Phenotypic Characterization of Circulating Tumor Cells Isolated from Non-Small and Small Cell Lung Cancer Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cancer Cell Culture
2.2. Patients’ Characteristics and Patients’ Blood Specimen Collection
2.3. ISET Isolation
2.4. Cytospin Preparation
2.5. Spiking Experiments
2.6. Triple Immunofluorescence Staining
2.7. Statistical Analysis of the Clinical Data
3. Results
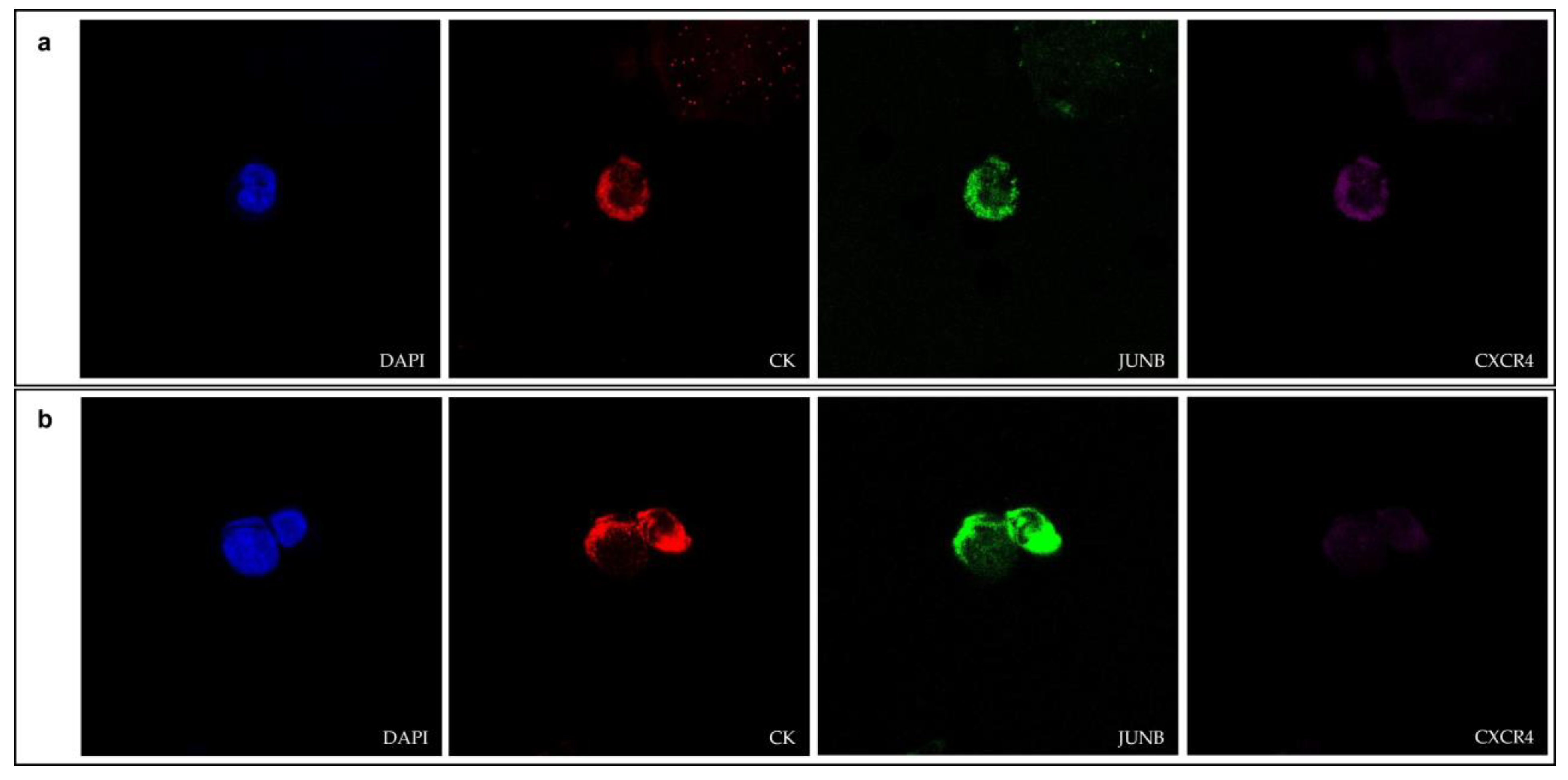
3.1. JUNB and CXCR4 Expression in CTCs Derived from NSCLC Patients
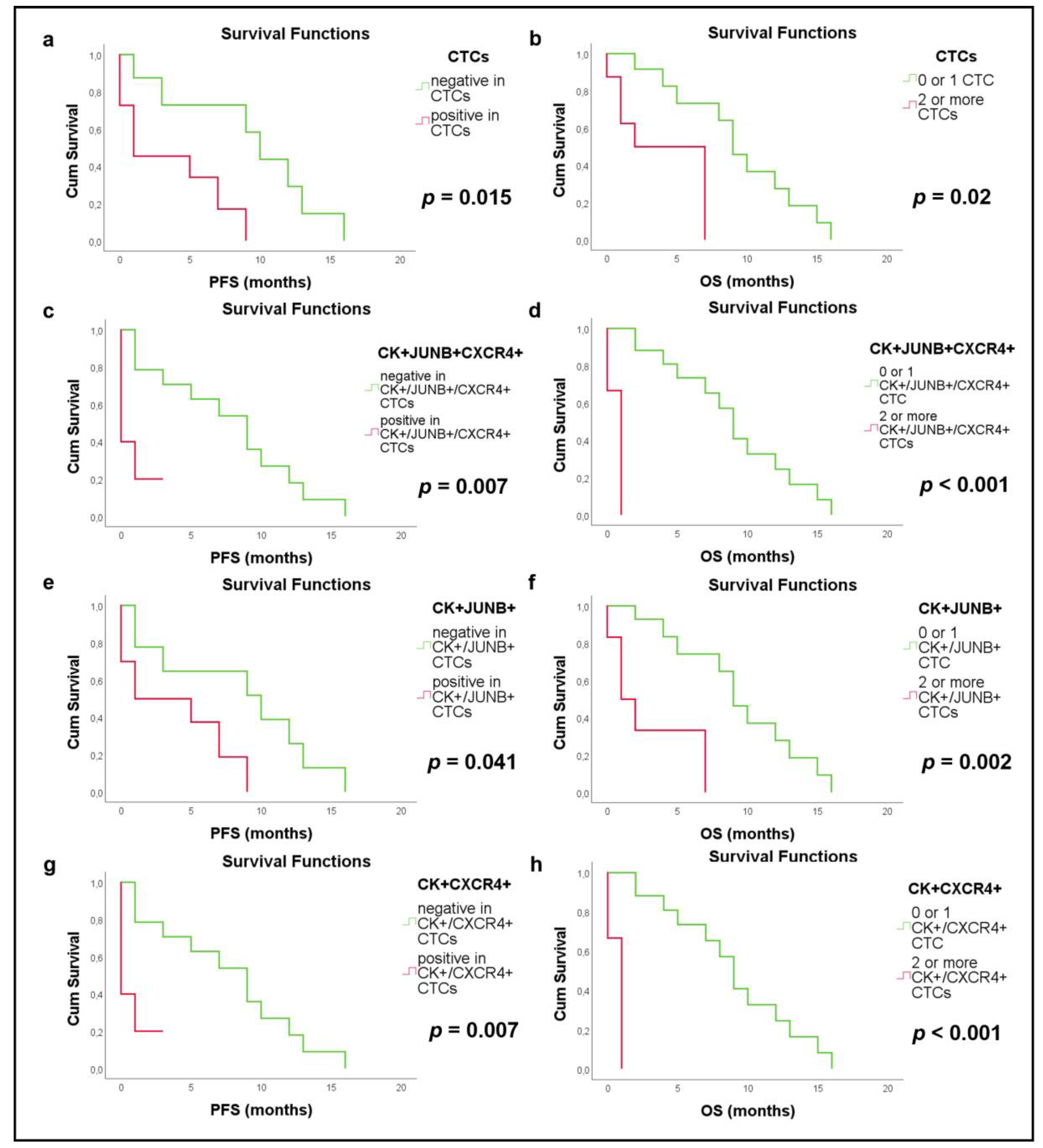
3.2. Clinical Relevance in NSCLC Patients
3.3. JUNB and CXCR4 Expression in CTCs Derived from SCLC Patients
3.4. Clinical Relevance in SCLC Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pantazaka, E.; Vardas, V.; Roumeliotou, A.; Kakavogiannis, S.; Kallergi, G. Clinical Relevance of Mesenchymal- and Stem-Associated Phenotypes in Circulating Tumor Cells Isolated from Lung Cancer Patients. Cancers 2021, 13, 2158. [Google Scholar] [CrossRef] [PubMed]
- Maly, V.; Maly, O.; Kolostova, K.; Bobek, V. Circulating Tumor Cells in Diagnosis and Treatment of Lung Cancer. In Vivo 2019, 33, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Oser, M.G.; Niederst, M.J.; Sequist, L.V.; Engelman, J.A. Transformation from Non-Small-Cell Lung Cancer to Small-Cell Lung Cancer: Molecular Drivers and Cells of Origin. Lancet Oncol. 2015, 16, e165–e172. [Google Scholar] [CrossRef]
- Obermayr, E.; Koppensteiner, N.; Heinzl, N.; Schuster, E.; Holzer, B.; Fabikan, H.; Weinlinger, C.; Illini, O.; Hochmair, M.; Zeillinger, R. Cancer Stem Cell-Like Circulating Tumor Cells Are Prognostic in Non-Small Cell Lung Cancer. J. Pers. Med. 2021, 11, 1225. [Google Scholar] [CrossRef]
- Messaritakis, I.; Politaki, E.; Kotsakis, A.; Dermitzaki, E.-K.; Koinis, F.; Lagoudaki, E.; Koutsopoulos, A.; Kallergi, G.; Souglakos, J.; Georgoulias, V. Phenotypic Characterization of Circulating Tumor Cells in the Peripheral Blood of Patients with Small Cell Lung Cancer. PLoS ONE 2017, 12, e0181211. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Bao, X.; Chen, M.; Lin, R.; Zhuyan, J.; Zhen, T.; Xing, K.; Zhou, W.; Zhu, S. Mechanisms and Future of Non-Small Cell Lung Cancer Metastasis. Front. Oncol. 2020, 10, 585284. [Google Scholar] [CrossRef]
- Heidrich, I.; Ačkar, L.; Mossahebi Mohammadi, P.; Pantel, K. Liquid Biopsies: Potential and Challenges. Int. J. Cancer 2021, 148, 528–545. [Google Scholar] [CrossRef]
- Esagian, S.M.; Grigoriadou, G.Ι.; Nikas, I.P.; Boikou, V.; Sadow, P.M.; Won, J.-K.; Economopoulos, K.P. Comparison of Liquid-Based to Tissue-Based Biopsy Analysis by Targeted next Generation Sequencing in Advanced Non-Small Cell Lung Cancer: A Comprehensive Systematic Review. J. Cancer Res. Clin. Oncol. 2020, 146, 2051–2066. [Google Scholar] [CrossRef]
- Lianidou, E.S.; Strati, A.; Markou, A. Circulating Tumor Cells as Promising Novel Biomarkers in Solid Cancers. Crit. Rev. Clin. Lab. Sci. 2014, 51, 160–171. [Google Scholar] [CrossRef]
- Kallergi, G.; Vetsika, E.-K.; Aggouraki, D.; Lagoudaki, E.; Koutsopoulos, A.; Koinis, F.; Katsarlinos, P.; Trypaki, M.; Messaritakis, I.; Stournaras, C.; et al. Evaluation of PD-L1/PD-1 on Circulating Tumor Cells in Patients with Advanced Non-Small Cell Lung Cancer. Ther. Adv. Med. Oncol. 2018, 10, 175883401775012. [Google Scholar] [CrossRef]
- Katsarou, S.D.; Messaritakis, I.; Voumvouraki, A.; Kakavogiannis, S.; Κotsakis, A.; Alkahtani, S.; Stournaras, C.; Martin, S.S.; Georgoulias, V.; Kallergi, G. Detyrosinated α-Tubulin, Vimentin and PD-L1 in Circulating Tumor Cells (CTCs) Isolated from Non-Small Cell Lung Cancer (NSCLC) Patients. J. Pers. Med. 2022, 12, 154. [Google Scholar] [CrossRef] [PubMed]
- Vardas, V.; Politaki, E.; Pantazaka, E.; Georgoulias, V.; Kallergi, G. Epithelial-to-Mesenchymal Transition of Tumor Cells: Cancer Progression and Metastasis. Int. J. Dev. Biol. 2022, 66, 277–283. [Google Scholar] [CrossRef]
- Kallergi, G.; Papadaki, M.A.; Politaki, E.; Mavroudis, D.; Georgoulias, V.; Agelaki, S. Epithelial to Mesenchymal Transition Markers Expressed in Circulating Tumour Cells of Early and Metastatic Breast Cancer Patients. Breast Cancer Res. 2011, 13, R59. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, G.; Aggouraki, D.; Zacharopoulou, N.; Stournaras, C.; Georgoulias, V.; Martin, S.S. Evaluation of α-Tubulin, Detyrosinated α-Tubulin, and Vimentin in CTCs: Identification of the Interaction between CTCs and Blood Cells through Cytoskeletal Elements. Breast Cancer Res. 2018, 20, 67. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Wang, Y.; Yin, H.; Chen, W.; Jin, G.; Ma, H.; Dai, J.; Chen, J.; Jiang, Y.; Wang, H.; et al. Circulating Tumor Cells Enriched by the Depletion of Leukocytes with Bi-Antibodies in Non-Small Cell Lung Cancer: Potential Clinical Application. PLoS ONE 2015, 10, e0137076. [Google Scholar] [CrossRef] [PubMed]
- Ntzifa, A.; Strati, A.; Kallergi, G.; Kotsakis, A.; Georgoulias, V.; Lianidou, E. Gene Expression in Circulating Tumor Cells Reveals a Dynamic Role of EMT and PD-L1 during Osimertinib Treatment in NSCLC Patients. Sci. Rep. 2021, 11, 2313. [Google Scholar] [CrossRef] [PubMed]
- Kallergi, G.; Tsintari, V.; Sfakianakis, S.; Bei, E.; Lagoudaki, E.; Koutsopoulos, A.; Zacharopoulou, N.; Alkahtani, S.; Alarifi, S.; Stournaras, C.; et al. The Prognostic Value of JUNB-Positive CTCs in Metastatic Breast Cancer: From Bioinformatics to Phenotypic Characterization. Breast Cancer Res. 2019, 21, 86. [Google Scholar] [CrossRef]
- Kallergi, G.; Hoffmann, O.; Bittner, A.-K.; Papadimitriou, L.; Katsarou, S.D.; Zacharopoulou, N.; Zervakis, M.; Sfakianakis, S.; Stournaras, C.; Georgoulias, V.; et al. CXCR4 and JUNB Double-Positive Disseminated Tumor Cells Are Detected Frequently in Breast Cancer Patients at Primary Diagnosis. Ther. Adv. Med. Oncol. 2020, 12, 175883591989575. [Google Scholar] [CrossRef]
- Teixidó, J.; Martínez-Moreno, M.; Díaz-Martínez, M.; Sevilla-Movilla, S. The Good and Bad Faces of the CXCR4 Chemokine Receptor. Int. J. Biochem. Cell. Biol. 2018, 95, 121–131. [Google Scholar] [CrossRef]
- Mukherjee, D.; Zhao, J. The Role of Chemokine Receptor CXCR4 in Breast Cancer Metastasis. Am. J. Cancer Res. 2013, 3, 46–57. [Google Scholar] [PubMed]
- Gokulnath, M.; Swetha, R.; Thejaswini, G.; Shilpa, P.; Selvamurugan, N. Transforming Growth Factor-Β1 Regulation of ATF-3, c-Jun and JunB Proteins for Activation of Matrix Metalloproteinase-13 Gene in Human Breast Cancer Cells. Int. J. Biol. Macromol. 2017, 94, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.; Guo, Z.; Wang, Z.; Dai, Y.; Zheng, L.; Zhu, L.; Zhang, J.; Hu, W.; Nie, J.; Mao, W.; et al. RAC2 Promotes Abnormal Proliferation of Quiescent Cells by Enhanced JUNB Expression via the MAL-SRF Pathway. Cell Cycle 2018, 17, 1115–1123. [Google Scholar] [CrossRef] [PubMed]
- Sundqvist, A.; Morikawa, M.; Ren, J.; Vasilaki, E.; Kawasaki, N.; Kobayashi, M.; Koinuma, D.; Aburatani, H.; Miyazono, K.; Heldin, C.-H.; et al. JUNB Governs a Feed-Forward Network of TGFβ Signaling That Aggravates Breast Cancer Invasion. Nucleic Acids Res. 2018, 46, 1180–1195. [Google Scholar] [CrossRef] [PubMed]
- Gervasi, M.; Bianchi-Smiraglia, A.; Cummings, M.; Zheng, Q.; Wang, D.; Liu, S.; Bakin, A.V. JunB Contributes to Id2 Repression and the Epithelial–Mesenchymal Transition in Response to Transforming Growth Factor–β. J. Cell Biol. 2012, 196, 589–603. [Google Scholar] [CrossRef]
- Lian, S.; Shao, Y.; Liu, H.; He, J.; Lu, W.; Zhang, Y.; Jiang, Y.; Zhu, J. PDK1 Induces JunB, EMT, Cell Migration and Invasion in Human Gallbladder Cancer. Oncotarget 2015, 6, 29076–29086. [Google Scholar] [CrossRef]
- Wanna-udom, S.; Terashima, M.; Lyu, H.; Ishimura, A.; Takino, T.; Sakari, M.; Tsukahara, T.; Suzuki, T. The M6A Methyltransferase METTL3 Contributes to Transforming Growth Factor-Beta-Induced Epithelial-Mesenchymal Transition of Lung Cancer Cells through the Regulation of JUNB. Biochem. Biophys. Res. Commun. 2020, 524, 150–155. [Google Scholar] [CrossRef]
- Reckamp, K.L.; Figlin, R.A.; Burdick, M.D.; Dubinett, S.M.; Elashoff, R.M.; Strieter, R.M. CXCR4 Expression on Circulating Pan-Cytokeratin Positive Cells Is Associated with Survival in Patients with Advanced Non-Small Cell Lung Cancer. BMC Cancer 2009, 9, 213. [Google Scholar] [CrossRef]
- Tu, Z.; Xie, S.; Xiong, M.; Liu, Y.; Yang, X.; Tembo, K.M.; Huang, J.; Hu, W.; Huang, X.; Pan, S.; et al. CXCR4 Is Involved in CD133-Induced EMT in Non-Small Cell Lung Cancer. Int. J. Oncol. 2017, 50, 505–514. [Google Scholar] [CrossRef]
- Salgia, R.; Weaver, R.W.; McCleod, M.; Stille, J.R.; Yan, S.B.; Roberson, S.; Polzer, J.; Flynt, A.; Raddad, E.; Peek, V.L.; et al. Prognostic and Predictive Value of Circulating Tumor Cells and CXCR4 Expression as Biomarkers for a CXCR4 Peptide Antagonist in Combination with Carboplatin-Etoposide in Small Cell Lung Cancer: Exploratory Analysis of a Phase II Study. Investig. New Drugs 2017, 35, 334–344. [Google Scholar] [CrossRef]
- Kallergi, G.; Politaki, E.; Alkahtani, S.; Stournaras, C.; Georgoulias, V. Evaluation of Isolation Methods for Circulating Tumor Cells (CTCs). Cell. Physiol. Biochem. 2016, 40, 411–419. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Tripathy, D.; Frenkel, E.P.; Shete, S.; Naftalis, E.Z.; Huth, J.F.; Beitsch, P.D.; Leitch, M.; Hoover, S.; Euhus, D.; et al. Circulating Tumor Cells in Patients with Breast Cancer Dormancy. Clin. Cancer Res. 2004, 10, 8152–8162. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.-M.; Krebs, M.G.; Lancashire, L.; Sloane, R.; Backen, A.; Swain, R.K.; Priest, L.J.C.; Greystoke, A.; Zhou, C.; Morris, K.; et al. Clinical Significance and Molecular Characteristics of Circulating Tumor Cells and Circulating Tumor Microemboli in Patients With Small-Cell Lung Cancer. J. Clin. Oncol. 2012, 30, 525–532. [Google Scholar] [CrossRef] [PubMed]
- Messaritakis, I.; Nikolaou, M.; Politaki, E.; Koinis, F.; Lagoudaki, E.; Koutsopoulos, A.; Georgoulia, N.; Georgoulias, V.; Kotsakis, A. Bcl-2 Expression in Circulating Tumor Cells (CTCs) of Patients with Small Cell Lung Cancer (SCLC) Receiving Front-Line Treatment. Lung Cancer 2018, 124, 270–278. [Google Scholar] [CrossRef]
- Gallo, M.; de Luca, A.; Maiello, M.R.; D’Alessio, A.; Esposito, C.; Chicchinelli, N.; Forgione, L.; Piccirillo, M.C.; Rocco, G.; Morabito, A.; et al. Clinical Utility of Circulating Tumor Cells in Patients with Non-Small-Cell Lung Cancer. Transl. Lung Cancer Res. 2017, 6, 486–498. [Google Scholar] [CrossRef]
- Sinoquet, L.; Jacot, W.; Gauthier, L.; Pouderoux, S.; Viala, M.; Cayrefourcq, L.; Quantin, X.; Alix-Panabières, C. Programmed Cell Death Ligand 1-Expressing Circulating Tumor Cells: A New Prognostic Biomarker in Non-Small Cell Lung Cancer. Clin. Chem. 2021, 67, 1503–1512. [Google Scholar] [CrossRef]
- Wang, T.; Han, S.; Wu, Z.; Han, Z.; Yan, W.; Liu, T.; Wei, H.; Song, D.; Zhou, W.; Yang, X.; et al. XCR1 Promotes Cell Growth and Migration and Is Correlated with Bone Metastasis in Non-Small Cell Lung Cancer. Biochem. Biophys. Res. Commun. 2015, 464, 635–641. [Google Scholar] [CrossRef]
- Pozzobon, T.; Goldoni, G.; Viola, A.; Molon, B. CXCR4 Signaling in Health and Disease. Immunol. Lett. 2016, 177, 6–15. [Google Scholar] [CrossRef]
- Kim, J.-Y.; Kim, H.-J.; Jung, C.-W.; Lee, T.S.; Kim, E.H.; Park, M.-J. CXCR4 Uses STAT3-Mediated Slug Expression to Maintain Radioresistance of Non-Small Cell Lung Cancer Cells: Emerges as a Potential Prognostic Biomarker for Lung Cancer. Cell Death Dis. 2021, 12, 48. [Google Scholar] [CrossRef]
- Hyakusoku, H.; Sano, D.; Takahashi, H.; Hatano, T.; Isono, Y.; Shimada, S.; Ito, Y.; Myers, J.N.; Oridate, N. JunB Promotes Cell Invasion, Migration and Distant Metastasis of Head and Neck Squamous Cell Carcinoma. J. Exp. Clin. Cancer Res. 2016, 35, 6. [Google Scholar] [CrossRef]
- Wang, H.; Xie, F.; Hu, Z.; Chen, L. Elevated Expression of CXCR4 and Correlation with Clinicopathological Features and Prognosis of Non-Small Cell Lung Cancer Patients: A Meta-Analysis. Genet. Mol. Res. 2015, 14, 17893–17903. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.-X.; Gao, W.; Liang, Y.; Zhou, X.-M. Chemokine Receptor CXCR4 Expression and Lung Cancer Prognosis: A Meta-Analysis. Int. J. Clin. Exp. Med. 2015, 8, 5163–5174. [Google Scholar] [PubMed]
- Zhao, H.; Guo, L.; Zhao, H.; Zhao, J.; Weng, H.; Zhao, B. CXCR4 Over-Expression and Survival in Cancer: A System Review and Meta-Analysis. Oncotarget 2015, 6, 5022–5040. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Xu, Y.; Xu, H.; Yu, B. The Clinicopathological and Prognostic Value of CXCR4 Expression in Patients with Lung Cancer: A Meta-Analysis. BMC Cancer 2022, 22, 681. [Google Scholar] [CrossRef]
- Li, Y.; Shen, Y.; Miao, Y.; Luan, Y.; Sun, B.; Qiu, X. Co-Expression of UPAR and CXCR4 Promotes Tumor Growth and Metastasis in Small Cell Lung Cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 3771–3780. [Google Scholar]
- Hou, J.-M.; Krebs, M.; Ward, T.; Sloane, R.; Priest, L.; Hughes, A.; Clack, G.; Ranson, M.; Blackhall, F.; Dive, C. Circulating Tumor Cells as a Window on Metastasis Biology in Lung Cancer. Am. J. Pathol. 2011, 178, 989–996. [Google Scholar] [CrossRef]
- Messaritakis, I.; Nikolaou, M.; Koinis, F.; Politaki, E.; Koutsopoulos, A.; Lagoudaki, E.; Vetsika, E.-K.; Georgoulias, V.; Kotsakis, A. Characterization of DLL3-Positive Circulating Tumor Cells (CTCs) in Patients with Small Cell Lung Cancer (SCLC) and Evaluation of Their Clinical Relevance during Front-Line Treatment. Lung Cancer 2019, 135, 33–39. [Google Scholar] [CrossRef]
- Hodgkinson, C.L.; Morrow, C.J.; Li, Y.; Metcalf, R.L.; Rothwell, D.G.; Trapani, F.; Polanski, R.; Burt, D.J.; Simpson, K.L.; Morris, K.; et al. Tumorigenicity and Genetic Profiling of Circulating Tumor Cells in Small-Cell Lung Cancer. Nat. Med. 2014, 20, 897–903. [Google Scholar] [CrossRef]




| Characteristics | Sub-Categories | Values |
|---|---|---|
| Median age | 66 years (range 44–82 years) | |
| Stage | III | 6 (20%) |
| IV | 24 (80%) | |
| Histology | Adenocarcinoma | 18 (60%) |
| Squamous cell carcinoma | 12 (40%) | |
| EGFR | WT | 11 (37%) |
| ND | 19 (63%) |
| Characteristics | Sub-Categories | Values |
|---|---|---|
| Median age | 68 years (range 44–79 years) | |
| Stage | Limited | 5 (14%) |
| Extensive | 20 (54%) | |
| Unknown | 12 (32%) | |
| Smoking | Yes | 29 (78%) |
| No | 0 (0%) | |
| Unknown | 8 (22%) | |
| Family history of cancer | Yes | 8 (22%) |
| No | 21 (57%) | |
| Unknown | 8 (22%) | |
| Metastasis | CNS | 10 (27%) |
| Liver | 8 (22%) | |
| Pancreas | 1 (3%) | |
| Bones | 5 (14%) | |
| Adrenal glands | 3 (8%) | |
| LNs | 5 (14%) | |
| Unknown | 14 (38%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roumeliotou, A.; Pantazaka, E.; Xagara, A.; Dimitrakopoulos, F.-I.; Koutras, A.; Christopoulou, A.; Kourelis, T.; Aljarba, N.H.; Alkahtani, S.; Koinis, F.; et al. Phenotypic Characterization of Circulating Tumor Cells Isolated from Non-Small and Small Cell Lung Cancer Patients. Cancers 2023, 15, 171. https://doi.org/10.3390/cancers15010171
Roumeliotou A, Pantazaka E, Xagara A, Dimitrakopoulos F-I, Koutras A, Christopoulou A, Kourelis T, Aljarba NH, Alkahtani S, Koinis F, et al. Phenotypic Characterization of Circulating Tumor Cells Isolated from Non-Small and Small Cell Lung Cancer Patients. Cancers. 2023; 15(1):171. https://doi.org/10.3390/cancers15010171
Chicago/Turabian StyleRoumeliotou, Argyro, Evangelia Pantazaka, Anastasia Xagara, Foteinos-Ioannis Dimitrakopoulos, Angelos Koutras, Athina Christopoulou, Theodoros Kourelis, Nada H. Aljarba, Saad Alkahtani, Filippos Koinis, and et al. 2023. "Phenotypic Characterization of Circulating Tumor Cells Isolated from Non-Small and Small Cell Lung Cancer Patients" Cancers 15, no. 1: 171. https://doi.org/10.3390/cancers15010171
APA StyleRoumeliotou, A., Pantazaka, E., Xagara, A., Dimitrakopoulos, F.-I., Koutras, A., Christopoulou, A., Kourelis, T., Aljarba, N. H., Alkahtani, S., Koinis, F., Kotsakis, A., & Kallergi, G. (2023). Phenotypic Characterization of Circulating Tumor Cells Isolated from Non-Small and Small Cell Lung Cancer Patients. Cancers, 15(1), 171. https://doi.org/10.3390/cancers15010171








