Oral Microbiota as Novel Biomarkers for Colorectal Cancer Screening
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Oral Sampling
2.2. Human Fecal Sampling
2.3. Inclusion and Exclusion Criteria
2.4. DNA Extraction and Purification from Oral and Fecal Specimens
2.5. PCR Amplification and Sequencing
2.6. 16S rRNA Sequence Preprocessing and Analysis
3. Results
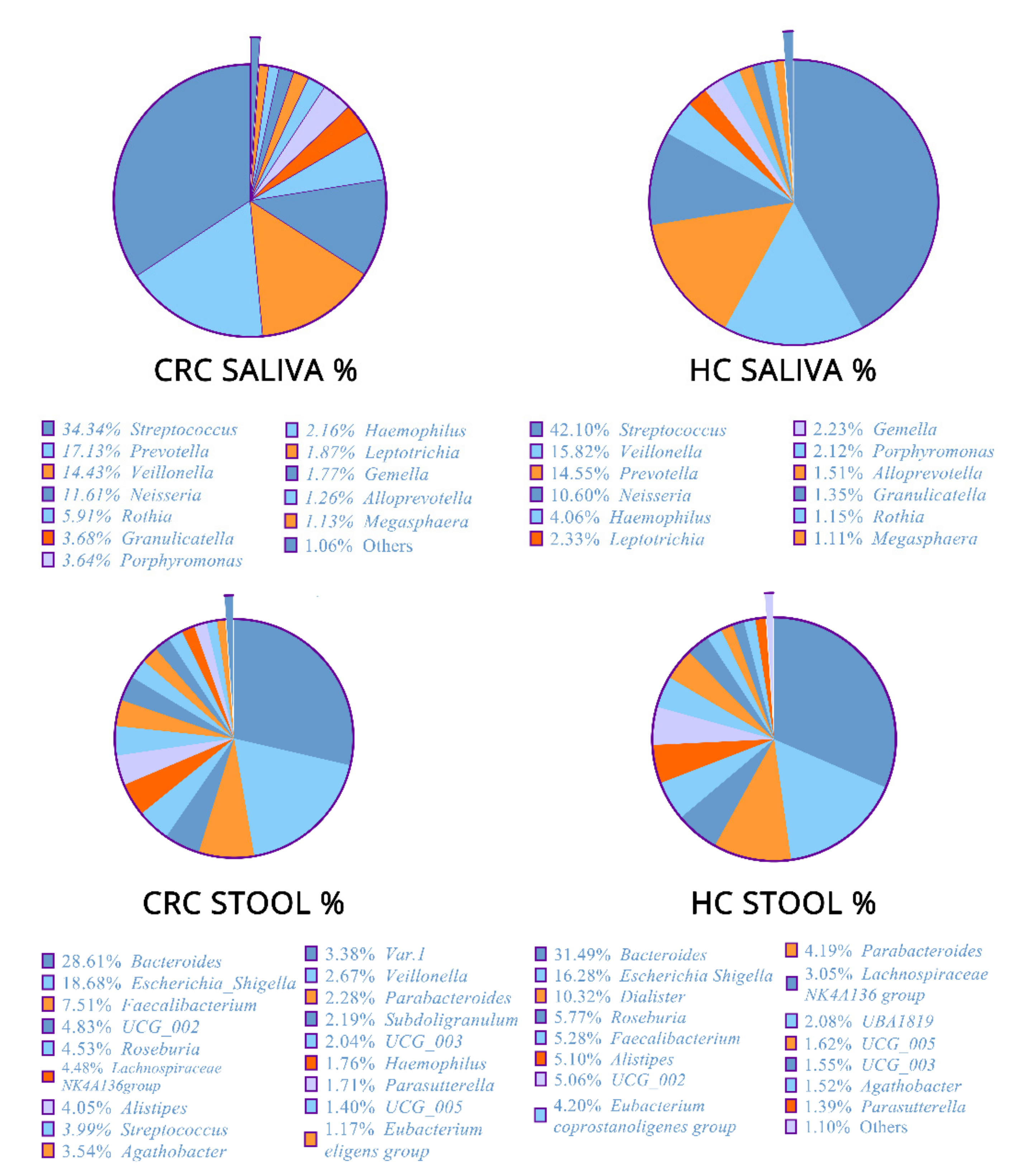
3.1. Microbial Community Composition
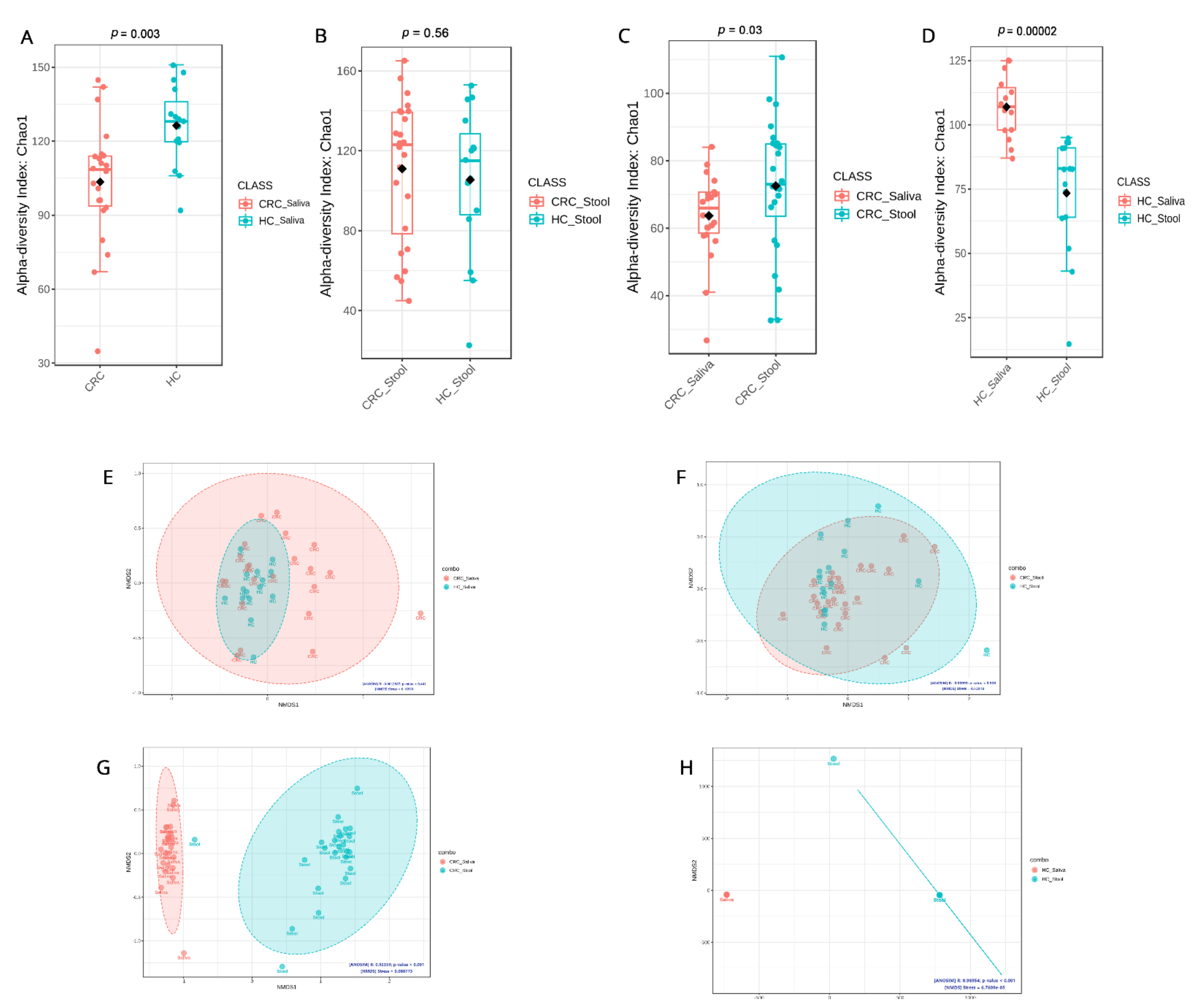
3.2. Microbial Community Diversity
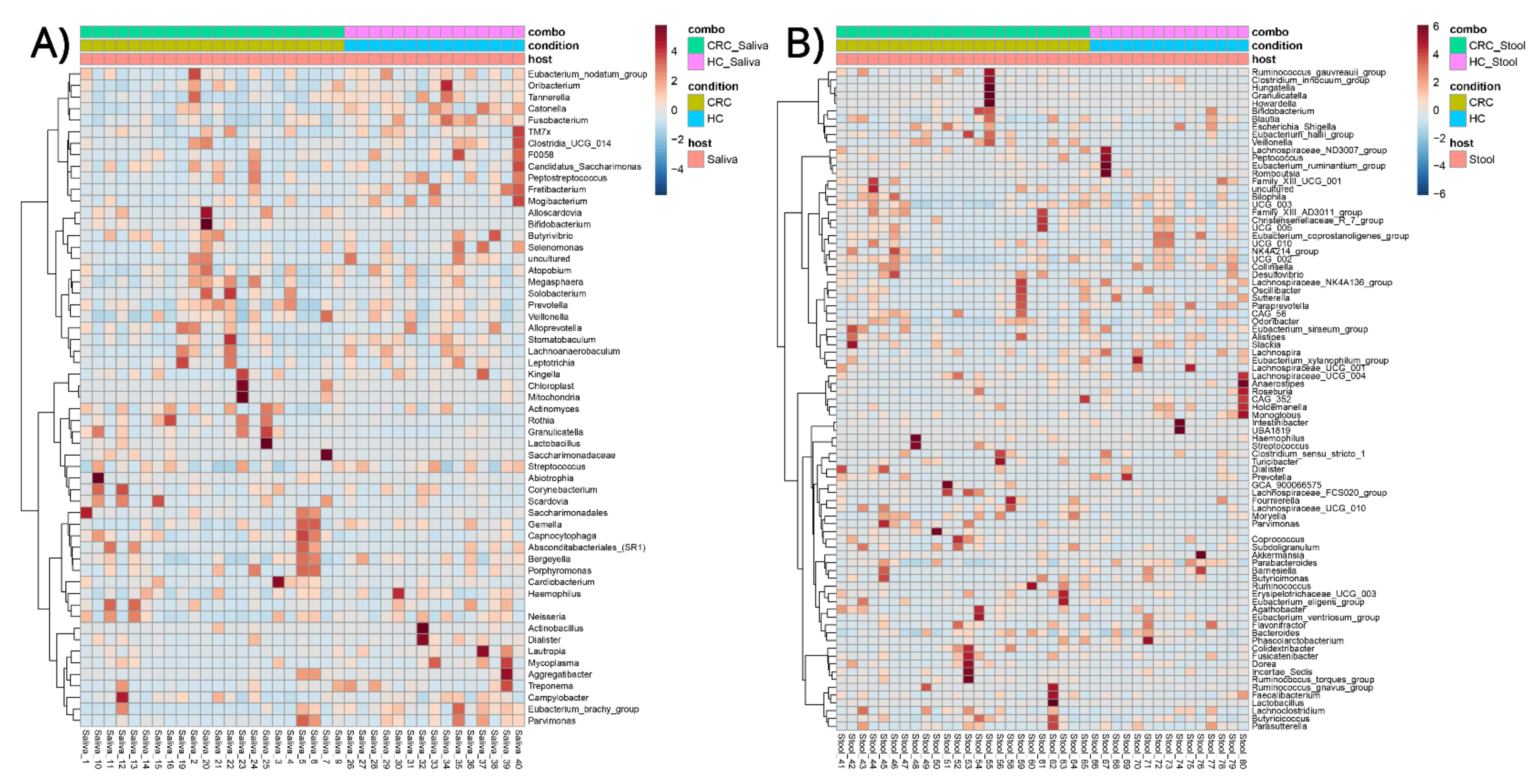
3.3. Phenotype–Microbial Associations
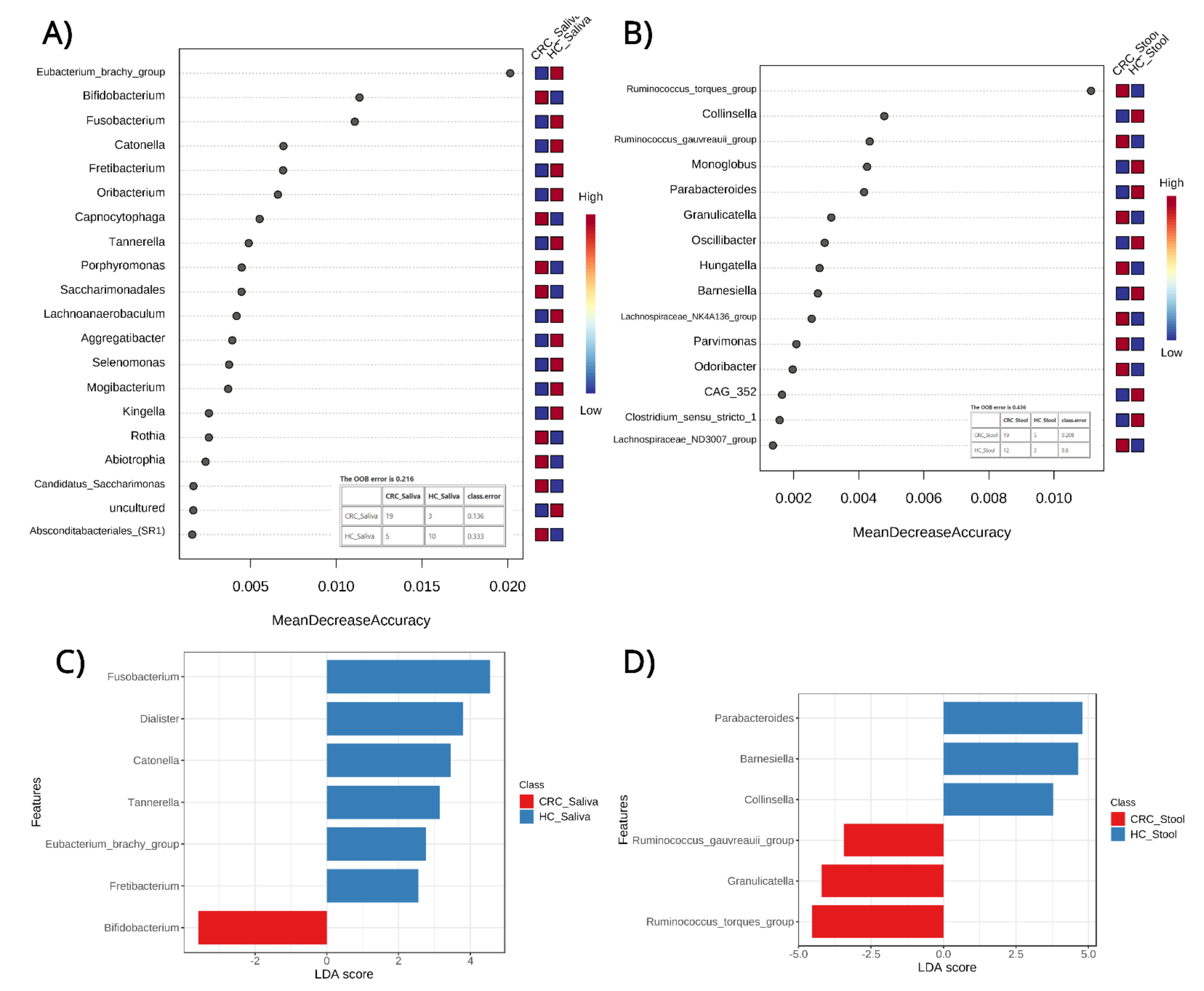
3.4. Classification and Biomarker Discovery
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Douaiher, J.; Ravipati, A.; Grams, B.; Chowdhury, S.; Alatise, O.; Are, C. Colorectal cancer-global burden, trends, and geographical variations. J. Surg. Oncol. 2017, 115, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Onyoh, E.F.; Hsu, W.-F.; Chang, L.-C.; Lee, Y.-C.; Wu, M.-S.; Chiu, H.-M. The Rise of Colorectal Cancer in Asia: Epidemiology, Screening, and Management. Curr. Gastroenterol. Rep. 2019, 21, 36. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, J.B.; Maggard, M.A.; Ko, C.Y. Colon cancer survival rates with the new American Joint Committee on Cancer sixth edition staging. J. Natl. Cancer Inst. 2004, 96, 1420–1425. [Google Scholar] [CrossRef]
- Niedermaier, T.; Tikk, K.; Gies, A.; Bieck, S.; Brenner, H. Sensitivity of Fecal Immunochemical Test for Colorectal Cancer Detection Differs According to Stage and Location. Clin. Gastroenterol. Hepatol. 2020, 18, 2920–2928.e6. [Google Scholar] [CrossRef]
- Durack, J.; Lynch, S.V. The gut microbiome: Relationships with disease and opportunities for therapy. J. Exp. Med. 2019, 216, 20–40. [Google Scholar] [CrossRef]
- Bu, F.; Yao, X.; Lu, Z.; Yuan, X.; Chen, C.; Li, L.; Li, Y.; Jiang, F.; Zhu, L.; Shi, G.; et al. Pathogenic or Therapeutic: The Mediating Role of Gut Microbiota in Non-Communicable Diseases. Front Cell Infect Microbiol. 2022, 12, 906349. [Google Scholar] [CrossRef]
- Jia, W.; Rajani, C.; Xu, H.; Zheng, X. Gut microbiota alterations are distinct for primary colorectal cancer and hepatocellular carcinoma. Protein Cell 2021, 12, 374–393. [Google Scholar] [CrossRef]
- Megat Mohd Azlan, P.-I.-H.; Chin, S.-F.; Low, T.Y.; Neoh, H.-M.; Jamal, R. Analyzing the Secretome of Gut Microbiota as the Next Strategy For Early Detection of Colorectal Cancer. Proteomics 2019, 19, 1800176. [Google Scholar] [CrossRef]
- Liang, Q.; Chiu, J.; Chen, Y.; Huang, Y.; Higashimori, A.; Fang, J.; Brim, H.; Ashktorab, H.; Ng, S.C.; Ng, S.S.M.; et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin. Cancer Res. 2017, 23, 2061–2070. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Sharafkhah, M.; Asadzadeh Aghdaei, H.; Nazemalhosseini Mojarad, E.; Dabiri, H.; Akhavan Sepahi, A.; Modarressi, M.H.; Feizabadi, M.M.; Zali, M.R. Applying simple linear combination, multiple logistic and factor analysis methods for candidate fecal bacteria as novel biomarkers for early detection of adenomatous polyps and colon cancer. J. Microbiol. Methods 2018, 155, 82–88. [Google Scholar] [CrossRef]
- Flemer, B.; Lynch, D.B.; Brown, J.M.R.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef]
- Baxter, N.T.; Ruffin, M.T.; Rogers, M.A.M.; Schloss, P.D. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016, 8, 37. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Asadzadeh Aghdaei, H.; Dabiri, H.; Akhavan Sepahi, A.; Modarressi, M.H.; Nazemalhosseini Mojarad, E. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb. Pathog. 2018, 124, 244–249. [Google Scholar] [CrossRef]
- Kostic, A.D.; Gevers, D.; Pedamallu, C.S.; Michaud, M.; Duke, F.; Earl, A.M.; Ojesina, A.I.; Jung, J.; Bass, A.J.; Tabernero, J.; et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012, 22, 292–298. [Google Scholar] [CrossRef]
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306. [Google Scholar] [CrossRef]
- Rezasoltani, S.; Ghanbari, R.; Looha, M.A.; Mojarad, E.N.; Yadegar, A.; Stewart, D.; Aghdaei, H.A.; Zali, M.R. Expression of Main Toll-Like Receptors in Patients with Different Types of Colorectal Polyps and Their Relationship with Gut Microbiota. Int. J. Mol. Sci. 2020, 21, 8968. [Google Scholar] [CrossRef]
- Pushalkar, S.; Ji, X.; Li, Y.; Estilo, C.; Yegnanarayana, R.; Singh, B.; Li, X.; Saxena, D. Comparison of oral microbiota in tumor and non-tumor tissues of patients with oral squamous cell carcinoma. BMC Microbiol. 2012, 12, 144. [Google Scholar] [CrossRef]
- Schmidt, B.L.; Kuczynski, J.; Bhattacharya, A.; Huey, B.; Corby, P.M.; Queiroz, E.L.S.; Nightingale, K.; Kerr, A.R.; DeLacure, M.D.; Veeramachaneni, R.; et al. Changes in abundance of oral microbiota associated with oral cancer. PLoS ONE 2014, 9, e98741. [Google Scholar] [CrossRef]
- Chen, X.; Winckler, B.; Lu, M.; Cheng, H.; Yuan, Z.; Yang, Y.; Jin, L.; Ye, W. Oral Microbiota and Risk for Esophageal Squamous Cell Carcinoma in a High-Risk Area of China. PLoS ONE 2015, 10, e0143603. [Google Scholar] [CrossRef]
- Farrell, J.J.; Zhang, L.; Zhou, H.; Chia, D.; Elashoff, D.; Akin, D.; Paster, B.J.; Joshipura, K.; Wong, D.T.W. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012, 61, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Torres, P.J.; Fletcher, E.M.; Gibbons, S.M.; Bouvet, M.; Doran, K.S.; Kelley, S.T. Characterization of the salivary microbiome in patients with pancreatic cancer. PeerJ 2015, 3, e1373. [Google Scholar] [CrossRef] [PubMed]
- Flemer, B.; Warren, R.D.; Barrett, M.P.; Cisek, K.; Das, A.; Jeffery, I.B.; Hurley, E.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 2018, 67, 1454–1463. [Google Scholar] [CrossRef] [PubMed]
- Kato, I.; Vasquez, A.A.; Moyerbrailean, G.; Land, S.; Sun, J.; Lin, H.-S.; Ram, J.L. Oral microbiome and history of smoking and colorectal cancer. J. Epidemiol. Res. 2016, 2, 92–101. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Pedregosa, F.V.G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Louppe, G.; Prettenhofer, P.; Weiss, R.; Weiss, R.J.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Pruesse, E.; Quast, C.; Knittel, K.; Fuchs, B.M.; Ludwig, W.; Peplies, J.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Chang, F.; He, S.; Dang, C. Assisted Selection of Biomarkers by Linear Discriminant Analysis Effect Size (LEfSe) in Microbiome Data. J. Vis. Exp. 2022, 183, e61715. [Google Scholar] [CrossRef]
- Ramosaj, B.; Pauly, M. Consistent estimation of residual variance with random forest Out-Of-Bag errors. Stat. Probab. Lett. 2019, 151, 49–57. [Google Scholar] [CrossRef]
- Wang, Y.; Wan, X.; Wu, X.; Zhang, C.; Liu, J.; Hou, S. Eubacterium rectale contributes to colorectal cancer initiation via promoting colitis. Gut Pathog. 2021, 13, 2. [Google Scholar] [CrossRef]
- Faghfoori, Z.; Faghfoori, M.H.; Saber, A.; Izadi, A.; Yari Khosroushahi, A. Anticancer effects of bifidobacteria on colon cancer cell lines. Cancer Cell Int. 2021, 21, 258. [Google Scholar] [CrossRef]
- Park, J.; Kim, N.-E.; Yoon, H.; Shin, C.M.; Kim, N.; Lee, D.H.; Park, J.Y.; Choi, C.H.; Kim, J.G.; Kim, Y.-K.; et al. Fecal Microbiota and Gut Microbe-Derived Extracellular Vesicles in Colorectal Cancer. Front. Oncol. 2021, 11, 650026. [Google Scholar] [CrossRef]
- Sobhani, I.; Bergsten, E.; Couffin, S.; Amiot, A.; Nebbad, B.; Barau, C.; de’Angelis, N.; Rabot, S.; Canoui-Poitrine, F.; Mestivier, D.; et al. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc. Natl. Acad. Sci. USA 2019, 116, 24285–24295. [Google Scholar] [CrossRef]
- Sheng, Q.; Du, H.; Cheng, X.; Cheng, X.; Tang, Y.; Pan, L.; Wang, Q.; Lin, J. Characteristics of fecal gut microbiota in patients with colorectal cancer at different stages and different sites. Oncol. Lett. 2019, 18, 4834–4844. [Google Scholar] [CrossRef]
- Guven, D.C.; Dizdar, O.; Alp, A.; Kittana, F.N.A.; Karakoc, D.; Hamaloglu, E.; Lacin, S.; Karakas, Y.; Kilickap, S.; Hayran, M.; et al. Analysis of Fusobacterium nucleatum and Streptococcus gallolyticus in saliva of colorectal cancer patients. Biomark. Med. 2019, 13, 725–735. [Google Scholar] [CrossRef]
- Segata, N.; Haake, S.K.; Mannon, P.; Lemon, K.P.; Waldron, L.; Gevers, D.; Huttenhower, C.; Izard, J. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012, 13, R42. [Google Scholar] [CrossRef] [PubMed]
- Bassis, C.M.; Erb-Downward, J.R.; Dickson, R.P.; Freeman, C.M.; Schmidt, T.M.; Young, V.B.; Beck, J.M.; Curtis, J.L.; Huffnagle, G.B. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. mBio 2015, 6, e00037. [Google Scholar] [CrossRef]
- Boleij, A.; van Gelder, M.M.H.J.; Swinkels, D.W.; Tjalsma, H. Clinical Importance of Streptococcus gallolyticus Infection Among Colorectal Cancer Patients: Systematic Review and Meta-analysis. Clin. Infect. Dis. 2011, 53, 870–878. [Google Scholar] [CrossRef]
- Abdulamir, A.S.; Hafidh, R.R.; Bakar, F.A. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: Inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol. Cancer 2010, 9, 249. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Herold, J.L.; Taylor, J.; Xu, J.; Xu, Y. Variations among Streptococcus gallolyticus subsp. gallolyticus strains in connection with colorectal cancer. Sci. Rep. 2018, 8, 1514. [Google Scholar] [CrossRef] [PubMed]
- Aymeric, L.; Donnadieu, F.; Mulet, C.; du Merle, L.; Nigro, G.; Saffarian, A.; Bérard, M.; Poyart, C.; Robine, S.; Regnault, B.; et al. Colorectal cancer specific conditions promote Streptococcus gallolyticus gut colonization. Proc. Natl. Acad. Sci. USA 2018, 115, E283–E291. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Qian, Y.; Xie, Y.-H.; Jiang, S.-S.; Kang, Z.-R.; Chen, Y.-X.; Chen, Z.-F.; Fang, J.-Y. Alterations in the oral and gut microbiome of colorectal cancer patients and association with host clinical factors. Int. J. Cancer 2021, 149, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Gao, X.; Wu, L.; Yan, B.; Wang, Z.; Zhang, X.; Peng, L.; Yu, J.; Sun, G.; Yang, Y. Salivary Microbiota for Gastric Cancer Prediction: An Exploratory Study. Front. Cell. Infect. Microbiol. 2021, 11, 640309. [Google Scholar] [CrossRef]
- Abdi, H.; Kordi-Tamandani, D.M.; Lagzian, M.; Bakhshipour, A. Microbiome Analysis in Patients with Colorectal Cancer by 16S Ribosomal RNA Sequencing in the Southeast of Iran. Jundishapur J. Microbiol. 2022, 15. [Google Scholar] [CrossRef]
- Salgia, N.J.; Bergerot, P.G.; Maia, M.C.; Dizman, N.; Hsu, J.; Gillece, J.D.; Folkerts, M.; Reining, L.; Trent, J.; Highlander, S.K.; et al. Stool Microbiome Profiling of Patients with Metastatic Renal Cell Carcinoma Receiving Anti–PD-1 Immune Checkpoint Inhibitors. Eur. Urol. 2020, 78, 498–502. [Google Scholar] [CrossRef]
- Zeber-Lubecka, N.; Kulecka, M.; Lindner, B.; Krynicki, R.; Paziewska, A.; Nowakowski, A.; Bidzinski, M.; Ostrowski, J. Increased diversity of a cervical microbiome associates with cervical cancer. Front. Oncol. 2022, 12, 1005537. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, Q.; Shu, X.-O.; Steinwandel, M.D.; Blot, W.J.; Zheng, W.; Long, J. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int. J. Cancer 2019, 144, 2381–2389. [Google Scholar] [CrossRef]
- Ringel, Y.; Maharshak, N.; Ringel-Kulka, T.; Wolber, E.A.; Sartor, R.B.; Carroll, I.M. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes 2015, 6, 173–181. [Google Scholar] [CrossRef]
- Shaukat, A.; Levin, T.R. Current and future colorectal cancer screening strategies. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 521–531. [Google Scholar] [CrossRef]
- Ferrari, A.; Neefs, I.; Hoeck, S.; Peeters, M.; Van Hal, G. Towards Novel Non-Invasive Colorectal Cancer Screening Methods: A Comprehensive Review. Cancers 2021, 13, 1820. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rezasoltani, S.; Aghdaei, H.A.; Jasemi, S.; Gazouli, M.; Dovrolis, N.; Sadeghi, A.; Schlüter, H.; Zali, M.R.; Sechi, L.A.; Feizabadi, M.M. Oral Microbiota as Novel Biomarkers for Colorectal Cancer Screening. Cancers 2023, 15, 192. https://doi.org/10.3390/cancers15010192
Rezasoltani S, Aghdaei HA, Jasemi S, Gazouli M, Dovrolis N, Sadeghi A, Schlüter H, Zali MR, Sechi LA, Feizabadi MM. Oral Microbiota as Novel Biomarkers for Colorectal Cancer Screening. Cancers. 2023; 15(1):192. https://doi.org/10.3390/cancers15010192
Chicago/Turabian StyleRezasoltani, Sama, Hamid Asadzadeh Aghdaei, Seyedesomaye Jasemi, Maria Gazouli, Nikolas Dovrolis, Amir Sadeghi, Hartmut Schlüter, Mohammad Reza Zali, Leonardo Antonio Sechi, and Mohammad Mehdi Feizabadi. 2023. "Oral Microbiota as Novel Biomarkers for Colorectal Cancer Screening" Cancers 15, no. 1: 192. https://doi.org/10.3390/cancers15010192
APA StyleRezasoltani, S., Aghdaei, H. A., Jasemi, S., Gazouli, M., Dovrolis, N., Sadeghi, A., Schlüter, H., Zali, M. R., Sechi, L. A., & Feizabadi, M. M. (2023). Oral Microbiota as Novel Biomarkers for Colorectal Cancer Screening. Cancers, 15(1), 192. https://doi.org/10.3390/cancers15010192








