The Role of the Microbiome on the Pathogenesis and Treatment of Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Microbiome and CRC Pathogenesis
2.1. Dysbiosis and Colorectal Cancer
2.2. Inflammation and Toxins from Dysbiosis
2.3. Evidence from Biofilm
2.4. Dysbiosis and Tumor Microenvironment
2.5. Mitochondria
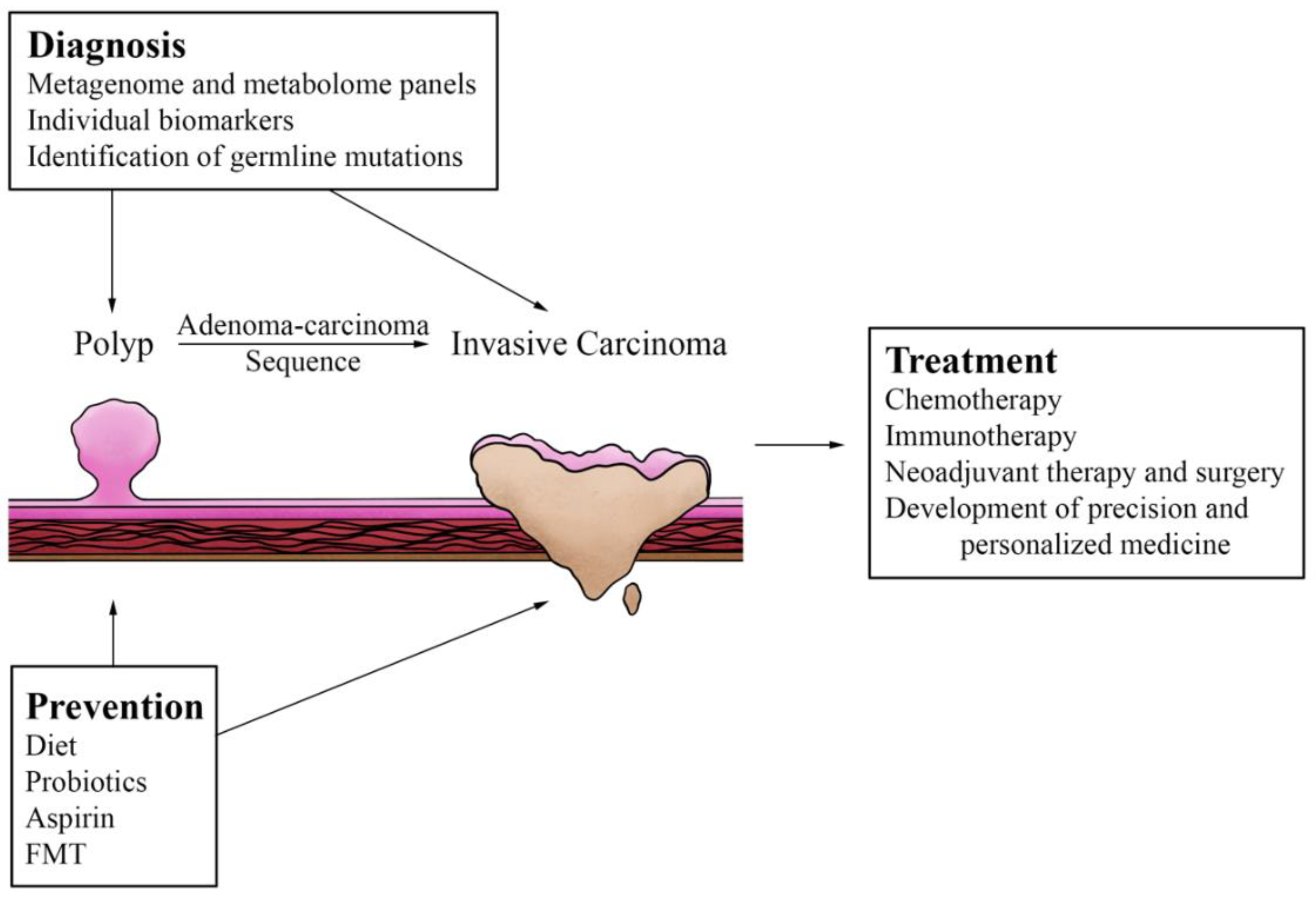
3. Diagnosis and Treatment Implications
3.1. Prediction of Cancer Development
3.2. Germline Mutations and Early-Onset CRC
3.3. Microbiome Effects on Chemotherapy
3.4. Potentiation of Radiation Therapy
3.5. Immunotherapy
3.6. Neoadjuvant Immunotherapy and Surgery
3.7. Precision and Personalized Medicine
4. Preventative Strategies
4.1. Diet
4.2. Probiotics
4.3. Aspirin and Other Medications
4.4. Fecal Microbiota Transplantation
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- NIH-National Cancer Institute. Cancer Stat Facts: Colorectal Cancer. Available online: https://seer.cancer.gov/statfacts/html/colorect.html (accessed on 8 August 2022).
- National Cancer Comprehensive Network. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines)-Colon Cancer, Version 3. 2021. Available online: https://policycommons.net/artifacts/1694254/nccn-clinical-practice-guidelines-in-oncology-nccn-guidelinesr-non-hodgkins-lymphomas/2425902/ (accessed on 7 August 2022).
- Hold, G.L. Gastrointestinal Microbiota and Colon Cancer. Dig. Dis. 2016, 34, 244–250. [Google Scholar] [CrossRef]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, R.; Shu, R.; Yu, J.; Li, H.; Long, H.; Jin, S.; Li, S.; Hu, Q.; Yao, F.; et al. Study of the Relationship between Microbiome and Colorectal Cancer Susceptibility Using 16SrRNA Sequencing. BioMed Res. Int. 2020, 2020, 7828392. [Google Scholar] [CrossRef]
- Fidelle, M.; Yonekura, S.; Picard, M.; Cogdill, A.; Hollebecque, A.; Roberti, M.P.; Zitvogel, L. Resolving the Paradox of Colon Cancer Through the Integration of Genetics, Immunology, and the Microbiota. Front. Immunol. 2020, 11, 600886. [Google Scholar] [CrossRef] [PubMed]
- Saus, E.; Iraola-Guzmán, S.; Willis, J.R.; Brunet-Vega, A.; Gabaldón, T. Microbiome and colorectal cancer: Roles in carcinogenesis and clinical potential. Mol. Asp. Med. 2019, 69, 93–106. [Google Scholar] [CrossRef] [PubMed]
- Knippel, R.J.; Sears, C.L. The Microbiome Colorectal Cancer Puzzle: Initiator, Propagator, and Avenue for Treatment and Research. J. Natl. Compr. Cancer Netw. 2021, 19, 986–992. [Google Scholar] [CrossRef] [PubMed]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef]
- Brennan, C.A.; Garrett, W.S. Fusobacterium nucleatum-symbiont, opportunist and oncobacterium. Nat. Rev. Microbiol. 2019, 17, 156–166. [Google Scholar] [CrossRef]
- Koliarakis, I.; Messaritakis, I.; Nikolouzakis, T.K.; Hamilos, G.; Souglakos, J.; Tsiaoussis, J. Oral Bacteria and Intestinal Dysbiosis in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4146. [Google Scholar] [CrossRef]
- Zhang, Z.; Aung, K.M.; Uhlin, B.E.; Wai, S.N. Reversible senescence of human colon cancer cells after blockage of mitosis/cytokinesis caused by the CNF1 cyclomodulin from Escherichia coli. Sci. Rep. 2018, 8, 17780. [Google Scholar] [CrossRef] [PubMed]
- Sears, C.L.; Geis, A.L.; Housseau, F. Bacteroides fragilis subverts mucosal biology: From symbiont to colon carcinogenesis. J. Clin. Investig. 2014, 124, 4166–4172. [Google Scholar] [CrossRef]
- DeGruttola, A.K.; Low, D.; Mizoguchi, A.; Mizoguchi, E. Current Understanding of Dysbiosis in Disease in Human and Animal Models. Inflamm. Bowel Dis. 2016, 22, 1137–1150. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Capurso, G.; Lahner, E. The interaction between smoking, alcohol and the gut microbiome. Best Pract. Research. Clin. Gastroenterol. 2017, 31, 579–588. [Google Scholar] [CrossRef] [PubMed]
- Emlet, C.; Ruffin, M.; Lamendella, R. Enteric Virome and Carcinogenesis in the Gut. Dig. Dis. Sci. 2020, 65, 852–864. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, G.; Zhou, H.; Wu, W.K.K.; Wong, S.H.; Coker, O.O.; Dai, Z.; Li, X.; Szeto, C.H.; Sugimura, N.; Lam, T.Y.; et al. Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology 2018, 155, 529–541.e5. [Google Scholar] [CrossRef] [PubMed]
- Coker, O.O.; Nakatsu, G.; Dai, R.Z.; Wu, W.K.K.; Wong, S.H.; Ng, S.C.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019, 68, 654–662. [Google Scholar] [CrossRef]
- Mola, S.; Pandolfo, C.; Sica, A.; Porta, C. The Macrophages-Microbiota Interplay in Colorectal Cancer (CRC)-Related Inflammation: Prognostic and Therapeutic Significance. Int. J. Mol. Sci. 2020, 21, 6866. [Google Scholar] [CrossRef]
- Meyer, J.E.; Narang, T.; Schnoll-Sussman, F.H.; Pochapin, M.B.; Christos, P.J.; Sherr, D.L. Increasing incidence of rectal cancer in patients aged younger than 40 years: An analysis of the surveillance, epidemiology, and end results database. Cancer 2010, 116, 4354–4359. [Google Scholar] [CrossRef]
- Cercek, A.; Chatila, W.K.; Yaeger, R.; Walch, H.; Fernandes, G.D.S.; Krishnan, A.; Palmaira, L.; Maio, A.; Kemel, Y.; Srinivasan, P.; et al. A Comprehensive Comparison of Early-Onset and Average-Onset Colorectal Cancers. J. Natl. Cancer Inst. 2021, 113, 1683–1692. [Google Scholar] [CrossRef]
- Chuang, L.C.; Chen, H.C.; You, S.L.; Lin, C.Y.; Pan, M.H.; Chou, Y.C.; Hsieh, C.Y.; Chen, C.J. Association between human papillomavirus and adenocarcinoma of rectum and recto-sigmoid junction: A cohort study of 10,612 women in Taiwan. Cancer Causes Control. 2010, 21, 2123–2128. [Google Scholar] [CrossRef]
- Marongiu, L.; Allgayer, H. Viruses in colorectal cancer. Mol. Oncol. 2022, 16, 1423–1450. [Google Scholar] [CrossRef]
- Oleynikova, N.A.; Danilova, N.V.; Grimuta, M.O.; Malkov, P.G. Epstein-Barr Virus in the Development of Colorectal Cancer (Review). Sovrem. Tekhnologii V Meditsine 2021, 13, 82–91. [Google Scholar] [CrossRef] [PubMed]
- Gupta, I.; Al Farsi, H.; Jabeen, A.; Skenderi, F.; Al-Thawadi, H.; AlAhmad, Y.M.; Al Moustafa, A.E.; Vranic, S. High-Risk Human Papillomaviruses and Epstein-Barr Virus in Colorectal Cancer and Their Association with Clinicopathological Status. Pathogens 2020, 9, 452. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, J.M. Nutrition and gut health: The impact of specific dietary components-it’s not just five-a-day. Proc. Nutr. Soc. 2021, 80, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Nagahashi, M.; Kim, E.Y.; Harikumar, K.B.; Yamada, A.; Huang, W.C.; Hait, N.C.; Allegood, J.C.; Price, M.M.; Avni, D.; et al. Sphingosine-1-phosphate links persistent STAT3 activation, chronic intestinal inflammation, and development of colitis-associated cancer. Cancer Cell 2013, 23, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Zeng, H.; Umar, S.; Rust, B.; Lazarova, D.; Bordonaro, M. Secondary Bile Acids and Short Chain Fatty Acids in the Colon: A Focus on Colonic Microbiome, Cell Proliferation, Inflammation, and Cancer. Int. J. Mol. Sci. 2019, 20, 1214. [Google Scholar] [CrossRef] [PubMed]
- Santhanam, S.; Alvarado, D.M.; Ciorba, M.A. Therapeutic targeting of inflammation and tryptophan metabolism in colon and gastrointestinal cancer. Transl. Res. J. Lab. Clin. Med. 2016, 167, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450. [Google Scholar] [CrossRef]
- McGarr, S.E.; Ridlon, J.M.; Hylemon, P.B. Diet, anaerobic bacterial metabolism, and colon cancer: A review of the literature. J. Clin. Gastroenterol. 2005, 39, 98–109. [Google Scholar] [PubMed]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed]
- O’Keefe, S.J.; Li, J.V.; Lahti, L.; Ou, J.; Carbonero, F.; Mohammed, K.; Posma, J.M.; Kinross, J.; Wahl, E.; Ruder, E.; et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015, 6, 6342. [Google Scholar] [CrossRef] [PubMed]
- Nagahashi, M.; Yuza, K.; Hirose, Y.; Nakajima, M.; Ramanathan, R.; Hait, N.C.; Hylemon, P.B.; Zhou, H.; Takabe, K.; Wakai, T. The roles of bile acids and sphingosine-1-phosphate signaling in the hepatobiliary diseases. J. Lipid Res. 2016, 57, 1636–1643. [Google Scholar] [CrossRef]
- Degirolamo, C.; Modica, S.; Palasciano, G.; Moschetta, A. Bile acids and colon cancer: Solving the puzzle with nuclear receptors. Trends Mol. Med. 2011, 17, 564–572. [Google Scholar] [CrossRef]
- Wu, S.; Rhee, K.J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F.; et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Montalban-Arques, A.; Scharl, M. Intestinal microbiota and colorectal carcinoma: Implications for pathogenesis, diagnosis, and therapy. EBioMedicine 2019, 48, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, A.C.; Destefano Shields, C.E.; Wu, S.; Huso, D.L.; Wu, X.; Murray-Stewart, T.R.; Hacker-Prietz, A.; Rabizadeh, S.; Woster, P.M.; Sears, C.L.; et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl. Acad. Sci. USA 2011, 108, 15354–15359. [Google Scholar] [CrossRef]
- Veziant, J.; Villéger, R.; Barnich, N.; Bonnet, M. Gut Microbiota as Potential Biomarker and/or Therapeutic Target to Improve the Management of Cancer: Focus on Colibactin-Producing Escherichia coli in Colorectal Cancer. Cancers 2021, 13, 2215. [Google Scholar] [CrossRef] [PubMed]
- Strakova, N.; Korena, K.; Karpiskova, R. Klebsiella pneumoniae producing bacterial toxin colibactin as a risk of colorectal cancer development-A systematic review. Toxicon Off. J. Int. Soc. Toxinology 2021, 197, 126–135. [Google Scholar] [CrossRef] [PubMed]
- Arthur, J.C.; Perez-Chanona, E.; Mühlbauer, M.; Tomkovich, S.; Uronis, J.M.; Fan, T.J.; Campbell, B.J.; Abujamel, T.; Dogan, B.; Rogers, A.B.; et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 2012, 338, 120–123. [Google Scholar] [CrossRef]
- Dejea, C.M.; Wick, E.C.; Hechenbleikner, E.M.; White, J.R.; Mark Welch, J.L.; Rossetti, B.J.; Peterson, S.N.; Snesrud, E.C.; Borisy, G.G.; Lazarev, M.; et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 18321–18326. [Google Scholar] [CrossRef]
- Dejea, C.M.; Fathi, P.; Craig, J.M.; Boleij, A.; Taddese, R.; Geis, A.L.; Wu, X.; DeStefano Shields, C.E.; Hechenbleikner, E.M.; Huso, D.L.; et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018, 359, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Tomkovich, S.; Dejea, C.M.; Winglee, K.; Drewes, J.L.; Chung, L.; Housseau, F.; Pope, J.L.; Gauthier, J.; Sun, X.; Mühlbauer, M.; et al. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J. Clin. Investig. 2019, 129, 1699–1712. [Google Scholar] [CrossRef] [PubMed]
- Arneth, B. Tumor Microenvironment. Medicina 2019, 56, 15. [Google Scholar] [CrossRef] [PubMed]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. CB 2020, 30, R921–R925. [Google Scholar] [CrossRef]
- Roma-Rodrigues, C.; Mendes, R.; Baptista, P.V.; Fernandes, A.R. Targeting Tumor Microenvironment for Cancer Therapy. Int. J. Mol. Sci. 2019, 20, 840. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Sarkar, J.; Wu, R.; Tokumaru, Y.; Yan, L.; Nakagawa, K.; Ishibe, A.; Matsuyama, R.; Endo, I.; Takabe, K. Intratumoral density of regulatory T cells is a predictor of host immune response and chemotherapy response in colorectal cancer. Am. J. Cancer Res. 2022, 12, 490–503. [Google Scholar]
- Oshi, M.; Huyser, M.R.; Le, L.; Tokumaru, Y.; Yan, L.; Matsuyama, R.; Endo, I.; Takabe, K. Abundance of Microvascular Endothelial Cells Is Associated with Response to Chemotherapy and Prognosis in Colorectal Cancer. Cancers 2021, 13, 1477. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef]
- Oshi, M.; Tokumaru, Y.; Asaoka, M.; Yan, L.; Satyananda, V.; Matsuyama, R.; Matsuhashi, N.; Futamura, M.; Ishikawa, T.; Yoshida, K.; et al. M1 Macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci. Rep. 2020, 10, 16554. [Google Scholar] [CrossRef] [PubMed]
- Mills, C.D.; Kincaid, K.; Alt, J.M.; Heilman, M.J.; Hill, A.M. M-1/M-2 macrophages and the Th1/Th2 paradigm. J. Immunol. 2000, 164, 6166–6173. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef]
- Väyrynen, J.P.; Haruki, K.; Lau, M.C.; Väyrynen, S.A.; Zhong, R.; Dias Costa, A.; Borowsky, J.; Zhao, M.; Fujiyoshi, K.; Arima, K.; et al. The Prognostic Role of Macrophage Polarization in the Colorectal Cancer Microenvironment. Cancer Immunol. Res. 2021, 9, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S.; Kawaguchi, T.; Peng, X.; Qi, Q.; Liu, S.; Yan, L.; Takabe, K. Tumor Infiltrating Lymphocytes and Macrophages Improve Survival in Microsatellite Unstable Colorectal Cancer. Sci. Rep. 2019, 9, 13455. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S.; Kawaguchi, T.; Yan, L.; Peng, X.; Qi, Q.; Takabe, K. Cytolytic Activity Score to Assess Anticancer Immunity in Colorectal Cancer. Ann. Surg. Oncol. 2018, 25, 2323–2331. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhang, S. Tumor-Associated Macrophages and Their Functional Transformation in the Hypoxic Tumor Microenvironment. Front. Immunol. 2021, 12, 741305. [Google Scholar] [CrossRef] [PubMed]
- Edin, S.; Wikberg, M.L.; Dahlin, A.M.; Rutegård, J.; Öberg, Å.; Oldenborg, P.A.; Palmqvist, R. The distribution of macrophages with a M1 or M2 phenotype in relation to prognosis and the molecular characteristics of colorectal cancer. PLoS ONE 2012, 7, e47045. [Google Scholar] [CrossRef] [PubMed]
- Rhee, I. Diverse macrophages polarization in tumor microenvironment. Arch. Pharm. Res. 2016, 39, 1588–1596. [Google Scholar] [CrossRef]
- Kikuchi, T.; Mimura, K.; Ashizawa, M.; Okayama, H.; Endo, E.; Saito, K.; Sakamoto, W.; Fujita, S.; Endo, H.; Saito, M.; et al. Characterization of tumor-infiltrating immune cells in relation to microbiota in colorectal cancers. Cancer Immunol. Immunother. CII 2020, 69, 23–32. [Google Scholar] [CrossRef]
- Kim, J.; Bae, J.S. Tumor-Associated Macrophages and Neutrophils in Tumor Microenvironment. Mediators Inflamm. 2016, 2016, 6058147. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, R.; Kawada, K.; Itatani, Y.; Ogawa, R.; Kiyasu, Y.; Sakai, Y. The Role of Tumor-Associated Neutrophils in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 529. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, L.; Li, X.; Zhuo, W. Neutrophil Extracellular Traps in Tumor Metastasis: Pathological Functions and Clinical Applications. Cancers 2021, 13, 2832. [Google Scholar] [CrossRef]
- Ronchetti, L.; Boubaker, N.S.; Barba, M.; Vici, P.; Gurtner, A.; Piaggio, G. Neutrophil extracellular traps in cancer: Not only catching microbes. J. Exp. Clin. Cancer Res. CR 2021, 40, 231. [Google Scholar] [CrossRef] [PubMed]
- Albrengues, J.; Shields, M.A.; Ng, D.; Park, C.G.; Ambrico, A.; Poindexter, M.E.; Upadhyay, P.; Uyeminami, D.L.; Pommier, A.; Küttner, V.; et al. Neutrophil extracellular traps produced during inflammation awaken dormant cancer cells in mice. Science 2018, 361, eaao4227. [Google Scholar] [CrossRef] [PubMed]
- Efrimescu, C.I.; Buggy, P.M.; Buggy, D.J. Neutrophil Extracellular Trapping Role in Cancer, Metastases, and Cancer-Related Thrombosis: A Narrative Review of the Current Evidence Base. Curr. Oncol. Rep. 2021, 23, 118. [Google Scholar] [CrossRef] [PubMed]
- Governa, V.; Trella, E.; Mele, V.; Tornillo, L.; Amicarella, F.; Cremonesi, E.; Muraro, M.G.; Xu, H.; Droeser, R.; Däster, S.R.; et al. The Interplay Between Neutrophils and CD8(+) T Cells Improves Survival in Human Colorectal Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 3847–3858. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Hu, P.; Donskov, F.; Wang, G.; Liu, Q.; Du, J. Tumor-associated neutrophils as a new prognostic factor in cancer: A systematic review and meta-analysis. PLoS ONE 2014, 9, e98259. [Google Scholar] [CrossRef] [PubMed]
- Yan, B.; Dai, X.; Ma, Q.; Wu, X. Stromal Neutrophil Extracellular Trap Density Is an Independent Prognostic Factor for Cervical Cancer Recurrence. Front. Oncol. 2021, 11, 659445. [Google Scholar] [CrossRef] [PubMed]
- Kondĕlková, K.; Vokurková, D.; Krejsek, J.; Borská, L.; Fiala, Z.; Ctirad, A. Regulatory T cells (TREG) and their roles in immune system with respect to immunopathological disorders. Acta Med. 2010, 53, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Waniczek, D.; Lorenc, Z.; Śnietura, M.; Wesecki, M.; Kopec, A.; Muc-Wierzgoń, M. Tumor-Associated Macrophages and Regulatory T Cells Infiltration and the Clinical Outcome in Colorectal Cancer. Arch. Immunol. Ther. Exp. 2017, 65, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhang, T.; Kang, Z.; Guo, G.; Sun, Y.; Lin, K.; Huang, Q.; Shi, X.; Ni, Z.; Ding, N.; et al. Tumor-Infiltrating Immune Cells Act as a Marker for Prognosis in Colorectal Cancer. Front. Immunol. 2019, 10, 2368. [Google Scholar] [CrossRef] [PubMed]
- Sarrabayrouse, G.; Alameddine, J.; Altare, F.; Jotereau, F. Microbiota-Specific CD4CD8αα Tregs: Role in Intestinal Immune Homeostasis and Implications for IBD. Front. Immunol. 2015, 6, 522. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Siles, M.; Duncan, S.H.; Garcia-Gil, L.J.; Martinez-Medina, M. Faecalibacterium prausnitzii: From microbiology to diagnostics and prognostics. ISME J. 2017, 11, 841–852. [Google Scholar] [CrossRef]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Katsuta, E.; Rashid, O.M.; Takabe, K. Fibroblasts as a Biological Marker for Curative Resection in Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 3890. [Google Scholar] [CrossRef]
- Martinez-Outschoorn, U.E.; Lisanti, M.P.; Sotgia, F. Catabolic cancer-associated fibroblasts transfer energy and biomass to anabolic cancer cells, fueling tumor growth. Semin. Cancer Biol. 2014, 25, 47–60. [Google Scholar] [CrossRef]
- Érsek, B.; Silló, P.; Cakir, U.; Molnár, V.; Bencsik, A.; Mayer, B.; Mezey, E.; Kárpáti, S.; Pós, Z.; Németh, K. Melanoma-associated fibroblasts impair CD8+ T cell function and modify expression of immune checkpoint regulators via increased arginase activity. Cell. Mol. Life Sci. CMLS 2021, 78, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.; He, S. Multi-faceted role of cancer-associated adipocytes in the tumor microenvironment (Review). Mol. Med. Rep. 2021, 24, 866. [Google Scholar] [CrossRef]
- Schwartz, B.; Yehuda-Shnaidman, E. Putative role of adipose tissue in growth and metabolism of colon cancer cells. Front. Oncol. 2014, 4, 164. [Google Scholar] [CrossRef] [PubMed]
- Himbert, C.; Delphan, M.; Scherer, D.; Bowers, L.W.; Hursting, S.; Ulrich, C.M. Signals from the Adipose Microenvironment and the Obesity-Cancer Link-A Systematic Review. Cancer Prev. Res. 2017, 10, 494–506. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Oshi, M.; Katsuta, E.; Yan, L.; Huang, J.L.; Nagahashi, M.; Matsuhashi, N.; Futamura, M.; Yoshida, K.; Takabe, K. Intratumoral Adipocyte-High Breast Cancer Enrich for Metastatic and Inflammation-Related Pathways but Associated with Less Cancer Cell Proliferation. Int. J. Mol. Sci. 2020, 21, 5744. [Google Scholar] [CrossRef] [PubMed]
- Sabharwal, S.S.; Schumacker, P.T. Mitochondrial ROS in cancer: Initiators, amplifiers or an Achilles’ heel? Nat. Rev. Cancer 2014, 14, 709–721. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, L.B.; Gui, D.Y.; Vander Heiden, M.G. Altered metabolite levels in cancer: Implications for tumour biology and cancer therapy. Nat. Rev. Cancer 2016, 16, 680–693. [Google Scholar] [CrossRef] [PubMed]
- Izzo, V.; Bravo-San Pedro, J.M.; Sica, V.; Kroemer, G.; Galluzzi, L. Mitochondrial Permeability Transition: New Findings and Persisting Uncertainties. Trends Cell Biol. 2016, 26, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Rai, N.K.; Mathur, S.; Singh, S.K.; Tiwari, M.; Singh, V.K.; Haque, R.; Tiwari, S.; Kumar Sharma, L. Differential regulation of mitochondrial complex I and oxidative stress based on metastatic potential of colorectal cancer cells. Oncol. Lett. 2020, 20, 313. [Google Scholar] [CrossRef]
- Al Tameemi, W.; Dale, T.P.; Al-Jumaily, R.M.K.; Forsyth, N.R. Hypoxia-Modified Cancer Cell Metabolism. Front. Cell Dev. Biol. 2019, 7, 4. [Google Scholar] [CrossRef]
- Jackson, D.N.; Theiss, A.L. Gut bacteria signaling to mitochondria in intestinal inflammation and cancer. Gut Microbes 2020, 11, 285–304. [Google Scholar] [CrossRef]
- Strickertsson, J.A.B.; Desler, C.; Rasmussen, L.J. Bacterial infection increases risk of carcinogenesis by targeting mitochondria. Semin. Cancer Biol. 2017, 47, 95–100. [Google Scholar] [CrossRef]
- Tokumaru, Y.; Oshi, M.; Patel, A.; Tian, W.; Yan, L.; Matsuhashi, N.; Futamura, M.; Yoshida, K.; Takabe, K. Organoids Are Limited in Modeling the Colon Adenoma-Carcinoma Sequence. Cells 2021, 10, 488. [Google Scholar] [CrossRef] [PubMed]
- Aprile, F.; Bruno, G.; Palma, R.; Mascellino, M.T.; Panetta, C.; Scalese, G.; Oliva, A.; Severi, C.; Pontone, S. Microbiota Alterations in Precancerous Colon Lesions: A Systematic Review. Cancers 2021, 13, 3061. [Google Scholar] [CrossRef] [PubMed]
- Rezasoltani, S.; Asadzadeh Aghdaei, H.; Dabiri, H.; Akhavan Sepahi, A.; Modarressi, M.H.; Nazemalhosseini Mojarad, E. The association between fecal microbiota and different types of colorectal polyp as precursors of colorectal cancer. Microb. Pathog. 2018, 124, 244–249. [Google Scholar] [CrossRef]
- Liang, S.; Mao, Y.; Liao, M.; Xu, Y.; Chen, Y.; Huang, X.; Wei, C.; Wu, C.; Wang, Q.; Pan, X.; et al. Gut microbiome associated with APC gene mutation in patients with intestinal adenomatous polyps. Int. J. Biol. Sci. 2020, 16, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Koido, S.; Odamaki, T.; Kajihara, M.; Kato, K.; Horiuchi, S.; Adachi, S.; Arakawa, H.; Yoshida, S.; Akasu, T.; et al. Metagenomic analyses of the gut microbiota associated with colorectal adenoma. PLoS ONE 2019, 14, e0212406. [Google Scholar] [CrossRef] [PubMed]
- McCoy, A.N.; Araújo-Pérez, F.; Azcárate-Peril, A.; Yeh, J.J.; Sandler, R.S.; Keku, T.O. Fusobacterium is associated with colorectal adenomas. PLoS ONE 2013, 8, e53653. [Google Scholar] [CrossRef]
- Yachida, S.; Mizutani, S.; Shiroma, H.; Shiba, S.; Nakajima, T.; Sakamoto, T.; Watanabe, H.; Masuda, K.; Nishimoto, Y.; Kubo, M.; et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019, 25, 968–976. [Google Scholar] [CrossRef]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef]
- Yan, Y.; Drew, D.A.; Markowitz, A.; Lloyd-Price, J.; Abu-Ali, G.; Nguyen, L.H.; Tran, C.; Chung, D.C.; Gilpin, K.K.; Meixell, D.; et al. Structure of the Mucosal and Stool Microbiome in Lynch Syndrome. Cell Host Microbe 2020, 27, 585–600.e4. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.-J.; Kang, Q.; Jin, P.; Sheng, J.-q. The Interactions Between Gut Microbiota and Lynch Syndrome. Clin. Gastroenterol. Hepatol. 2017, 15, e48. [Google Scholar] [CrossRef]
- Mori, G.; Orena, B.S.; Cultrera, I.; Barbieri, G.; Albertini, A.M.; Ranzani, G.N.; Carnevali, I.; Tibiletti, M.G.; Pasca, M.R. Gut Microbiota Analysis in Postoperative Lynch Syndrome Patients. Front. Microbiol. 2019, 10, 1746. [Google Scholar] [CrossRef] [PubMed]
- Mori, G.; Pasca, M.R. Gut Microbial Signatures in Sporadic and Hereditary Colorectal Cancer. Int. J. Mol. Sci. 2021, 22, 1312. [Google Scholar] [CrossRef]
- Hofseth, L.J.; Hebert, J.R.; Chanda, A.; Chen, H.; Love, B.L.; Pena, M.M.; Murphy, E.A.; Sajish, M.; Sheth, A.; Buckhaults, P.J.; et al. Early-onset colorectal cancer: Initial clues and current views. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, M.; Sukartini, N.; Nursyirwan, S.A.; Pribadi, R.R.; Maulahela, H.; Utari, A.P.; Muzellina, V.N.; Wiraatmadja, A.; Renaldi, K. Gut Microbiota Profiles in Early- and Late-Onset Colorectal Cancer: A Potential Diagnostic Biomarker in the Future. Digestion 2021, 102, 823–832. [Google Scholar] [CrossRef]
- Yang, Y.; Du, L.; Shi, D.; Kong, C.; Liu, J.; Liu, G.; Li, X.; Ma, Y. Dysbiosis of human gut microbiome in young-onset colorectal cancer. Nat. Commun. 2021, 12, 6757. [Google Scholar] [CrossRef]
- Akimoto, N.; Ugai, T.; Zhong, R.; Hamada, T.; Fujiyoshi, K.; Giannakis, M.; Wu, K.; Cao, Y.; Ng, K.; Ogino, S. Rising incidence of early-onset colorectal cancer-a call to action. Nat. Rev. Clin. Oncol. 2021, 18, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Mukherji, R.; Weinberg, B.A. The gut microbiome and potential implications for early-onset colorectal cancer. Color. Cancer 2020, 9, CRC25. [Google Scholar] [CrossRef]
- Mima, K.; Nishihara, R.; Qian, Z.R.; Cao, Y.; Sukawa, Y.; Nowak, J.A.; Yang, J.; Dou, R.; Masugi, Y.; Song, M.; et al. Fusobacterium nucleatum in colorectal carcinoma tissue and patient prognosis. Gut 2016, 65, 1973–1980. [Google Scholar] [CrossRef]
- Archambault, A.N.; Jeon, J.; Lin, Y.; Thomas, M.; Harrison, T.A.; Bishop, D.T.; Brenner, H.; Casey, G.; Chan, A.T.; Chang-Claude, J.; et al. Risk Stratification for Early-Onset Colorectal Cancer Using a Combination of Genetic and Environmental Risk Scores: An International Multi-Center Study. J. Natl. Cancer Inst. 2022, 114, 528–539. [Google Scholar] [CrossRef]
- Vivarelli, S.; Salemi, R.; Candido, S.; Falzone, L.; Santagati, M.; Stefani, S.; Torino, F.; Banna, G.L.; Tonini, G.; Libra, M. Gut Microbiota and Cancer: From Pathogenesis to Therapy. Cancers 2019, 11, 38. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Mao, Q.; Xia, W.; Dong, G.; Yu, C.; Jiang, F. Gut Microbiota Shapes the Efficiency of Cancer Therapy. Front. Microbiol. 2019, 10, 1050. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Guo, F.; Yu, Y.; Sun, T.; Ma, D.; Han, J.; Qian, Y.; Kryczek, I.; Sun, D.; Nagarsheth, N.; et al. Fusobacterium nucleatum Promotes Chemoresistance to Colorectal Cancer by Modulating Autophagy. Cell 2017, 170, 548–563.e16. [Google Scholar] [CrossRef]
- Song, J.; Qu, Z.; Guo, X.; Zhao, Q.; Zhao, X.; Gao, L.; Sun, K.; Shen, F.; Wu, M.; Wei, L. Hypoxia-induced autophagy contributes to the chemoresistance of hepatocellular carcinoma cells. Autophagy 2009, 5, 1131–1144. [Google Scholar] [CrossRef] [PubMed]
- Roberti, M.P.; Yonekura, S.; Duong, C.P.M.; Picard, M.; Ferrere, G.; Tidjani Alou, M.; Rauber, C.; Iebba, V.; Lehmann, C.H.K.; Amon, L.; et al. Chemotherapy-induced ileal crypt apoptosis and the ileal microbiome shape immunosurveillance and prognosis of proximal colon cancer. Nat. Med. 2020, 26, 919–931. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef] [PubMed]
- Brandi, G.; Dabard, J.; Raibaud, P.; Di Battista, M.; Bridonneau, C.; Pisi, A.M.; Morselli Labate, A.M.; Pantaleo, M.A.; De Vivo, A.; Biasco, G. Intestinal microflora and digestive toxicity of irinotecan in mice. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2006, 12, 1299–12307. [Google Scholar] [CrossRef] [PubMed]
- Harb, A.H.; Abou Fadel, C.; Sharara, A.I. Radiation enteritis. Curr. Gastroenterol. Rep. 2014, 16, 383. [Google Scholar] [CrossRef]
- Crawford, P.A.; Gordon, J.I. Microbial regulation of intestinal radiosensitivity. Proc. Natl. Acad. Sci. USA 2005, 102, 13254–13259. [Google Scholar] [CrossRef] [PubMed]
- González-Mercado, V.J.; Henderson, W.A.; Sarkar, A.; Lim, J.; Saligan, L.N.; Berk, L.; Dishaw, L.; McMillan, S.; Groer, M.; Sepehri, F.; et al. Changes in Gut Microbiome Associated With Co-Occurring Symptoms Development During Chemo-Radiation for Rectal Cancer: A Proof of Concept Study. Biol. Res. Nurs. 2021, 23, 31–41. [Google Scholar] [CrossRef]
- Manichanh, C.; Varela, E.; Martinez, C.; Antolin, M.; Llopis, M.; Doré, J.; Giralt, J.; Guarner, F.; Malagelada, J.R. The gut microbiota predispose to the pathophysiology of acute postradiotherapy diarrhea. Am. J. Gastroenterol. 2008, 103, 1754–1761. [Google Scholar] [CrossRef]
- Egan, L.J.; Eckmann, L.; Greten, F.R.; Chae, S.; Li, Z.W.; Myhre, G.M.; Robine, S.; Karin, M.; Kagnoff, M.F. IkappaB-kinasebeta-dependent NF-kappaB activation provides radioprotection to the intestinal epithelium. Proc. Natl. Acad. Sci. USA 2004, 101, 2452–2457. [Google Scholar] [CrossRef] [PubMed]
- Packey, C.D.; Ciorba, M.A. Microbial influences on the small intestinal response to radiation injury. Curr. Opin. Gastroenterol. 2010, 26, 88–94. [Google Scholar] [CrossRef] [PubMed]
- Pepoyan, A.Z.; Manvelyan, A.M.; Balayan, M.H.; McCabe, G.; Tsaturyan, V.V.; Melnikov, V.G.; Chikindas, M.L.; Weeks, R.; Karlyshev, A.V. The Effectiveness of Potential Probiotics Lactobacillus rhamnosus Vahe and Lactobacillus delbrueckii IAHAHI in Irradiated Rats Depends on the Nutritional Stage of the Host. Probiotics Antimicrob. Proteins 2020, 12, 1439–1450. [Google Scholar] [CrossRef] [PubMed]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Sumransub, N.; Vantanasiri, K.; Prakash, A.; Lou, E. Advances and new frontiers for immunotherapy in colorectal cancer: Setting the stage for neoadjuvant success? Mol. Ther. Oncolytics 2021, 22, 1–12. [Google Scholar] [CrossRef]
- Cercek, A.; Lumish, M.; Sinopoli, J.; Weiss, J.; Shia, J.; Lamendola-Essel, M.; El Dika, I.H.; Segal, N.; Shcherba, M.; Sugarman, R.; et al. PD-1 Blockade in Mismatch Repair-Deficient, Locally Advanced Rectal Cancer. N. Engl. J. Med. 2022, 386, 2363–2376. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Pu, Y.; Cron, K.; Deng, L.; Kline, J.; Frazier, W.A.; Xu, H.; Peng, H.; Fu, Y.X.; Xu, M.M. CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nat. Med. 2015, 21, 1209–1215. [Google Scholar] [CrossRef]
- Shi, Y.; Zheng, W.; Yang, K.; Harris, K.G.; Ni, K.; Xue, L.; Lin, W.; Chang, E.B.; Weichselbaum, R.R.; Fu, Y.X. Intratumoral accumulation of gut microbiota facilitates CD47-based immunotherapy via STING signaling. J. Exp. Med. 2020, 217, e20192282. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Mei, J.; Yi, S.; Feng, C.; Ma, Y.; Liu, Y.; Liu, Y.; Chen, C. Tumor associated macrophage and microbe: The potential targets of tumor vaccine delivery. Adv. Drug Deliv. Rev. 2021, 180, 114046. [Google Scholar] [CrossRef]
- Shahnazari, M.; Samadi, P.; Pourjafar, M.; Jalali, A. Therapeutic vaccines for colorectal cancer: The progress and future prospect. Int. Immunopharmacol. 2020, 88, 106944. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.J.; Rifkin, R.F.; Cowan, D.A.; Potgieter, M. The Healthy Human Blood Microbiome: Fact or Fiction? Front. Cell. Infect. Microbiol. 2019, 9, 148. [Google Scholar] [CrossRef]
- Yang, D.; Wang, X.; Zhou, X.; Zhao, J.; Yang, H.; Wang, S.; Morse, M.A.; Wu, J.; Yuan, Y.; Li, S.; et al. Blood microbiota diversity determines response of advanced colorectal cancer to chemotherapy combined with adoptive T cell immunotherapy. Oncoimmunology 2021, 10, 1976953. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.C.S.; Chia, M.; Ko, K.K.K.; Chen, H.; Liu, J.; Loh, M.; Nagarajan, N. No evidence for a common blood microbiome based on a population study of 9,770 healthy humans. bioRxiv 2022. [Google Scholar] [CrossRef]
- Coukos, G. Neoadjuvant immune-checkpoint blockade in resectable colon cancer. Nat. Med. 2020, 26, 473–474. [Google Scholar] [CrossRef] [PubMed]
- Chalabi, M.; Fanchi, L.F.; Dijkstra, K.K.; van den Berg, J.G.; Aalbers, A.G.; Sikorska, K.; Lopez-Yurda, M.; Grootscholten, C.; Beets, G.L.; Snaebjornsson, P.; et al. Neoadjuvant immunotherapy leads to pathological responses in MMR-proficient and MMR-deficient early-stage colon cancers. Nat. Med. 2020, 26, 566–576. [Google Scholar] [CrossRef] [PubMed]
- Chalabi, M.; Verschoor, Y.L.; van den Berg, J.; Sikorska, K.; Beets, G.; Lent, A.V.; Grootscholten, M.C.; Aalbers, A.; Buller, N.; Marsman, H.; et al. LBA7 Neoadjuvant immune checkpoint inhibition in locally advanced MMR-deficient colon cancer: The NICHE-2 study. Ann. Oncol. 2022, 33, S1389. [Google Scholar] [CrossRef]
- Zhang, Z.; Cheng, S.; Gong, J.; Lu, M.; Zhou, J.; Zhang, X.; Li, J.; Shen, L.; Peng, Z. Efficacy and safety of neoadjuvant immunotherapy in patients with microsatellite instability-high gastrointestinal malignancies: A case series. Eur. J. Surg. Oncol. J. Eur. Soc. Surg. Oncol. Br. Assoc. Surg. Oncol. 2020, 46, e33–e39. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Nowak, J.A.; Hamada, T.; Milner, D.A., Jr.; Nishihara, R. Insights into Pathogenic Interactions Among Environment, Host, and Tumor at the Crossroads of Molecular Pathology and Epidemiology. Annu. Rev. Pathol. 2019, 14, 83–103. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Dai, X.; Zhou, C.C.; Li, K.X.; Zhang, Y.J.; Lou, X.Y.; Zhu, Y.M.; Sun, Y.L.; Peng, B.X.; Cui, W. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut 2022, 71, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Li, S.; Gan, R.Y.; Zhao, C.N.; Meng, X.; Li, H.B. Targeting gut microbiota with dietary components on cancer: Effects and potential mechanisms of action. Crit. Rev. Food Sci. Nutr. 2020, 60, 1025–1037. [Google Scholar] [CrossRef] [PubMed]
- Malla, R.R. Microbiome Conundrum in Colon Cancer: Development, Progression, and Therapeutics. Crit. Rev. Oncog. 2020, 25, 129–139. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.S.; Nishihara, R.; Cao, Y.; Song, M.; Mima, K.; Qian, Z.R.; Nowak, J.A.; Kosumi, K.; Hamada, T.; Masugi, Y.; et al. Association of Dietary Patterns With Risk of Colorectal Cancer Subtypes Classified by Fusobacterium nucleatum in Tumor Tissue. JAMA Oncol. 2017, 3, 921–927. [Google Scholar] [CrossRef]
- Arima, K.; Zhong, R.; Ugai, T.; Zhao, M.; Haruki, K.; Akimoto, N.; Lau, M.C.; Okadome, K.; Mehta, R.S.; Väyrynen, J.P.; et al. Western-Style Diet, pks Island-Carrying Escherichia coli, and Colorectal Cancer: Analyses From Two Large Prospective Cohort Studies. Gastroenterology 2022, 163, 862–874. [Google Scholar] [CrossRef]
- O’Keefe, S.J. Diet, microorganisms and their metabolites, and colon cancer. Nat. Reviews. Gastroenterol. Hepatol. 2016, 13, 691–706. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Gao, C.; Hou, J.; Gu, J.; Law, B.Y.K.; Xu, Y. Non-Digestible Carbohydrate and the Risk of Colorectal Neoplasia: A Systematic Review. Nutr. Cancer 2021, 73, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Hullings, A.G.; Sinha, R.; Liao, L.M.; Freedman, N.D.; Graubard, B.I.; Loftfield, E. Whole grain and dietary fiber intake and risk of colorectal cancer in the NIH-AARP Diet and Health Study cohort. Am. J. Clin. Nutr. 2020, 112, 603–612. [Google Scholar] [CrossRef]
- Aykan, N.F. Red Meat and Colorectal Cancer. Oncol. Rev. 2015, 9, 288. [Google Scholar] [CrossRef]
- Wang, F.; Chandler, P.D.; Zeleznik, O.A.; Wu, K.; Wu, Y.; Yin, K.; Song, R.; Avila-Pacheco, J.; Clish, C.B.; Meyerhardt, J.A.; et al. Plasma Metabolite Profiles of Red Meat, Poultry, and Fish Consumption, and Their Associations with Colorectal Cancer Risk. Nutrients 2022, 14, 978. [Google Scholar] [CrossRef]
- Abu-Ghazaleh, N.; Chua, W.J.; Gopalan, V. Intestinal microbiota and its association with colon cancer and red/processed meat consumption. J. Gastroenterol. Hepatol. 2021, 36, 75–88. [Google Scholar] [CrossRef] [PubMed]
- Malesza, I.J.; Malesza, M.; Walkowiak, J.; Mussin, N.; Walkowiak, D.; Aringazina, R.; Bartkowiak-Wieczorek, J.; Mądry, E. High-Fat, Western-Style Diet, Systemic Inflammation, and Gut Microbiota: A Narrative Review. Cells 2021, 10, 3164. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Sun, J.; Li, Z.; Yao, F.; Lin, K.; Jiao, X. Food intake and its effect on the species and abundance of intestinal flora in colorectal cancer and healthy individuals. Korean J. Intern. Med. 2021, 36, 568–583. [Google Scholar] [CrossRef] [PubMed]
- Caini, S.; Chioccioli, S.; Pastore, E.; Fontana, M.; Tortora, K.; Caderni, G.; Masala, G. Fish Consumption and Colorectal Cancer Risk: Meta-Analysis of Prospective Epidemiological Studies and Review of Evidence from Animal Studies. Cancers 2022, 14, 640. [Google Scholar] [CrossRef] [PubMed]
- Jafari Nasab, S.; Ghanavati, M.; Bahrami, A.; Rafiee, P.; Sadeghi, A.; Clark, C.C.T.; Sohrab, G.; Hoshyarrad, A.; Hejazi, E. Dietary nutrient patterns and the risk of colorectal cancer and colorectal adenomas: A case-control study. Eur. J. Cancer Prev. Off. J. Eur. Cancer Prev. Organ. 2021, 30, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Um, C.Y.; Prizment, A.; Hong, C.P.; Lazovich, D.; Bostick, R.M. Associations of Calcium, Vitamin D, and Dairy Product Intakes with Colorectal Cancer Risk among Older Women: The Iowa Women’s Health Study. Nutr. Cancer 2019, 71, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Barrubés, L.; Babio, N.; Mena-Sánchez, G.; Toledo, E.; Ramírez-Sabio, J.B.; Estruch, R.; Ros, E.; Fitó, M.; Arós, F.; Fiol, M.; et al. Dairy product consumption and risk of colorectal cancer in an older mediterranean population at high cardiovascular risk. Int. J. Cancer 2018, 143, 1356–1366. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Hur, J.; Wu, K.; Song, M.; Wang, M.; Smith-Warner, S.A.; Zhang, X.; Giovannucci, E.L. Total calcium, dairy foods and risk of colorectal cancer: A prospective cohort study of younger US women. Int. J. Epidemiol. 2022, dyac202. Available online: https://academic.oup.com/ije/advance-article-abstract/doi/10.1093/ije/dyac202/6782936?redirectedFrom=fulltext (accessed on 31 October 2022). [CrossRef]
- Alegria-Lertxundi, I.; Bujanda, L.; Arroyo-Izaga, M. Role of Dairy Foods, Fish, White Meat, and Eggs in the Prevention of Colorectal Cancer: A Systematic Review of Observational Studies in 2018-2022. Nutrients 2022, 14, 3430. [Google Scholar] [CrossRef] [PubMed]
- Hidaka, A.; Harrison, T.A.; Cao, Y.; Sakoda, L.C.; Barfield, R.; Giannakis, M.; Song, M.; Phipps, A.I.; Figueiredo, J.C.; Zaidi, S.H.; et al. Intake of Dietary Fruit, Vegetables, and Fiber and Risk of Colorectal Cancer According to Molecular Subtypes: A Pooled Analysis of 9 Studies. Cancer Res. 2020, 80, 4578–4590. [Google Scholar] [CrossRef] [PubMed]
- Michels, K.B.; Edward, G.; Joshipura, K.J.; Rosner, B.A.; Stampfer, M.J.; Fuchs, C.S.; Colditz, G.A.; Speizer, F.E.; Willett, W.C. Prospective study of fruit and vegetable consumption and incidence of colon and rectal cancers. J. Natl. Cancer Inst. 2000, 92, 1740–1752. [Google Scholar] [CrossRef]
- Tabung, F.K.; Brown, L.S.; Fung, T.T. Dietary Patterns and Colorectal Cancer Risk: A Review of 17 Years of Evidence (2000-2016). Curr. Colorectal Cancer Rep. 2017, 13, 440–454. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Chan, A.T. Diet, Gut Microbiota, and Colorectal Cancer Prevention: A Review of Potential Mechanisms and Promising Targets for Future Research. Curr. Colorectal Cancer Rep. 2017, 13, 429–439. [Google Scholar] [CrossRef]
- Molska, M.; Reguła, J. Potential Mechanisms of Probiotics Action in the Prevention and Treatment of Colorectal Cancer. Nutrients 2019, 11, 2453. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Lee, J.; Kim, J.H.; Cho, N.; Lee, S.H.; Park, S.B.; Koh, B.; Kang, D.; Kim, S.; Yoo, H.M. Characterization of the Anti-Cancer Activity of the Probiotic Bacterium Lactobacillus fermentum Using 2D vs. 3D Culture in Colorectal Cancer Cells. Biomolecules 2019, 9, 557. [Google Scholar] [CrossRef]
- Tiptiri-Kourpeti, A.; Spyridopoulou, K.; Santarmaki, V.; Aindelis, G.; Tompoulidou, E.; Lamprianidou, E.E.; Saxami, G.; Ypsilantis, P.; Lampri, E.S.; Simopoulos, C.; et al. Lactobacillus casei Exerts Anti-Proliferative Effects Accompanied by Apoptotic Cell Death and Up-Regulation of TRAIL in Colon Carcinoma Cells. PLoS ONE 2016, 11, e0147960. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Wang, Y.; Xu, H.; Mei, X.; Yu, D.; Wang, Y.; Li, W. Antioxidant Properties of Probiotic Bacteria. Nutrients 2017, 9, 521. [Google Scholar] [CrossRef]
- Bahmani, S.; Azarpira, N.; Moazamian, E. Anti-colon cancer activity of Bifidobacterium metabolites on colon cancer cell line SW742. Turk. J. Gastroenterol. Off. J. Turk. Soc. Gastroenterol. 2019, 30, 835–842. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, C.Y.; Osaka, T.; Moriyama, E.; Date, Y.; Kikuchi, J.; Tsuneda, S. Strengthening of the intestinal epithelial tight junction by Bifidobacterium bifidum. Physiol. Rep. 2015, 3, e12327. [Google Scholar] [CrossRef] [PubMed]
- Bibbins-Domingo, K. Aspirin Use for the Primary Prevention of Cardiovascular Disease and Colorectal Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann. Intern. Med. 2016, 164, 836–845. [Google Scholar] [CrossRef]
- Cederlund, H.; Mårdh, P.A. Antibacterial activities of non-antibiotic drugs. J. Antimicrob. Chemother. 1993, 32, 355–365. [Google Scholar] [CrossRef]
- Brennan, C.A.; Nakatsu, G.; Gallini Comeau, C.A.; Drew, D.A.; Glickman, J.N.; Schoen, R.E.; Chan, A.T.; Garrett, W.S. Aspirin Modulation of the Colorectal Cancer-Associated Microbe Fusobacterium nucleatum. mBio 2021, 12, e00547-21. [Google Scholar] [CrossRef]
- Burn, J.; Sheth, H.; Elliott, F.; Reed, L.; Macrae, F.; Mecklin, J.P.; Möslein, G.; McRonald, F.E.; Bertario, L.; Evans, D.G.; et al. Cancer prevention with aspirin in hereditary colorectal cancer (Lynch syndrome), 10-year follow-up and registry-based 20-year data in the CAPP2 study: A double-blind, randomised, placebo-controlled trial. Lancet 2020, 395, 1855–1863. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Q.; Halfdanarson, Ó.Ö.; Zoega, H.; Sadr-Azodi, O.; Engstrand, L.; Fall, K.; Brusselaers, N. Proton pump inhibitors and survival in patients with colorectal cancer: A Swedish population-based cohort study. Br. J. Cancer 2021, 125, 893–900. [Google Scholar] [CrossRef]
- Maniewska, J.; Jeżewska, D. Non-Steroidal Anti-Inflammatory Drugs in Colorectal Cancer Chemoprevention. Cancers 2021, 13, 594. [Google Scholar] [CrossRef]
- Fritz, I.; Wagner, P.; Olsson, H. Improved survival in several cancers with use of H(1)-antihistamines desloratadine and loratadine. Transl. Oncol. 2021, 14, 101029. [Google Scholar] [CrossRef]
- Forslund, S.K.; Chakaroun, R.; Zimmermann-Kogadeeva, M.; Markó, L.; Aron-Wisnewsky, J.; Nielsen, T.; Moitinho-Silva, L.; Schmidt, T.S.B.; Falony, G.; Vieira-Silva, S.; et al. Combinatorial, additive and dose-dependent drug–microbiome associations. Nature 2021, 600, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Nooij, S.; Ducarmon, Q.R.; Laros, J.F.J.; Zwittink, R.D.; Norman, J.M.; Smits, W.K.; Verspaget, H.W.; Keller, J.J.; Terveer, E.M.; Kuijper, E.J. Fecal Microbiota Transplantation Influences Procarcinogenic Escherichia coli in Recipient Recurrent Clostridioides difficile Patients. Gastroenterology 2021, 161, 1218–1228.e5. [Google Scholar] [CrossRef] [PubMed]
- Parker, K.D.; Maurya, A.K.; Ibrahim, H.; Rao, S.; Hove, P.R.; Kumar, D.; Kant, R.; Raina, B.; Agarwal, R.; Kuhn, K.A.; et al. Dietary Rice Bran-Modified Human Gut Microbial Consortia Confers Protection against Colon Carcinogenesis Following Fecal Transfaunation. Biomedicines 2021, 9, 144. [Google Scholar] [CrossRef]
- Zhang, W.; Zou, G.; Li, B.; Du, X.; Sun, Z.; Sun, Y.; Jiang, X. Fecal Microbiota Transplantation (FMT) Alleviates Experimental Colitis in Mice by Gut Microbiota Regulation. J. Microbiol. Biotechnol. 2020, 30, 1132–1141. [Google Scholar] [CrossRef] [PubMed]
- Weingarden, A.R.; Vaughn, B.P. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes 2017, 8, 238–252. [Google Scholar] [CrossRef] [PubMed]
- Alang, N.; Kelly, C.R. Weight gain after fecal microbiota transplantation. Open Forum Infect. Dis. 2015, 2, ofv004. [Google Scholar] [CrossRef] [PubMed]
- Woodworth, M.H.; Carpentieri, C.; Sitchenko, K.L.; Kraft, C.S. Challenges in fecal donor selection and screening for fecal microbiota transplantation: A review. Gut Microbes 2017, 8, 225–237. [Google Scholar] [CrossRef]
- Shanahan, F.; Ghosh, T.S.; O’Toole, P.W. The Healthy Microbiome-What Is the Definition of a Healthy Gut Microbiome? Gastroenterology 2021, 160, 483–494. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, I.; Wu, R.; Tokumaru, Y.; Terracina, K.P.; Takabe, K. The Role of the Microbiome on the Pathogenesis and Treatment of Colorectal Cancer. Cancers 2022, 14, 5685. https://doi.org/10.3390/cancers14225685
Yu I, Wu R, Tokumaru Y, Terracina KP, Takabe K. The Role of the Microbiome on the Pathogenesis and Treatment of Colorectal Cancer. Cancers. 2022; 14(22):5685. https://doi.org/10.3390/cancers14225685
Chicago/Turabian StyleYu, Irene, Rongrong Wu, Yoshihisa Tokumaru, Krista P. Terracina, and Kazuaki Takabe. 2022. "The Role of the Microbiome on the Pathogenesis and Treatment of Colorectal Cancer" Cancers 14, no. 22: 5685. https://doi.org/10.3390/cancers14225685
APA StyleYu, I., Wu, R., Tokumaru, Y., Terracina, K. P., & Takabe, K. (2022). The Role of the Microbiome on the Pathogenesis and Treatment of Colorectal Cancer. Cancers, 14(22), 5685. https://doi.org/10.3390/cancers14225685





