Store-Operated Calcium Entry in Breast Cancer Cells Is Insensitive to Orai1 and STIM1 N-Linked Glycosylation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Cell Culture and Transfections
2.3. Determination of Cytosolic Free-Ca2+ Concentration
2.4. Western Blotting
2.5. Biotinylation of Cell Surface Proteins
2.6. Caspase-3 Activity Assay
2.7. CRISPR/Cas9 Mediated Generation of STIM1 and Orai1 KO MDA-MB-231 and MCF-7 Cells
2.8. Statistical Analysis
3. Results
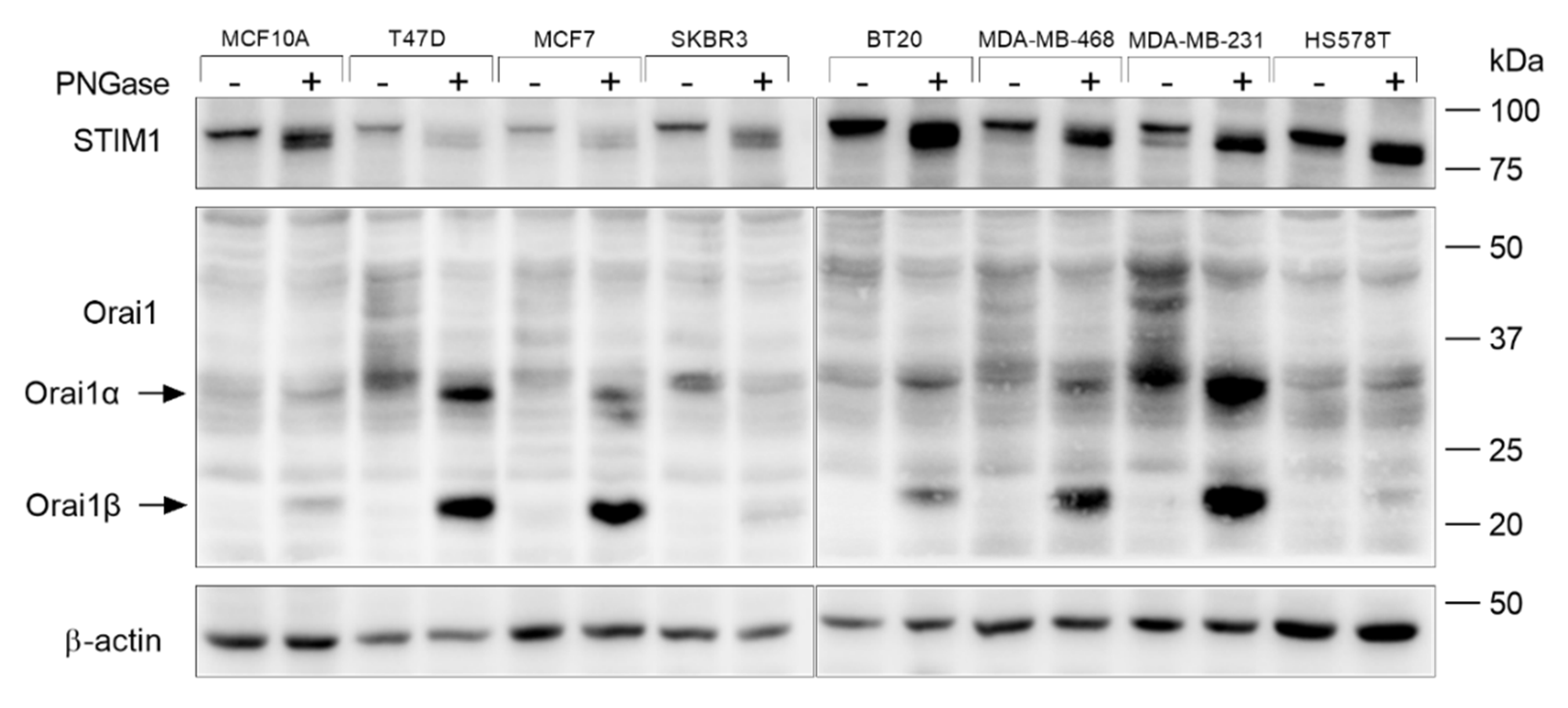
3.1. Orai1 and STIM1 Glycosylation in Breast Cancer and Non-Tumoral Breast Epithelial Cells
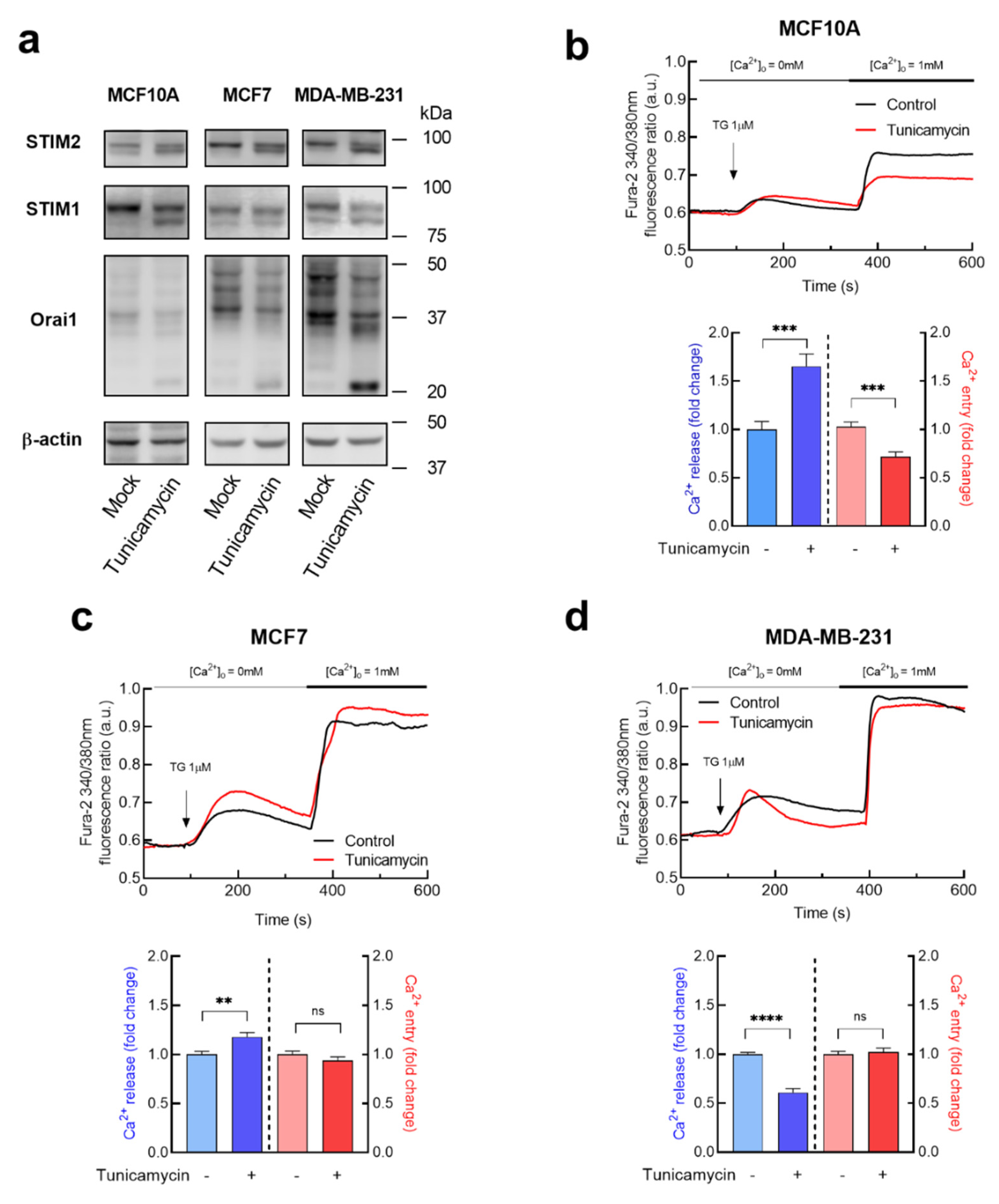
3.2. Effect of Tunicamycin in TG-Induced Ca2+ Release and Entry in Breast Cancer and Non-Tumoral Breast Epithelial Cells
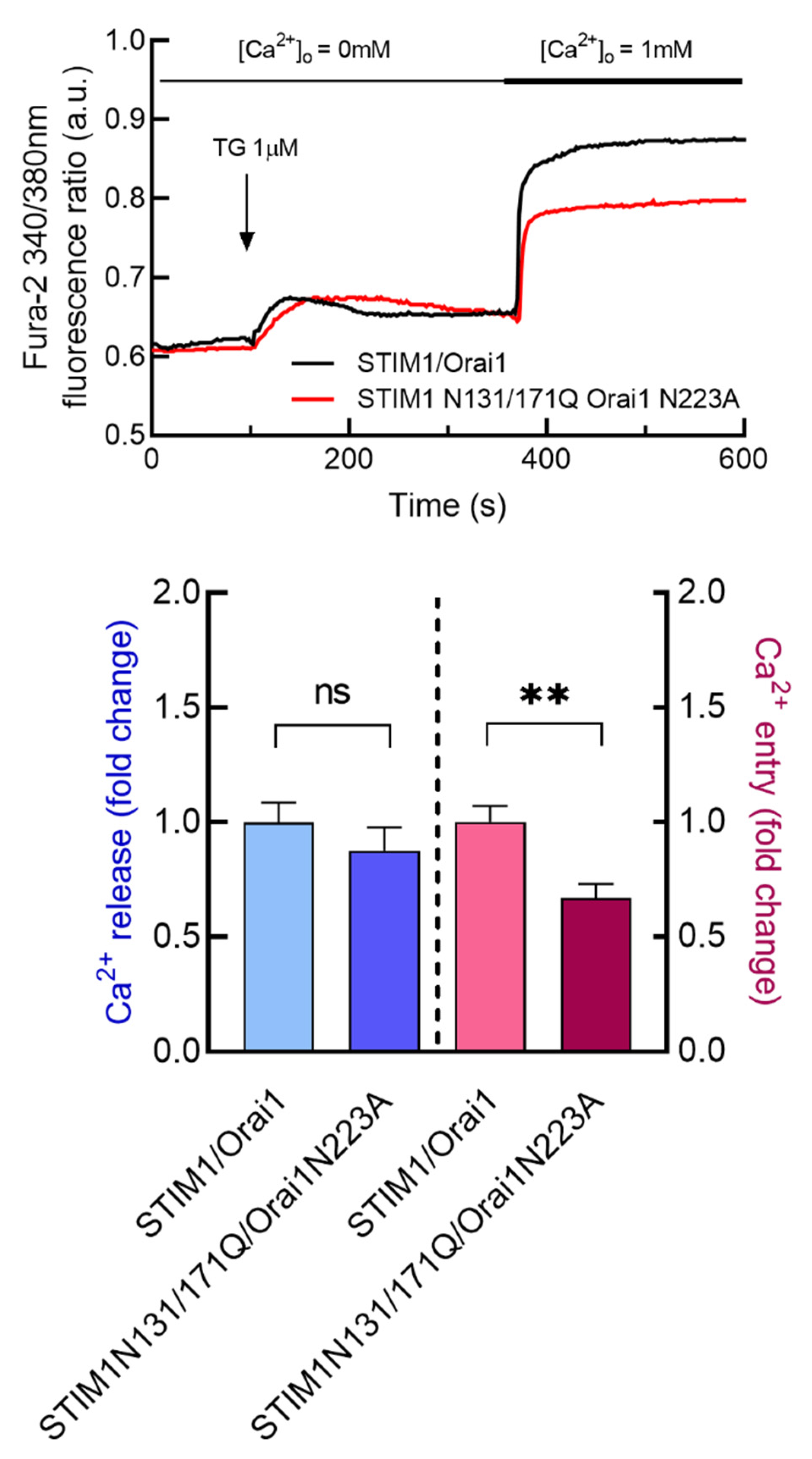
3.3. Characterization of the Functional Role of Orai1 and STIM1 N-Linked Glycosylation in TG-Induced Ca2+ Release and Entry in Breast Cancer and Non-Tumoral Breast Epithelial Cells
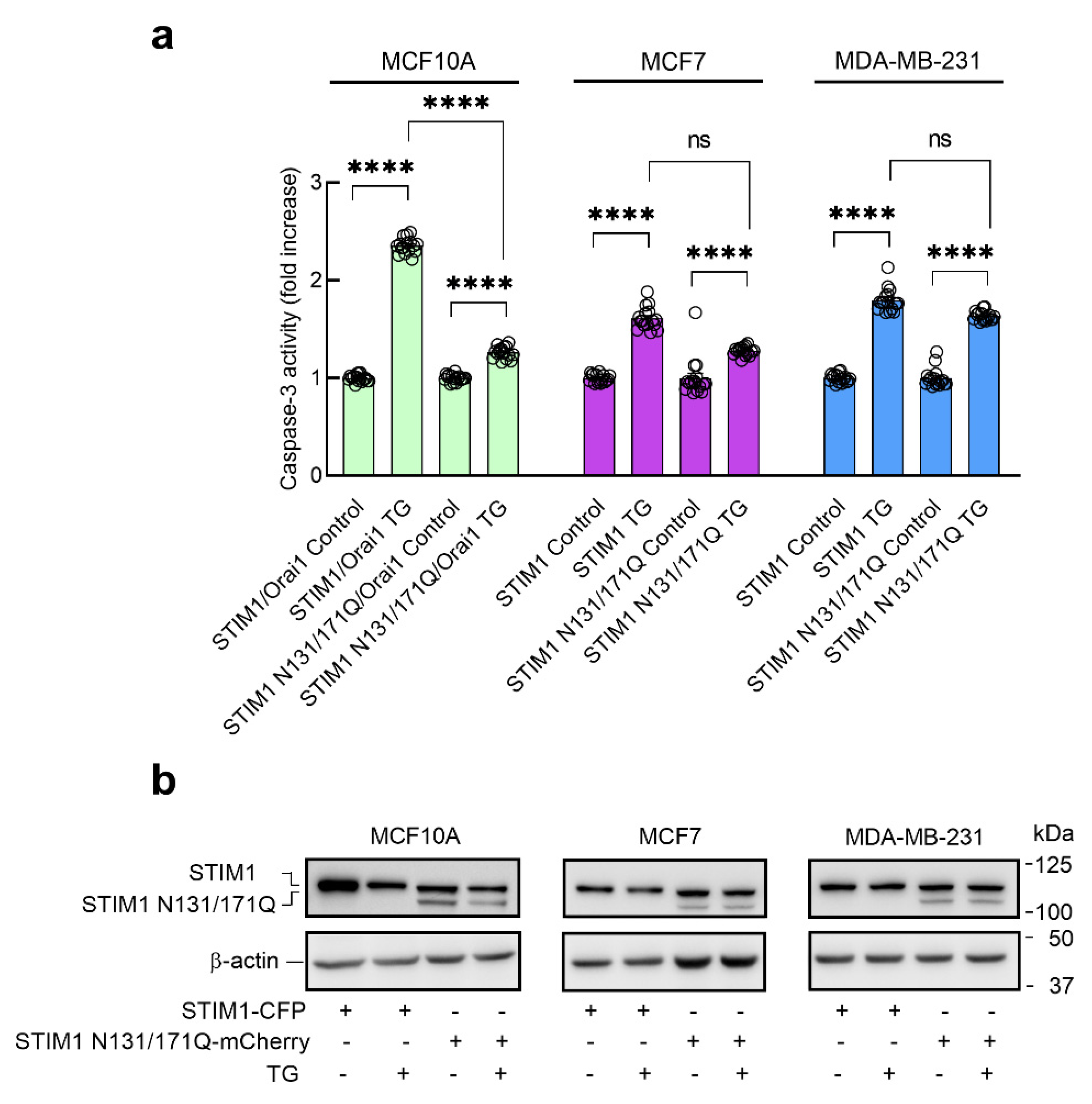
3.4. Role of STIM1 Glycosylation in TG-Evoked Apoptosis in Breast Cancer and Non-Tumoral Breast Epithelial Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roos, J.; Digregorio, P.J.; Yeromin, A.V.; Ohlsen, K.; Lioudyno, M.; Zhang, S.; Safrina, O.; Kozak, J.A.; Wagner, S.L.; Cahalan, M.D.; et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 2005, 169, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.L.; Yu, Y.; Roos, J.; Kozak, J.A.; Deerinck, T.J.; Ellisman, M.H.; Stauderman, K.A.; Cahalan, M.D. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature 2005, 437, 902–905. [Google Scholar] [CrossRef] [PubMed]
- Feske, S.; Gwack, Y.; Prakriya, M.; Srikanth, S.; Puppel, S.-H.; Tanasa, B.; Hogan, P.G.; Lewis, R.S.; Daly, M.; Rao, A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 2006, 441, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Mercer, J.C.; DeHaven, W.I.; Smyth, J.T.; Wedel, B.; Boyles, R.R.; Bird, G.S.; Putney, J.W., Jr. Large Store-operated Calcium Selective Currents Due to Co-expression of Orai1 or Orai2 with the Intracellular Calcium Sensor, Stim. J. Biol. Chem. 2006, 281, 24979–24990. [Google Scholar] [CrossRef] [PubMed]
- Peinelt, C.; Vig, M.; Koomoa, D.L.; Beck, A.; Nadler, M.J.S.; Koblan-Huberson, M.; Lis, A.; Fleig, A.; Penner, R.; Kinet, J.-P. Amplification of CRAC current by STIM1 and CRACM1 (Orai1). Nature 2006, 8, 771–773. [Google Scholar] [CrossRef]
- Prakriya, M.; Feske, S.; Gwack, Y.; Srikanth, S.; Rao, A.; Hogan, P.G. Orai1 is an essential pore subunit of the CRAC channel. Nature 2006, 443, 230–233. [Google Scholar] [CrossRef]
- Yoast, R.E.; Emrich, S.M.; Zhang, X.; Xin, P.; Johnson, M.T.; Fike, A.J.; Walter, V.; Hempel, N.; Yule, D.I.; Sneyd, J.; et al. The native ORAI channel trio underlies the diversity of Ca2+ signaling events. Nat. Commun. 2020, 11, 2444. [Google Scholar] [CrossRef]
- Berna-Erro, A.; Jardin, I.; Salido, G.M.; Rosado, J.A. Role of STIM2 in cell function and physiopathology. J. Physiol. 2017, 595, 3111–3128. [Google Scholar] [CrossRef]
- Emrich, S.M.; Yoast, R.E.; Xin, P.; Zhang, X.; Pathak, T.; Nwokonko, R.; Gueguinou, M.F.; Subedi, K.P.; Zhou, Y.; Ambudkar, I.S.; et al. Cross-talk between N-terminal and C-terminal domains in stromal interaction molecule 2 (STIM2) determines enhanced STIM2 sensitivity. J. Biol. Chem. 2019, 294, 6318–6332. [Google Scholar] [CrossRef]
- Ahmad, M.; Ong, H.L.; Saadi, H.; Son, G.-Y.; Shokatian, Z.; Terry, L.E.; Trebak, M.; Yule, D.I.; Ambudkar, I. Functional communication between IP 3 R and STIM2 at subthreshold stimuli is a critical checkpoint for initiation of SOCE. Proc. Natl. Acad. Sci. USA 2022, 119, e2114928118. [Google Scholar] [CrossRef]
- Jha, A.; Ahuja, M.; Maléth, J.; Moreno, C.M.; Yuan, J.P.; Kim, M.S.; Muallem, S. The STIM1 CTID domain determines access of SARAF to SOAR to regulate Orai1 channel function. J. Cell Biol. 2013, 202, 71–79. [Google Scholar] [CrossRef]
- Jardín, I.; Albarran, L.; Salido, G.M.; López, J.J.; Sage, S.O.; Rosado, J.A. Fine-tuning of store-operated calcium entry by fast and slow Ca2+-dependent inactivation: Involvement of SARAF. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Zomot, E.; Cohen, H.A.; Dagan, I.; Militsin, R.; Palty, R. Bidirectional regulation of calcium release–activated calcium (CRAC) channel by SARAF. J. Cell Biol. 2021, 220, e202104007. [Google Scholar] [CrossRef]
- Lopez, J.J.; Albarran, L.; Gómez, L.J.; Smani, T.; Salido, G.M.; Rosado, J.A. Molecular modulators of store-operated calcium entry. Biochim. et Biophys. Acta 2016, 1863, 2037–2043. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, T.; Ueyama, T.; Lange, I.; Feske, S.; Saito, N. Protein Kinase C-induced Phosphorylation of Orai1 Regulates the Intracellular Ca2+ Level via the Store-operated Ca2+ Channel. J. Biol. Chem. 2010, 285, 25720–25730. [Google Scholar] [CrossRef]
- Zhang, X.; Pathak, T.; Yoast, R.; Emrich, S.; Xin, P.; Nwokonko, R.M.; Johnson, M.; Wu, S.; Delierneux, C.; Gueguinou, M.; et al. A calcium/cAMP signaling loop at the ORAI1 mouth drives channel inactivation to shape NFAT induction. Nat. Commun. 2019, 10, 1971. [Google Scholar] [CrossRef] [PubMed]
- Lopez, E.; Frischauf, I.; Jardin, I.; Derler, I.; Muik, M.; Cantonero, C.; Salido, G.M.; Smani, T.; Rosado, J.; Redondo, P.C. STIM1 phosphorylation at Y316 modulates its interaction with SARAF and the activation of SOCE and ICRAC. J. Cell Sci. 2019, 132, jcs.226019. [Google Scholar] [CrossRef] [PubMed]
- Smyth, J.T.; Petranka, J.G.; Boyles, R.R.; DeHaven, W.I.; Fukushima, M.; Johnson, K.L.; Williams, J.G.; Putney, J.W., Jr. Phosphorylation of STIM1 underlies suppression of store-operated calcium entry during mitosis. Nat. Cell Biol. 2009, 11, 1465–1472. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, B.J.; Irrinki, K.M.; Mallilankaraman, K.; Lien, Y.-C.; Wang, Y.; Bhanumathy, C.D.; Subbiah, R.; Ritchie, M.F.; Soboloff, J.; Baba, Y.; et al. S-glutathionylation activates STIM1 and alters mitochondrial homeostasis. J. Cell Biol. 2010, 190, 391–405. [Google Scholar] [CrossRef]
- Johnson, J.; Blackman, R.; Gross, S.; Soboloff, J. Control of STIM and Orai function by post-translational modifications. Cell Calcium 2022, 103, 102544. [Google Scholar] [CrossRef]
- Kilch, T.; Alansary, D.; Peglow, M.; Dörr, K.; Rychkov, G.; Rieger, H.; Peinelt, C.; Niemeyer, B.A. Mutations of the Ca2+-sensing Stromal Interaction Molecule STIM1 Regulate Ca2+ Influx by Altered Oligomerization of STIM1 and by Destabilization of the Ca2+ Channel Orai. J. Biol. Chem. 2013, 288, 1653–1664. [Google Scholar] [CrossRef] [PubMed]
- Gwack, Y.; Srikanth, S.; Feske, S.; Cruz-Guilloty, F.; Oh-Hora, M.; Neems, D.S.; Hogan, P.G.; Rao, A. Biochemical and Functional Characterization of Orai Proteins. J. Biol. Chem. 2007, 282, 16232–16243. [Google Scholar] [CrossRef] [PubMed]
- Dörr, K.; Kilch, T.; Kappel, S.; Alansary, D.; Schwär, G.; Niemeyer, B.A.; Peinelt, C. Cell type–specific glycosylation of Orai1 modulates store-operated Ca 2+ entry. Sci. Signal. 2016, 9, ra25. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Zhao, Y.; Bhattacharya, M.; Stathopulos, P.B. Structural perturbations induced by Asn131 and Asn171 glycosylation converge within the EFSAM core and enhance stromal interaction molecule-1 mediated store operated calcium entry. Biochim. et Biophys. Acta Mol. Cell Res. 2017, 1864, 1054–1063. [Google Scholar] [CrossRef] [PubMed]
- Novello, M.J.; Zhu, J.; Feng, Q.; Ikura, M.; Stathopulos, P.B. Structural elements of stromal interaction molecule function. Cell Calcium 2018, 73, 88–94. [Google Scholar] [CrossRef]
- Ye, J.; Li, M.; Li, Q.; Jia, Z.; Hu, X.; Zhao, G.; Zhi, S.; Hong, G.; Lu, Z. Activation of STIM1/Orai1-mediated SOCE in sepsis-induced myocardial depression. Mol. Med. Rep. 2022, 26, 1–11. [Google Scholar] [CrossRef]
- Gang, Q.; Bettencourt, C.; Brady, S.; Holton, J.L.; Healy, E.G.; McConville, J.; Morrison, P.J.; Ripolone, M.; Violano, R.; Sciacco, M.; et al. Genetic defects are common in myopathies with tubular aggregates. Ann. Clin. Transl. Neurol. 2022, 9, 4–15. [Google Scholar] [CrossRef]
- Smani, T.; Shapovalov, G.; Skryma, R.; Prevarskaya, N.; Rosado, J.A. Functional and physiopathological implications of TRP channels. Biochim. et Biophys. Acta Mol. Cell Res. 2015, 1853, 1772–1782. [Google Scholar] [CrossRef]
- Motiani, R.K.; Abdullaev, I.F.; Trebak, M. A novel native store-operated calcium channel encoded by Orai3: Selective requirement of Orai3 versus Orai1 in estrogen receptor-positive versus estrogen receptor-negative breast cancer cells. J. Biol. Chem. 2010, 285, 19173–19183. [Google Scholar] [CrossRef]
- Jardin, I.; Diez-Bello, R.; Lopez, J.; Redondo, P.; Salido, G.; Smani, T.; Rosado, J. TRPC6 Channels Are Required for Proliferation, Migration and Invasion of Breast Cancer Cell Lines by Modulation of Orai1 and Orai3 Surface Exposure. Cancers 2018, 10, 331. [Google Scholar] [CrossRef]
- Robitaille, M.; Chan, S.M.; Peters, A.A.; Dai, L.; So, C.L.; Bong, A.H.L.; Sadras, F.; Roberts-Thomson, S.J.; Monteith, G.R. ORAI1-Regulated Gene Expression in Breast Cancer Cells: Roles for STIM1 Binding, Calcium Influx and Transcription Factor Translocation. Int. J. Mol. Sci. 2022, 23, 5867. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhang, J.J.; Huang, X.-Y. Orai1 and STIM1 Are Critical for Breast Tumor Cell Migration and Metastasis. Cancer Cell 2009, 15, 124–134. [Google Scholar] [CrossRef] [PubMed]
- McAndrew, D.; Grice, D.M.; Peters, A.A.; Davis, F.M.; Stewart, T.; Rice, M.; Smart, C.E.; Brown, M.A.; Kenny, P.A.; Roberts-Thomson, S.J.; et al. ORAI1-Mediated Calcium Influx in Lactation and in Breast Cancer. Mol. Cancer Ther. 2011, 10, 448–460. [Google Scholar] [CrossRef] [PubMed]
- Kappel, S.; Borgström, A.; Stokłosa, P.; Dörr, K.; Peinelt, C. Store-operated calcium entry in disease: Beyond STIM/Orai expression levels. Semin. Cell Dev. Biol. 2019, 94, 66–73. [Google Scholar] [CrossRef]
- Jardin, I.; Diez-Bello, R.; Falcon, D.; Alvarado, S.; Regodon, S.; Salido, G.M.; Smani, T.; Rosado, J.A. Melatonin downregulates TRPC6, impairing store-operated calcium entry in triple-negative breast cancer cells. J. Biol. Chem. 2021, 296, 100254. [Google Scholar] [CrossRef]
- Sanchez-Collado, J.; Lopez, J.J.; Cantonero, C.; Jardin, I.; Regodón, S.; Redondo, P.C.; Gordillo, J.; Smani, T.; Salido, G.M.; Rosado, J.A. Orai2 Modulates Store-Operated Ca2+ Entry and Cell Cycle Progression in Breast Cancer Cells. Cancers 2021, 14, 114. [Google Scholar] [CrossRef]
- Sanchez-Collado, J.; Lopez, J.J.; Jardin, I.; Berna-Erro, A.; Camello, P.J.; Cantonero, C.; Smani, T.; Salido, G.M.; Rosado, J.A. Orai1α, but not Orai1β, co-localizes with TRPC1 and is required for its plasma membrane location and activation in HeLa cells. Cell Mol. Life Sci. 2022, 79, 33. [Google Scholar] [CrossRef]
- Rosado, J.; Lopez, J.J.; Gomez-Arteta, E.; Redondo, P.C.; Salido, G.M.; Pariente, J.A. Early caspase-3 activation independent of apoptosis is required for cellular function. J. Cell Physiol. 2006, 209, 142–152. [Google Scholar] [CrossRef]
- Schild, A.; Bhardwaj, R.; Wenger, N.; Tscherrig, D.; Kandasamy, P.; Dernič, J.; Baur, R.; Peinelt, C.; Hediger, M.A.; Lochner, M. Synthesis and Pharmacological Characterization of 2-Aminoethyl Diphenylborinate (2-APB) Derivatives for Inhibition of Store-Operated Calcium Entry (SOCE) in MDA-MB-231 Breast Cancer Cells. Int. J. Mol. Sci. 2020, 21, 5604. [Google Scholar] [CrossRef]
- Bhardwaj, R.; Augustynek, B.S.; Ercan-Herbst, E.; Kandasamy, P.; Seedorf, M.; Peinelt, C.; Hediger, M.A. Ca2+/Calmodulin Binding to STIM1 Hydrophobic Residues Facilitates Slow Ca2+-Dependent Inactivation of the Orai1 Channel. Cell Physiol. Biochem. 2020, 54, 252–270. [Google Scholar] [CrossRef]
- Butorac, C.; Muik, M.; Derler, I.; Stadlbauer, M.; Lunz, V.; Krizova, A.; Lindinger, S.; Schober, R.; Frischauf, I.; Bhardwaj, R.; et al. A novel STIM1-Orai1 gating interface essential for CRAC channel activation. Cell Calcium 2019, 79, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Collado, J.; Lopez, J.J.; Jardin, I.; Camello, P.J.; Falcon, D.; Regodon, S.; Salido, G.M.; Smani, T.; Rosado, J.A. Adenylyl Cyclase Type 8 Overexpression Impairs Phosphorylation-Dependent Orai1 Inactivation and Promotes Migration in MDA-MB-231 Breast Cancer Cells. Cancers 2019, 11, 1624. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, M.; Tomita, T.; Janoshazi, A.; Putney, J.W. Alternative translation initiation gives rise to two isoforms of orai1 with distinct plasma membrane mobilities. J. Cell Sci. 2012, 125, 4354–4361. [Google Scholar] [CrossRef] [PubMed]
- Takatsuki, A.; Arima, K.; Tamura, G. Tunicamycin, a new antibiotic. I Isolation and characterization of tunicamycin. J. Antibiot. 1971, 24, 215–223. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Ke, Z.-J.; Comer, A.L.; Xu, M.; Frank, J.A.; Zhang, Z.; Shi, X.; Luo, J. Tunicamycin-induced unfolded protein response in the developing mouse brain. Toxicol. Appl. Pharmacol. 2015, 283, 157–167. [Google Scholar] [CrossRef]
- Kay, J.E.; Korner, A. Effect of cycloheximide on protein and ribonucleic acid synthesis in cultured human lymphocytes. Biochem. J. 1966, 100, 815–822. [Google Scholar] [CrossRef]
- Shiraishi, H.; Okamoto, H.; Yoshimura, A.; Yoshida, H. ER stress-induced apoptosis and caspase-12 activation occurs downstream of mitochondrial apoptosis involving Apaf-1. J. Cell Sci. 2006, 119, 3958–3966. [Google Scholar] [CrossRef]
- Sanchez-Collado, J.; Lopez, J.J.; Gonzalez-Gutierrez, L.; Cantonero, C.; Jardin, I.; Salido, G.M.; Rosado, J.A. Functional role of TRPC6 and STIM2 in cytosolic and endoplasmic reticulum Ca2+ content in resting estrogen receptor-positive breast cancer cells. Biochem. J. 2020, 477, 3183–3197. [Google Scholar] [CrossRef]
- Imperiali, B.; O’Connor, S.E. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr. Opin. Chem. Biol. 1999, 3, 643–649. [Google Scholar] [CrossRef]
- Williams, R.T.; Senior, P.V.; Van Stekelenburg, L.; Layton, J.E.; Smith, P.J.; Dziadek, M.A. Stromal interaction molecule 1 (STIM1), a transmembrane protein with growth suppressor activity, contains an extracellular SAM domain modified by N-linked glycosylation. Biochim. Biophys. Acta Protein Struct. Mol. Enzym. 2002, 1596, 131–137. [Google Scholar] [CrossRef]
- Gueder, N.; Allan, G.; Telliez, M.-S.; Hague, F.; Fernandez, J.M.; Sanchez-Fernandez, E.M.; Ortiz-Mellet, C.; Ahidouch, A.; Ouadid-Ahidouch, H. sp2 -Iminosugar α-glucosidase inhibitor 1-C -octyl-2-oxa-3-oxocastanospermine specifically affected breast cancer cell migration through Stim1, β1-integrin, and FAK signaling pathways. J. Cell Physiol. 2017, 232, 3631–3640. [Google Scholar] [CrossRef] [PubMed]
- López, J.; Siegfried, G.; Cantonero, C.; Soulet, F.; Descarpentrie, J.; Smani, T.; Badiola, I.; Pernot, S.; Evrard, S.; Rosado, J.; et al. Furin Prodomain ppFurin Enhances Ca2+ Entry Through Orai and TRPC6 Channels’ Activation in Breast Cancer Cells. Cancers 2021, 13, 1670. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, R.P.; Elmi, A.; Alcantara-Adap, E.; Hubrack, S.; Nader, N.; Yu, F.; Dib, M.; Ramachandran, V.; Shoushtari, H.N.; Machaca, K. miRNA-dependent regulation of STIM1 expression in breast cancer. Sci. Rep. 2019, 9, 13076. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, W.; Wang, X.; Gao, H.; Liu, R.; Shou, J.; Yan, J. Enhanced store-operated calcium entry (SOCE) exacerbates motor neurons apoptosis following spinal cord injury. Gen. Physiol. Biophys. 2021, 40, 61–69. [Google Scholar]
- Czyż, A.; Brutkowski, W.; Fronk, J.; Duszyński, J.; Zabłocki, K. Tunicamycin desensitizes store-operated Ca2+ entry to ATP and mitochondrial potential. Biochem. Biophys. Res. Commun. 2009, 381, 176–180. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchez-Collado, J.; Nieto-Felipe, J.; Jardin, I.; Bhardwaj, R.; Berna-Erro, A.; Salido, G.M.; Smani, T.; Hediger, M.A.; Lopez, J.J.; Rosado, J.A. Store-Operated Calcium Entry in Breast Cancer Cells Is Insensitive to Orai1 and STIM1 N-Linked Glycosylation. Cancers 2023, 15, 203. https://doi.org/10.3390/cancers15010203
Sanchez-Collado J, Nieto-Felipe J, Jardin I, Bhardwaj R, Berna-Erro A, Salido GM, Smani T, Hediger MA, Lopez JJ, Rosado JA. Store-Operated Calcium Entry in Breast Cancer Cells Is Insensitive to Orai1 and STIM1 N-Linked Glycosylation. Cancers. 2023; 15(1):203. https://doi.org/10.3390/cancers15010203
Chicago/Turabian StyleSanchez-Collado, Jose, Joel Nieto-Felipe, Isaac Jardin, Rajesh Bhardwaj, Alejandro Berna-Erro, Gines M. Salido, Tarik Smani, Matthias A Hediger, Jose J. Lopez, and Juan A. Rosado. 2023. "Store-Operated Calcium Entry in Breast Cancer Cells Is Insensitive to Orai1 and STIM1 N-Linked Glycosylation" Cancers 15, no. 1: 203. https://doi.org/10.3390/cancers15010203
APA StyleSanchez-Collado, J., Nieto-Felipe, J., Jardin, I., Bhardwaj, R., Berna-Erro, A., Salido, G. M., Smani, T., Hediger, M. A., Lopez, J. J., & Rosado, J. A. (2023). Store-Operated Calcium Entry in Breast Cancer Cells Is Insensitive to Orai1 and STIM1 N-Linked Glycosylation. Cancers, 15(1), 203. https://doi.org/10.3390/cancers15010203








