate Pathway, HK: Hexokinase, PFK: Phosphofructokinase 1, PGM: Phosphoglycerate mutase, PEP: Phosphoenolpyruvate. TCA cycle: Tricarboxylic acid cycle, PKM2: Pyruvate kinase M2.
Endogenous and exogenous activators and inhibitors can regulate the transition between the dimers and tetramers of PKM2. Tetramers have a seesaw-like pattern with allosteric regulatory domains, and they switch between the inactive T-state and active R-state [11]. Yuan et al. found that amino acids bind to an allosteric pocket (existing in PKM2 and PKM1); most of them stabilize PKM2 in an inactive T-state, and phenylalanine and alanine are almost 100% inhibited [12]. In addition, most amino acids show specificity for PKM2, with phenylalanine, arginine, glycine, and lysine showing a slight inhibitory effect (approximately 10%) on PKM1 [12,13]. Recently, novel naphthoquinone derivatives have been synthesized as selective inhibitors of PKM2, including compound 3K. The anticancer and PKM2 inhibitory effects of compound 3K were evaluated in vivo and in vitro. Currently, a novel 2, 3-didithiocarbamate-substituted naphthoquinone called compound 3h (half-maximal inhibitory concentration [IC50] = 0.96 ± 0.18 μM), which is synthesized based on compound 3K, provides potent inhibition of PKM2 that is superior to compound 3K (IC50 = 2.95 ± 0.53 μM) [14].
Activation of the protein kinase B (Akt)/mechanistic or mammalian target of rapamycin (mTOR) plays a vital role in regulating cell survival and suppressing autophagy in cancer cells [15,16]. It has been proven that autophagy in human prostate cancer cells is induced by PKM2 knockdown via the Akt/mTOR pathway [17], and a specific PKM2 inhibitor, compound 3K, also promoted the cell death of SK-OV-3 via restraining the Akt/mTOR pathway [18].
Although inhibiting PKM2 and glycolysis has been explored as an effective approach for cancer therapy, screening for novel PKM2 inhibitors and comparing their anticancer effects, thereby clarifying their action mechanisms, have not yet been reported; therefore, we found a specific PKM2 inhibitor called compound 3h, evaluated its anticancer effects on prostate cancer cells, and performed molecular docking to further understand the mechanism of PKM2 inhibition in this study. Our results suggest that compound 3h can suppress glycolysis by selectively inhibiting PKM2 expression and activity and then playing an anticancer role, which may provide a strategy for prostate cancer treatment.
2. Materials and Methods
2.1. Reagents
Compounds 3h and 3K were dissolved in dimethyl sulfoxide (DMSO) (Sigma-Aldrich, St. Louis, MO, USA). Compound 3h was synthesized by Professor In Su Kim (Department of Pharmacy, Sungkyunkwan University, Suwon, Republic of Korea) according to Ning’s method [14]. Compound 3K (Catalog No. S8616) was purchased from SelleckChem (Houston, TX, USA). The structures of compounds 3h and 3K are shown in Figure 2. Roswell Park Memorial Institute (RPMI) 1640 medium (LM011-01) and fetal bovine serum (FBS) were purchased from Gibco Invitrogen Corporation (Carlsbad, CA, USA). Compound 3h was diluted to an appropriate concentration in RPMI 1640 medium containing 10% FBS (S101-07, Welgene, Gyeongsan-si, Republic of Korea). Propidium iodide (PI) solutions and 4,6-diamidino-2-phenylindole (DAPI) were purchased from Sigma-Aldrich (St. Louis, MO, USA).
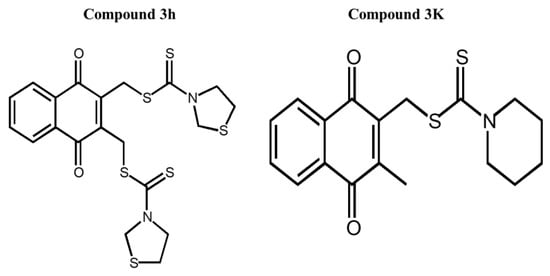
Figure 2.
Chemical structures of compounds 3h and 3K.
2.2. Antibodies
Primary antibodies against PKM2 (#4053S), PKM1 (#7067), poly (ADP-ribose) polymerase (PARP) (#9542), cleaved-PARP (#5625S), pro-caspase-7 (#9492), pro-caspase-3 (#9662), Cyclin-B1 (#4138), cell division cycle protein 2 homolog (Cdc2) (#77055), p-Cdc2 (#9111S), AMP-activated protein kinase (AMPK)-α (#2532), p-AMPKα (#2535), phosphatase and tensin homolog (PTEN) (#9559S), p-PTEN (#9551), Akt (#9272), p-Akt (#9271S), p-mTOR (#2971S), mTOR (#2972), ribosomal protein S6 kinase beta-1 (p70S6K) (#9202S), and p-p70S6K (#9206S) were purchased from Cell Signaling (Beverly, MA, USA). Primary antibodies against B-cell lymphoma 2 (Bcl-2) (sc-783), c-Myc (sc-40), Bcl-2-associated X (Bax) (sc-7480), GLUT1 (sc-7903), MCT4 (sc-376465), and β-actin (sc-47778) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Purchased primary antibodies against LC3B (ab51520), p62 (ab56416), and β-catenin (ab6302) were from Abcam (Abcam, Cambridge, UK). Beclin-1 (NB500-249) was purchased from Novus Biologicals (Littleton, CO, USA). Horseradish peroxidase (HRP)-conjugated secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, Dallas, TX, USA).
2.3. Cell Lines and Cell Culture
All three human prostate cancer cell lines (LNCaP, PC3, and DU145) were purchased from the American Type Culture Collection (Manassas, VA, USA). Cultured cells in RPMI 1640 medium were supplemented with 10% heat-inactivated FBS, antibiotic–antimycotic (15240062) in a humidified, 5% CO2 atmosphere at 37 °C.
2.4. PK Assay
A PK assay kit (ab83432, Abcam, Cambridge, UK) was used to detect PK assay activity in LNCaP cells. In short, according to the guide, protein was extracted from cells (50 × 104), pellets with four volumes of assay buffer and protein were added to a 96-well plate, then the PK assay buffer was added for a volume of 50 μL/well. The prepared reaction was mixed with the assay buffer, substrate mix, enzyme mix, and OxiRed™ probe provided in the PK assay kit, and the 50 μL/well reaction mix was added to the 96-well plate containing the protein. The standard linear curve was determined by serial dilutions of 1 nmol/mL standard pyruvate solution. After the reaction, the absorbance of all standards and samples was determined at a 570 nm wavelength using a VERSA Max Microplate Reader (Molecular Devices Corp., Sunnyvale, CA, USA).
2.5. Prepare Ligand and Receptor for Docking
All docking modeling procedures were performed using the Tripos Sybyl-X 2.1.1 (Tripos Inc., St Louis, MO, USA) molecular modeling package with a Windows 7 professional K operating system. First, compounds 3h and 3K were prepared with the sketch module and saved as mol2 format; we then assigned all atoms Gasteiger-Hückel charges. Before performing molecular docking, in order to obtain a stable conformation that converged to the maximum derivatives of 0.001 kcal mol−1.Å−1, we performed an energy minimization of ligands. The crystal structure of PKM2 in complex with phenylalanine (Protein Data Bank [PDB]:4FXJ) used in the experiment was downloaded from PDB. To prepare the receptor, the ligand phenylalanine was extracted, and all water molecules were deleted from the complex structure. The structure was then prepared with the protein preparation module of Sybyl-X 2.1.1 using the default parameters.
2.6. Molecular Docking of PKM2 and Compound 3K and 3h
In this study, we used Surflex-Dock embedded in Tripos Sybyl-X 2.1.1 to perform flexible docking. An SFXC file was constructed using a mol2 prepared protein structure. Phenylalanine was separated from the co-crystal structure of PKM2 (PDB: 4FXJ) to generate the binding site, and the binding site was saved as a protomol. The main setting was 20 solutions per compound, and other parameters accepted the Surflex-Dock Geom settings. Surflex-Dock scoring function was used to calculate the binding affinity (−log Kd) for each docking model. Subsequently, the consensus scores were calculated to compare the Total Score, PMF score, ChemScore, G score, and D score. Lastly, we selected the best docking model equipped with excellent binding affinity, consensus scores (≥3), and the number of intermolecular interactions.
2.7. Cytotoxicity Assay
LNCaP, PC3, and DU145 cells (1 × 104 or 5 × 103 cells) were seeded into a 96-well plate and after 24 h incubation, different concentrations of compounds 3h and 3K were made and then treated for 24 or 48 h. A total of 10 μL/well EZ-Cytox cell viability (WST) assay kit (EZ-BULK150, Daeillab, Republic of Korea) was added and incubated, followed by detecting the absorbance measurement at 450 nm with a VERSA Max Microplate Reader (Molecular Devices Corp., CA, USA) to determine cell viability. Finally, according to absorbance measurement, we calculated the IC50 using Sigma Plot 10.0 software.
2.8. Proliferation Assay
Plated LNCaP (5 × 103 cells/well) were placed into a 96-well plate and incubated until the confluency of cells achieved 50%. Then, we treated compound 3h (0, 2.5, 5, and 10 µM) and put the 96-well plate into IncuCyte® ZOOM 2016B software (Essen Bioscience, Ann Arbor, MI, USA) to acquire the images every 3h, and analyze the confluency.
2.9. Colony Forming Assay of LNCaP cells
A total of 1000 cells/well was seeded into a 6-well plate. It was treated with either compound 3h or control for 24 h after 5 days incubation, and then the cells were fixed and stained overnight with 4% paraformaldehyde (J19943, Thermo Fisher Scientific, Waltham, MA, USA) and 0.5% crystal violet (CR1035-100-00, Biosesang, Seongnam, Republic of Korea). ImageJ software 1.52a (NIH, Bethesda, MD, USA) was used to analyze the relative colony area.
2.10. Cell Cycle Analysis
LNCaP cells (30 × 104 cells/dish) were seeded into 100-π dishes for 24 h incubation. Amounts of 0, 2.5, 5, and 10 μM compound 3h were treated to LNCaP for 48 h, the attached cells were harvested and put into 70% cold ethyl alcohol (EtOH) to fix overnight at 4 °C. Then, cells were resuspended in phosphate-buffered saline (PBS), and stained with a mixed solution of 0.5 mL PBS and 5 μL of PI (P4864-10ML, Sigma-Aldrich, St. Louis, MO, USA) and 10 mg/mL RNase A. In the last step, we measured the effect of compound 3h on the cell cycle in LNCaP cells using a NovoCyte flow cytometer (ACEA Biosciences, Santa Clara, CA, USA).
2.11. Western Blot Analysis
LNCaP cells were seeded at a density of 80 × 104 cells/dish in 100-π dishes and compound 3h (0, 2.5, 5, and 10 μM) was treated afterwards. Cells were harvested and then lysed with PRO-PREPTM extraction solution (17081, iNtRON, Seongnam-shi, Republic of Korea) on ice for 30 min. Before Western blotting, we conducted a bicinchoninic acid (BCA) assay (23228, Thermo Fisher Scientific, Waltham, MA, USA) to measure protein concentrations in the different samples according to the manufacturer’s instructions. After electrophoresing 30 μg protein on 6–15% sodium dodecyl sulfate polyacrylamide gels for 30 min under 80 V, and for 1 h 30 min under 100 V, we transferred the protein to olyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA) for 1 h 10 min under 400 mA. We followed this by blocking with skimmed milk for 1 h, and then incubated the PVDF membranes with all kinds of primary antibodies for at least 8 h at 4 °C. The following day, horseradish peroxidase-conjugated anti-rabbit IgG (NB7176, 1:20000; Novus Biologicals, Littleton, CO, USA) or anti-mouse IgG (NB7561, 1:40000; Novus Biologicals, Littleton, CO, USA) secondary antibodies were incubated with PVDF membranes for 1 h at room temperature. At the last step, the Immobilon Forte Western HRP (Merck Millipore, Burlington, MA, USA) and WSE-6200 LuminoGraph Ⅱ Imaging System (ATTO, Tokyo, Japan) were used to visualize and detect the protein bands. The intensities of protein bands were quantified with ImageJ software 1.52a (NIH, Bethesda, MD, USA).
2.12. Apoptosis
After treatment for 48 h with compound 3h, LNCaP cells were harvested and stained with 2 μL Annexin V-FITC (556419, BD Biosciences, San Diego, CA, USA) and 4 μL PI in 100 μL of 1 × binding buffer for 30 min. After diluting cells with 1 × binding buffer, an apoptosis assay was performed using a NovoCyte flow cytometer (ACEA Biosciences, Santa Clara, CA, USA).
2.13. DAPI Nuclear Staining
Apoptosis can be identified based on the presence of broken, condensed, degenerated nuclei, which can be stained by DAPI. LNCaP cells (5 × 104) seeded in confocal dishes were treated with DMSO or compound 3h (2.5, 5, and 10 µM) for 48 h. Acetone was used for fixation, and 0.1 μg/mL DAPI (D9542, Sigma-Aldrich, St. Louis, MO, USA) was used to stain DNA. At the last step, a confocal K1-fluo microscope (Nanoscope Systems, Daejeon, Republic of Korea) was used to observe morphological changes in the nucleus.
2.14. Stain LNCaP Cells with Acridine Orange
Seeded LNCaP cells (5 × 104) in confocal dishes were treated with DMSO or compound 3h (2.5, 5, and 10 µM) for an extra 48 h. LNCaP was stained with 1 μg/mL acridine orange (00910250, Thermo Fisher Scientific, Waltham, MA, USA) for 15 min. Last, stained cells were observed under a Confocal K1-fluo microscope (Nanoscope Systems, Daejeon, Republic of Korea) at 400×/600× magnification.
2.15. Seahorse XFe96 Analysis of Cell Mito Stress Test and Glycolytic Rate Assay
The Agilent Seahorse Xfe cell mito stress test (Kit 103015-100) evaluated key parameters of mitochondrial function by directly measuring the OCR, and after mitochondrial inhibition, a glycolytic rate assay (Kit 103344-100) accurately measured glycolytic rates under basal glycolysis and compensatory glycolysis. During both experiments, the OCR and ECAR were continuously monitored and recorded by a Seahorse Xfe96 analyzer (Agilent, Santa Clara, CA, USA). Briefly, on the day before detecting with the Seahorse Xfe96 analyzer, LNCaP cells seeded into a Xfe96 cell culture microplate were treated with DMSO or compound 3h (2.5, 5, and 10 µM), and calibrated by an XFe cartridge with 200 μL XF calibration buffer overnight. On the day of assay, according to the user guide, XF assay medium was used to wash cells; different compounds (oligomycin, carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP), Rotenone and antimycin A (ROT/AA) for the cell mito stress test, and ROT/AA, 2-Deoxy-D-glucose (2-DG) for the glycolytic rate assay) were added to the injection ports for the cell mito stress test and glycolytic rate assay. After detection, raw data were analyzed automatically by Assay Generator.
2.16. Statistical Analysis
All data are expressed as the mean ± standard error (SEM) of at least three independent experiments. Our analysis was conducted using GraphPad Prism software 5.0 (GraphPad Software, San Diego, CA, USA). Statistical significance was analyzed using one-way analysis of variance followed by Bonferroni’s post hoc comparisons test. Statistical significance was set at p value < 0.05.
3. Results
3.1. Anti-Proliferation Effects of Compound 3h in Three Prostate Cancer Cell Lines
To evaluate the anticancer effects of compound 3h in three different human prostate cancer cell lines, the in vitro cytotoxicity of compound 3h was assessed in LNCaP, DU145, PC3 cells. WST results indicated that compound 3h had a higher inhibitory effect on LNCaP cells than on the other two cell lines, even though DU145 and PC3 cells had higher protein expressions of PKM2 (Figure 3A,B). Since compound 3K has been proven to be a specific PKM2 inhibitor and shows anticancer effects in various human cancer lines, including SK-OV-3, HCT116, and Hela [18,19], this study compared the anticancer effects of compound 3h and compound 3K on the LNCaP cell line. WST results indicated that both compounds had similar inhibitory effects on LNCaP after 48 h of treatment (Figure 3C,D). The results of the IncuCyte Zoom assay proved that compound 3h inhibited the growth of LNCaP in a concentration-dependent manner (Figure 3E). The effect of compound 3h on LNCaP cell morphology can be seen in Figure 3F. As shown in Figure 3G, colony formation was significantly inhibited after compound 3h treatment.
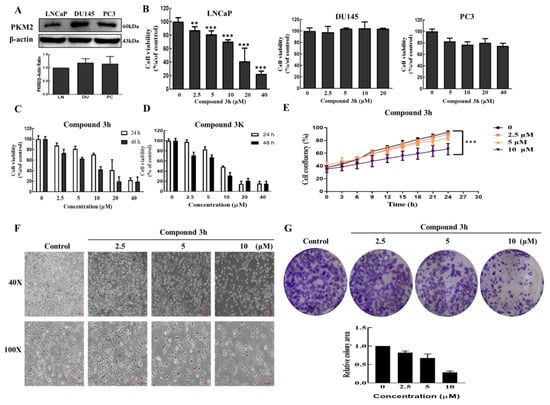
Figure 3.
Cytotoxicity of compounds 3h and 3K on three different prostate cancer cell lines. (A) The expression of PKM2 LNCaP, DU145, and PC3. (B) Cell viability of compound 3h. Compound 3h was treated in LNCaP, DU145, or PC3 cells for 24 h. (C) Anticancer effect of compound 3h on LNCaP cells. Cell viability of LNCaP decreased with the increase in compound 3h concentration (0, 2.5, 5, 10, 20, and 40 μM) for 24 and 48 h treatment. Cell viability was assessed using WST assay kit. (D) Cytotoxicity effect of compound 3K on LNCaP cells for 24 h and 48 h. (E) Compound 3h suppressed the proliferation of LNCaP cells. (F) Compound 3h treatment for 48 h caused morphological changes in LNCaP cells. Scale bar, 200 μm (40×), 100 μm (100×). (G) Compound 3h treatment inhibited colony formation in LNCaP and the quantitative analysis. The values represent the mean ± SD. ** p < 0.01 and *** p < 0.001. The uncropped blots are shown Supplementary Material.
3.2. PKM2 Expression Was Selectively Inhibited by Compound 3h, Rather Than PKM1
Both compound 3h (IC50 = 0.96 ± 0.18 μM) and compound 3K (IC50 = 2.95 ± 0.53 μM) showed high inhibitory activity against PKM2 [14]. To confirm how compounds 3h and 3K affected the expression of pyruvate kinase in LNCaP cells, we performed Western blotting with antibodies against PKM1 and PKM2. As shown in Figure 4A, from decreased PKM2 expression, compounds 3h and 3K had a specific selectivity for PKM2 rather than PKM1 in LNCaP cells. To confirm the effects of compounds 3h and 3K on the pyruvate kinase activity in LNCaP cells, we treated LNCaP cells with different concentrations of compounds 3h and 3K; both were able to impair the pyruvate kinase activity in a concentration-dependent manner (Figure 4B). In conclusion, compound 3h inhibited selectively the pyruvate kinase activity and expression of PKM2, but not PKM1 in LNCaP cells, similar to compound 3K.
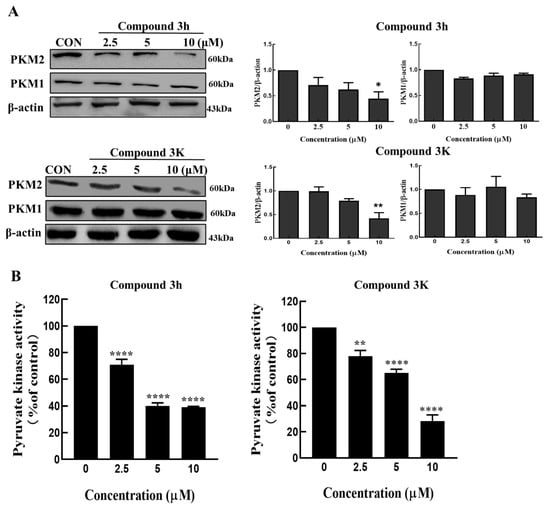
Figure 4.
Compound 3K and 3h selectively inhibit PKM2 rather than PKM1. (A) Compounds 3K and 3h have a specific selectivity in reducing PKM2 expression in LNCaP cells. (B) The activity of pyruvate kinase in LNCaP cells was inhibited by compound 3h and compound 3K depending on concentration. The values represent the mean ± SD. * p < 0.05, ** p < 0.01, and **** p < 0.0001. The uncropped blots are shown Supplementary Material.
3.3. Molecular Docking Analysis of Compounds 3h and 3K
To elucidate the molecular binding basis of the interaction between both compounds and PKM2, we investigated the possible binding modes of compounds 3h and 3K by docking stimulation using Surflex-Dock. The allosteric binding of small-molecule metabolites can enable PKM2 to exist in different isoforms, with equilibrium between monomers and tetramers. Among the metabolites, phenylalanine works as a PKM2 inhibitor that stabilizes it in an inactive T-state tetrameric conformer. Thus, the crystal structure of PKM2 complexed with phenylalanine (PDB ID: 4FXJ), that remains in an inactive T-state, was selected as the target receptor to carry out the docking stimulation of compounds 3K and 3h [12]. According to our docking results in Figure 5A–C, the binding modes of compounds 3h and 3K were almost identical and similar to the X-ray of phenylalanine, which was well-occupied in the allosteric pocket, further stabilizing PKM2 in the inactive T-state. X-ray structure analysis revealed a polar interaction network in the binding pocket of phenylalanine (carboxylate group of phenylalanine with Asn70 and Arg106), which is crucial for PKM2 inhibition activity. Likewise, in the docked models of PKM2 with 3h and 3K (Figure 5D,E), the carboxylate moiety of 3h also formed hydrogen bonds with Arg106, and 3K formed hydrogen bonds with Arg106. In addition, the S atom of the carbonyl sulfide of 3K interacted with Asn397 and Arg383 by forming an H-bond, whereas 3h only formed hydrogen bonds with Asn397. We also identified unique T-shaped π-π and cation-π interactions between the phenyl ring of 3h and Phe470 and Arg43, respectively, in the backbone of PKM2. Thus, these two unique interactions might contribute a higher binding affinity for the inhibitory effect of PKM2, consistent with the docking score (−log Kd) of 3h (5.6815) being higher than that of 3K (5.1002).
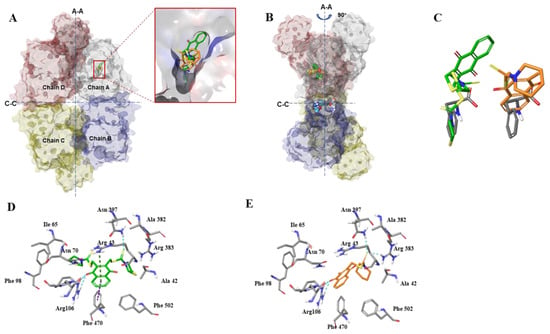
Figure 5.
Binding mode of compounds 3h and 3K in the effector phenylalanine-binding site of PKM2. (A) Superimposition and enlarged view of phenylalanine (gray sticks), compound 3h (green sticks), and compound 3K (faded orange sticks) over the X-ray of PKM2. PKM2 tetramer is shown with the effector site complexed with the original ligand phenylalanine (gray sticks), compound 3h (green sticks) and compound 3K (faded orange sticks). The chain is colored, light gray is for Chain A, cyan Chain is for B, green is for Chain C, magenta is for Chain D. The large (A-A) and small (C-C) interfaces between monomers are shown as dashed lines. (B) Side view (rotated 90° to that of A) of PKM2 tetramer complexed with the original ligand phenylalanine (gray sticks), and compound 3h (green sticks) and compound 3K (faded orange sticks). (C) Superimpositions of phenylalanine (gray) and compound 3h (green) or 3K (faded orange). (D,E) Close-up view of the key interactions involved in bonding with compound 3h (D, green sticks) and compound 3K (E, faded orange sticks) in effector-binding pocket. Key residues surrounding compound 3h and 3K are shown as gray sticks and labeled. π-π stacking is depicted by dotted purple lines. π-cation is shown as dotted dark green lines. Hydrogen bonds are depicted as dotted cyan lines.
3.4. Compound 3h Induces Apoptotic Cell Death in LNCaP Cells
Here, we elucidated the mechanisms involved in the anticancer activity of compound 3h after treatment on LNCaP. As shown in Figure 6A, the percentage of late apoptosis increased in the compound 3h-treated group by 2.3% (with 2.5 μM), 2.4% (with 5 μM), and 6.6% (with 10 μM) compared with 1.7% in the control group. Similarly, the necrosis percentage increased depending on concentration (1.8% in the control, 4.9% in the 2.5 μM, 6.9% in the 5 μM, and 14.1% in the 10 μM groups). As shown in Figure 6B, Bax and cleaved-PARP increased, whereas Bcl-2, pro-caspase-3, pro-caspase-7, and PARP decreased. The relative Bcl-2/Bax ratio decreased, and the relative cleaved-PARP/PARP ratio markedly increased (Figure 6C). DAPI staining was carried out to examine the nuclear morphology and apoptotic bodies of apoptosis via confocal microscopy, which were observed in the compound 3h-treated group (Figure 6D).
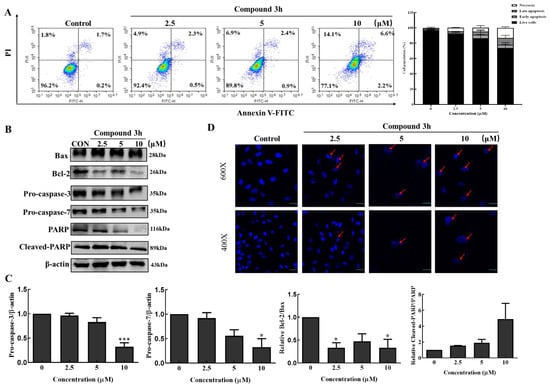
Figure 6.
Compound 3h induces LNCaP cellular apoptotic cell death. (A) Compound 3h caused apoptosis in LNCaP cells. Late apoptosis and necrosis percentage increased in the compound 3h-treated group. (B) Effect of compound 3h on the expression levels of apoptotic proteins. Compound 3h induced apoptotic proteins expression. (C) Quantitative data of apoptotic proteins. (D) Nuclear morphological changes and apoptotic bodies were detected in the compound 3h-treated group. The values represent the mean ± SD. * p < 0.05, and *** p < 0.001. The uncropped blots are shown Supplementary Material.
3.5. Compound 3h Slightly Induces LNCaP Cells Arrest at G2/M Phase
PKM2 inhibition or PKM2 knockdown is associated with G2/M arrest in various cancer cell lines, including DU145 and SK-O-V3 [17,18]. To explore whether compound 3h induced G2/M arrest in LNCaP, we analyzed DNA content using a NovoCyte flow cytometer after compound 3h treatment for 48 h. According to the relative distribution of cells in Figure 7, G2/M arrest was slightly induced (Figure 7A,B), cell cycle-related proteins, p-Cdc2 and Cyclin B1 were upregulated, and Cdc2 was markedly downregulated (Figure 7C,D).
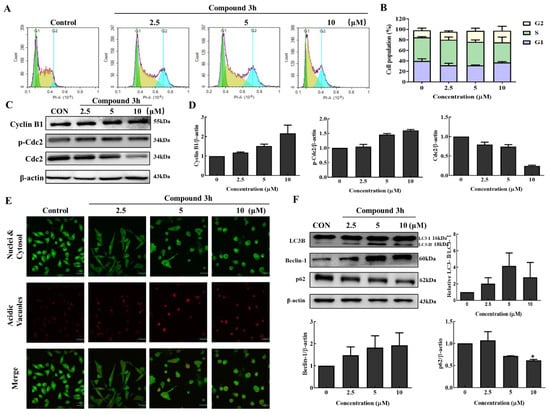
Figure 7.
Compound 3h induces G2/M arrest and autophagy in LNCaP cells. (A) Cell cycle results of LNCaP measured by NovoCyte after treating compound 3h (0, 2.5, 5, and 10 μM). (B) Bar graph indicates the cell population involved in G1, S, and G2/M phases in LNCaP cells after compound 3h treatment. (C) Expression changes in G2/M arrest-related proteins (Cyclin B1, p-Cdc2, Cdc2) in LNCaP cells after compound 3h treatment. (D) Representative histogram showing the expression levels of Cyclin B1, p-Cdc2, Cdc2, compared to β-actin in LNCaP. (E) Autophagy activation was clearly observed in LNCaP cells after compound 3h treatment. Red fluorescence intensity of autophagic vacuoles was observed in compound 3h-treated group in a concentration-dependent manner (Magnification 600×). (F) Expression of autophagic proteins in LNCaP cells after compound 3h treatment and LC3-Ⅱ/LC3-Ⅰ, relative Beclin-1, and relative p62 were calculated and shown as graphs. The values represent the mean ± SD. * p < 0.05. The uncropped blots are shown Supplementary Material.
3.6. Compound 3h Induces Autophagic Cell Death in LNCaP Cells
To analyze whether autophagy was induced in the compound 3h-treated group, the acridine orange staining assay was performed. The emission color of acridine orange changed from yellow to orange to red as the pH dropped in the acidic vacuole upon autophagy. Autophagic vacuoles with red fluorescence intensity were observed in the compound 3h-treated group in a concentration-dependent manner (Figure 7E). LC3-Ⅱ is the autophagic vacuole-related form, so the transformation of LC3-Ⅰ to LC3-Ⅱ is an important step in autophagosome formation. Beclin-1 and LC3-Ⅱ levels increased, while p62 decreased in the cells (Figure 7F).
3.7. Compound 3h Suppresses the Akt/AMPK/mTOR Pathway in LNCaP
Akt/AMPK/mTOR has been studied as a typical anticancer pathway involved in autophagy. To further understand the mechanism involved in the anticancer effect of compound 3h, according to our results in Figure 8A,B, the Akt/mTOR pathway was suppressed by compound 3h treatment in LNCaP. In addition, as we can see, after compound 3h treatment, the expression of p-PTEN increased and p-Akt decreased, which restrained mTOR phosphorylation; thereby, autophagy was promoted. Although the expression levels of total Akt and mTOR were similar between the experimental and control groups, increased p-AMPKα further inhibited mTOR phosphorylation. Increased AMPK phosphorylation curbed mTOR aivity and downregulated protein synthesis to inhibit cell proliferation. Therefore, p70S6K and p-p70S6K, as the main targets of mTOR, were analyzed. p-p70S6K levels decreased in response to decreased p-mTOR, and increased p-AMPKα levels after PKM2 was inhibited. These results indicated that compound 3h suppressed the prostate cancer cell line, LNCaP, by regulating the Akt/AMPK/mTOR pathway.
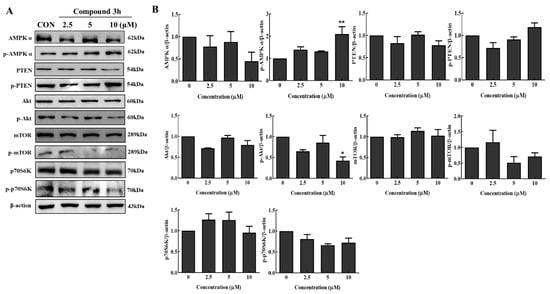
Figure 8.
Changes in protein expression in signaling pathways in LNCaP cells after PKM2 inhibition by compound 3h. (A) Expression levels of proteins related to Akt/AMPK/mTOR signaling pathway. Compound 3h treatment suppressed the activation of Akt/mTOR. (B) Representative histogram showing the expression levels of proteins. The values represent the mean ± SD. * p < 0.05, and ** p < 0.01. The uncropped blots are shown Supplementary Material.
3.8. Mitochondrial Activity and Glycolysis in LNCaP Cells Were Impaired by Compound 3h
The Seahorse analyzer was used to analyze the bioenergetic changes in LNCaP cells after compound 3h treatment. Cellular bioenergetic markers, including OCR and proton efflux rate (PER), were detected. As shown in Figure 9A, compound 3h inhibited OXPHOS, as indicated by the decreased OCR. Decreased basal respiration, spare respiratory capacity, and maximal respiration indicated that compound 3h regulated the mitochondrial dynamics (Figure 9B–E).
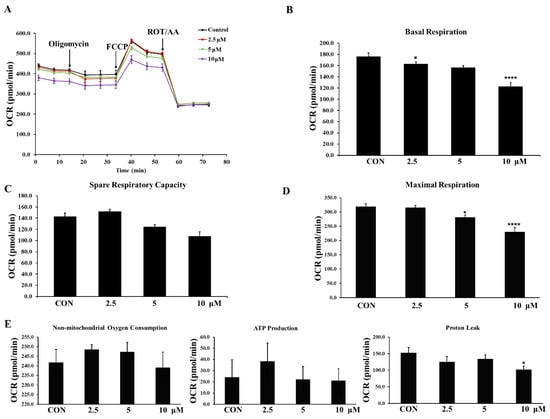
Figure 9.
PKM2 specific inhibitor, compound 3h, impairs the mitochondrial activity in LNCaP cells, leading to decreased basal respiration, spare respiratory capacity, and maximal respiration. (A) Profile of mito stress test data for oxygen consumption rate (OCR) (pmol/min) in LNCaP cells with arrows indicating injections into media of specific stressors. (B–E) Results of Seahorse XFe cell mito stress test generator report shown for OCR (pmol/min). B is for basal respiration, C is for spare respiration capacity, D is for maximal respiration, E is for non-mitochondrial oxygen consumption, ATP production, and proton leak. The values represent the mean ± SD. * p < 0.05 and **** p < 0.0001.
The compound 3h-treated group showed decreased basal glycolysis and compensatory glycolysis after measuring glycoPER (Figure 10A,B). Dimer PKM2 translocated into the nucleus to perform non-metabolic functions, where β-catenin expression was unregulated, thereby increasing the expression of c-Myc and GLUT1. As shown in Figure 10C,D, compound 3h not only decreased β-catenin and c-Myc expression levels, but it also significantly decreased the expression levels of GLUT1 and MCT4, thereby decreasing the rates of glucose uptake and lactate efflux, respectively. Therefore, compound 3h decreased the glycolytic activity in LNCaP cells by downregulating β-catenin and c-Myc levels.
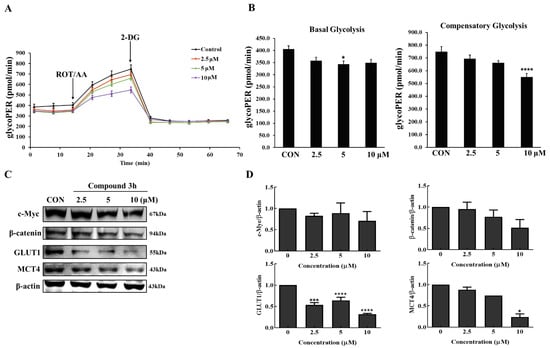
Figure 10.
Compound 3h impairs the glycolytic function in LNCaP cells. (A) Profile of glycolytic rate assay in LNCaP with specific injections into media. (B) Results of Seahorse XF glycolytic rate assay-generator report for basal and compensatory glycolysis (pmol/min). (C) Protein expression levels of c-Myc, β-catenin, GLUT1, MCT4 were evaluated by Western blot after compound 3h treatment. (D) Representative histograms showing the expression levels of c-Myc, β-catenin, GLUT1, MCT4. The values represent the mean ± SD. * p < 0.05, *** p < 0.001. and **** p < 0.0001. The uncropped blots are shown Supplementary Material.
4. Discussion
Prostate cancer is a common malignant cancer characterized by slow progression, with a gradually increasing incidence, and a high mortality rate in advanced cases. Although the 5-year survival rate of localized prostate cancer is >99%, advanced prostate cancer is regarded as incurable [20,21]. Although androgen deprivation therapy (ADT) is considered to be the primary treatment option for advanced prostate cancer, most patients develop recurrence and insensitivity after an initial positive response [22]. Docetaxel significantly improves the overall survival of castration-resistant prostate cancer (CRPC) patients, and has been approved as a first-line treatment option for CRPC [23]. However, many patients develop resistance to docetaxel. Therefore, the development of novel agents as new targets for prostate cancer treatment is essential to solve the problems associated with chemotherapy.
Currently, the Warburg effect, as a hallmark of cancer, may be a breakthrough for cancer therapy. PKM2 plays an important role in the regulation of anabolic metabolism, including the Warburg effect. High PKM2 expression was previously shown in prostate cancer tissues [24]; therefore, targeting PKM2 or glycolysis may be a potent strategy for prostate cancer treatment. Previously, it was shown that PKM2 knockdown in the PKM2-overexpressed prostate cancer cell line DU145 induced autophagic cell death by influencing cellular metabolism and the Akt/mTOR pathway [17]; however, PKM2 knockdown therapy is severely limited in clinical treatment. Therefore, exploring novel anticancer agents that target PKM2 remains a hotspot of research. Recently, the anticancer effect of several PKM2 inhibitors (shikonin, metformin) has been identified. In particular, the inhibitory effect of compound 3K, a PKM2 specific inhibitor, on SK-OV-3 has been proven in vitro and in vivo [18]. In this study, we evaluated the anticancer effect of compound 3h, which was synthesized based on the structure of compound 3K and its molecular mechanism in LNCaP. Our results provide a new perspective for the development of novel agents for prostate cancer treatment. We found that compound 3h treatment decreased the proliferation and colony formation of LNCaP cells in a dose- and time-dependent manner. According to a previous report, PKM2 knockdown was related to G2/M arrest in HeLa and SiHa cells [25]; therefore, we assessed the influence of compound 3h on the cell cycle in LNCaP cells. Compound 3h treatment led to a slight increase in the percentage of cells in the G2/M phase, which was mediated by increased p-Cdc2 expression.
Abundant glucose uptake and lactate elimination are essential features of aerobic glycolysis in cancer cells [4]. In this study, compound 3h inhibited PKM2 expression in LNCaP cells, which led to the inhibition of aerobic glycolysis, reduction of glucose intake and lactate elimination—consistent with our results of downregulated GLUT1 and MCT4 expression—and decreased basal glycolysis and compensatory glycolysis. It has been reported that the mTOR-MFN2-PKM2 signaling axis couples glycolysis and mitochondrial OXPHOS, which increases the phosphorylation of MFN2, leading to increased PKM2:MFN2 interaction and OXPHOS increase [26]. Therefore, compound 3h may impair OXPHOS and glycolysis by inhibiting PKM2 and interfering with the PKM2:MFN2 interaction.
PKM2 has dimeric and tetrameric forms. Dimeric PKM2 translocates to the nucleus and regulates transcription factors (including β-catenin), thus affecting a variety of signaling pathways that can promote tumor progression [27,28]. Tetrameric PKM2 has allosteric regulatory domains, and PKM2 can be stabilized in the inactive T-state when inhibitory phenylalanine binds to the binding pocket [12]. According to our molecular docking results, compound 3h can bind to the phenylalanine-binding pocket at a similar position; therefore, compound 3h also stabilizes PKM2 in the inactive T-state, which inhibits PKM2 activity and blocks dimer formation, causing inhibition of glycolysis and nuclear translocation of dimeric PKM2, further suppressing cancer cell proliferation.
Autophagic death is a common pathway of cell death. Activation of the Akt/mTOR pathway can inhibit autophagy, which is critical for cancer cell growth and survival [29]. Akt is a major regulator of cell survival under stressful conditions [30], which not only regulates nutrient intake, but also the expression of growth factors [31]. P70S6K is a downstream target of mTOR, and can be upregulated and phosphorylated by mTOR phosphorylation; thus, more downstream proteins associated with proliferation will be activated and induced [32]. In addition, AMPK phosphorylation induces mTOR inhibition, leading to autophagy inhibition [33]. It was reported that the PKM2 knockdown, and PKM2 inhibitor, compound 3K, inhibited mTOR phosphorylation and autophagy activation by activating AMPK phosphorylation in DU145 cells and SK-OV-3 cells, respectively [17,18]. In this study, according to the Western blot results, compound 3h increased the expression of p-AMPKα, and decreased that of PKM2, p-Akt, p-mTOR, and p-p70S6K; therefore, compound 3h inhibited PKM2, which caused autophagic cell death by inhibiting the Akt/mTOR signaling pathway in LNCaP.
Apoptosis and autophagy are key cellular processes that maintain cellular homeostasis and are connected in complex ways. Bcl-2 and Beclin-1 play important roles in the connection between apoptosis and autophagy [34]. Bcl-2 activation can be modulated by post-translational modifications such as phosphorylation of Bad. Bad is phosphorylated by Akt and p70S6K, which inhibits its interaction with anti-apoptotic Bcl-2 proteins, and leads to the inhibition of Bax-triggered apoptosis [35]. Beclin-1 is a lipid-kinase complex that is involved in autophagosome nucleation. Beclin-1 conjugates with members of the Bcl-2 family through the BH3 domain, and dissociation from Bcl-2 is critical for its autophagic activity [36]. PKM2 knockdown activates apoptotic and autophagic signaling pathways in different cancer cells, including DU145 and A549 cells [17,37]. So, PKM2 knockdown and PKM2 inhibition influence the interaction between Bcl-2 and Beclin-1, which may be accompanied by a decrease in Bcl-2 expression, and a Beclin-1 dissociation. In this study, compound 3h inhibited Akt and p70S6K phosphorylation, which may have promoted Bax-triggered apoptosis through the interaction between Bcl-2 and Bad. In addition, Bcl-2 decreased after compound 3h treatment, and more Beclin-1 was dissociated, which led to autophagic death. Our results suggested that compound 3h inhibited PKM2, and induced apoptosis and autophagy in LNCaP cells.
5. Conclusions
In conclusion, our study showed that compound 3h, which acts as a specific PKM2 inhibitor, inhibited PKM2 activity and expression, and further significantly inhibited the glycolysis and proliferation of LNCaP cells. Compound 3h decreased the nuclear translocation of dimeric PKM2, which decreased the expression of β-catenin, further decreasing the expression of GLUT1 and MCT4. Combined with the results of Western blotting, compound 3h exerted anti-tumor effects in LNCaP cells by inhibiting glycolysis, and inducing apoptosis and autophagy mediated by the Akt/mTOR signaling pathway.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers15010265/s1, The uncropped blots are shown Supplementary Material.
Author Contributions
C.J.: methodology, investigation, formal analysis, writing—original draft preparation. X.Z.: methodology, investigation. T.J., J.Y.K., J.H.P.: methodology, investigation. I.S.K. and H.S.K.: methodology, investigation, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the National Research Foundation (NRF) of Korea (grant number NRF-2019R1A2C2002923; NRF-2022R1A4A1018930). Chunxue Jiang was supported by the China Scholarship Council (CSC).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data and materials generated are relevant to the results that are included in this article. Other data are available from the corresponding author Kim upon reasonable request.
Acknowledgments
We thank In Su Kim (Department of Chemistry, School of Pharmacy, Sungkyunkwan University, Suwon, Republic of Korea) for his excellent technological help in synthesizing compound 3h.
Conflicts of Interest
The authors declare that they have no known competing financial interests or personal relationships that influenced the work reported in this paper.
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA A Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Heinzer, H.; Steuber, T. Prostate cancer in the elderly. Urol. Oncol. 2009, 27, 668–672. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Wind, F.; Negelein, E. THE METABOLISM OF TUMORS IN THE BODY. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Dombrauckas, J.D.; Santarsiero, B.D.; Mesecar, A.D. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry 2005, 44, 9417–9429. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zheng, Y.; Xia, Y.; Ji, H.; Chen, X.; Guo, F.; Lyssiotis, C.A.; Aldape, K.; Cantley, L.C.; Lu, Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat. Cell Biol. 2012, 14, 1295–1304. [Google Scholar] [CrossRef]
- Yang, W.; Lu, Z. Nuclear PKM2 regulates the Warburg effect. Cell Cycle 2013, 12, 3154–3158. [Google Scholar] [CrossRef]
- Ullah, M.S.; Davies, A.J.; Halestrap, A.P. The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1alpha-dependent mechanism. J. Biol. Chem. 2006, 281, 9030–9037. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Zou, Z.; Liu, X.; Ling, M.; Wang, Q.; Wang, Q.; Lu, L.; Shi, L.; Liu, Y.; Liu, Q.; et al. Enhanced glycolysis, regulated by HIF-1α via MCT-4, promotes inflammation in arsenite-induced carcinogenesis. Carcinogenesis 2017, 38, 615–626. [Google Scholar] [CrossRef]
- Wu, S.; Le, H. Dual roles of PKM2 in cancer metabolism. Acta Biochim. Biophys. Sin. 2013, 45, 27–35. [Google Scholar] [CrossRef]
- Wang, P.; Sun, C.; Zhu, T.; Xu, Y. Structural insight into mechanisms for dynamic regulation of PKM2. Protein Cell 2015, 6, 275–287. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; McNae, I.W.; Chen, Y.; Blackburn, E.A.; Wear, M.A.; Michels, P.A.M.; Fothergill-Gilmore, L.A.; Hupp, T.; Walkinshaw, M.D. An allostatic mechanism for M2 pyruvate kinase as an amino-acid sensor. Biochem. J. 2018, 475, 1821–1837. [Google Scholar] [CrossRef] [PubMed]
- Morgan, H.P.; O’Reilly, F.J.; Wear, M.A.; O’Neill, J.R.; Fothergill-Gilmore, L.A.; Hupp, T.; Walkinshaw, M.D. M2 pyruvate kinase provides a mechanism for nutrient sensing and regulation of cell proliferation. Proc. Natl. Acad. Sci. USA 2013, 110, 5881–5886. [Google Scholar] [CrossRef]
- Ning, X.; Qi, H.; Li, R.; Jin, Y.; McNutt, M.A.; Yin, Y. Synthesis and antitumor activity of novel 2, 3-didithiocarbamate substituted naphthoquinones as inhibitors of pyruvate kinase M2 isoform. J. Enzym. Inhib. Med. Chem. 2018, 33, 126–129. [Google Scholar] [CrossRef] [PubMed]
- He, C.L.; Bian, Y.Y.; Xue, Y.; Liu, Z.X.; Zhou, K.Q.; Yao, C.F.; Lin, Y.; Zou, H.F.; Luo, F.X.; Qu, Y.Y.; et al. Pyruvate Kinase M2 Activates mTORC1 by Phosphorylating AKT1S1. Sci. Rep. 2016, 6, 21524. [Google Scholar] [CrossRef]
- Gabriele, F.; Martinelli, C.; Comincini, S. Prostate cancer cells at a therapeutic gunpoint of the autophagy process. J. Cancer Metastasis Treat. 2018, 4, 17. [Google Scholar] [CrossRef]
- Dey, P.; Kundu, A.; Sachan, R.; Park, J.H.; Ahn, M.Y.; Yoon, K.; Lee, J.; Kim, N.D.; Kim, I.S.; Lee, B.M.; et al. PKM2 Knockdown Induces Autophagic Cell Death via AKT/mTOR Pathway in Human Prostate Cancer Cells. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2019, 52, 1535–1552. [Google Scholar] [CrossRef]
- Park, J.H.; Kundu, A.; Lee, S.H.; Jiang, C.; Lee, S.H.; Kim, Y.S.; Kyung, S.Y.; Park, S.H.; Kim, H.S. Specific Pyruvate Kinase M2 Inhibitor, Compound 3K, Induces Autophagic Cell Death through Disruption of the Glycolysis Pathway in Ovarian Cancer Cells. Int. J. Biol. Sci. 2021, 17, 1895–1908. [Google Scholar] [CrossRef]
- Ning, X.; Qi, H.; Li, R.; Li, Y.; Jin, Y.; McNutt, M.A.; Liu, J.; Yin, Y. Discovery of novel naphthoquinone derivatives as inhibitors of the tumor cell specific M2 isoform of pyruvate kinase. Eur. J. Med. Chem. 2017, 138, 343–352. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Moul, J.W. The evolving definition of advanced prostate cancer. Rev. Urol. 2004, 6 (Suppl. 8), S10–S17. [Google Scholar]
- Wang, G.; Zhao, D.; Spring, D.J.; DePinho, R.A. Genetics and biology of prostate cancer. Genes Dev. 2018, 32, 1105–1140. [Google Scholar] [CrossRef]
- Tannock, I.F.; de Wit, R.; Berry, W.R.; Horti, J.; Pluzanska, A.; Chi, K.N.; Oudard, S.; Théodore, C.; James, N.D.; Turesson, I.; et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N. Engl. J. Med. 2004, 351, 1502–1512. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Xu, Y.; Wang, X.; Jiang, C.; Han, S.; Dong, K.; Shen, M.; Xu, D. LincRNA-p21 suppresses development of human prostate cancer through inhibition of PKM2. Cell Prolif. 2017, 50, 12395. [Google Scholar] [CrossRef]
- Lin, Y.; Zhai, H.; Ouyang, Y.; Lu, Z.; Chu, C.; He, Q.; Cao, X. Knockdown of PKM2 enhances radiosensitivity of cervical cancer cells. Cancer Cell Int. 2019, 19, 129. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Han, J.; Jia, L.; Hu, X.; Chen, L.; Wang, Y. PKM2 coordinates glycolysis with mitochondrial fusion and oxidative phosphorylation. Protein Cell 2019, 10, 583–594. [Google Scholar] [CrossRef]
- Wong, N.; Ojo, D.; Yan, J.; Tang, D. PKM2 contributes to cancer metabolism. Cancer Lett. 2015, 356, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, H.; Taniguchi, K.; Kumazaki, M.; Yamada, N.; Ito, Y.; Otsuki, Y.; Uno, B.; Hayakawa, F.; Minami, Y.; Naoe, T.; et al. Anti-cancer fatty-acid derivative induces autophagic cell death through modulation of PKM isoform expression profile mediated by bcr-abl in chronic myeloid leukemia. Cancer Lett. 2015, 360, 28–38. [Google Scholar] [CrossRef]
- Porta, C.; Paglino, C.; Mosca, A. Targeting PI3K/Akt/mTOR Signaling in Cancer. Front. Oncol. 2014, 4, 64. [Google Scholar] [CrossRef]
- Datta, S.R.; Brunet, A.; Greenberg, M.E. Cellular survival: A play in three Akts. Genes Dev. 1999, 13, 2905–2927. [Google Scholar] [CrossRef] [PubMed]
- Facompre, N.D.; Sinha, I.; El-Bayoumy, K.; Pinto, J.T.; Sinha, R. Remarkable inhibition of mTOR signaling by the combination of rapamycin and 1,4-phenylenebis(methylene)selenocyanate in human prostate cancer cells. Int. J. Cancer 2012, 131, 2134–2142. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.Y.; White, E. Role of autophagy in cancer prevention. Cancer Prev. Res. 2011, 4, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Saud, S.M.; Young, M.R.; Chen, G.; Hua, B. Targeting AMPK for cancer prevention and treatment. Oncotarget 2015, 6, 7365–7378. [Google Scholar] [CrossRef] [PubMed]
- Maiuri, M.C.; Criollo, A.; Kroemer, G. Crosstalk between apoptosis and autophagy within the Beclin 1 interactome. EMBO J. 2010, 29, 515–516. [Google Scholar] [CrossRef] [PubMed]
- Johnstone, R.W.; Ruefli, A.A.; Lowe, S.W. Apoptosis: A link between cancer genetics and chemotherapy. Cell 2002, 108, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Colbert, C.L.; Becker, N.; Wei, Y.; Levine, B. Molecular basis of the regulation of Beclin 1-dependent autophagy by the gamma-herpesvirus 68 Bcl-2 homolog M11. Autophagy 2008, 4, 989–997. [Google Scholar] [CrossRef]
- Chu, B.; Wang, J.; Wang, Y.; Yang, G. Knockdown of PKM2 induces apoptosis and autophagy in human A549 alveolar adenocarcinoma cells. Mol. Med. Rep. 2015, 12, 4358–4363. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).